Abstract
CellCircuits (http://www.cellcircuits.org) is an open-access database of molecular network models, designed to bridge the gap between databases of individual pairwise molecular interactions and databases of validated pathways. CellCircuits captures the output from an increasing number of approaches that screen molecular interaction networks to identify functional subnetworks, based on their correspondence with expression or phenotypic data, their internal structure or their conservation across species. This initial release catalogs 2019 computationally derived models drawn from 11 journal articles and spanning five organisms (yeast, worm, fly, Plasmodium falciparum and human). Models are available either as images or in machine-readable formats and can be queried by the names of proteins they contain or by their enriched biological functions. We envision CellCircuits as a clearinghouse in which theorists may distribute or revise models in need of validation and experimentalists may search for models or specific hypotheses relevant to their interests. We demonstrate how such a repository of network models is a novel systems biology resource by performing several meta-analyses not currently possible with existing databases.
INTRODUCTION
At present, a great deal of biological information is represented as interactions between molecules. This information includes physical interactions that occur among proteins, DNA, RNA and small molecules (1–3); genetic interactions such as synthetic lethality, enhancement or suppression (4); and interactions due to co-expression (5) or co-citation (6). Modern analyses of interaction data typically accomplish two goals. The first goal is to clean the data, by filtering erroneous interactions that can be associated with high-throughput screens [false positives, e.g. (7,8)] or by predicting new interactions that may have been previously missed [false negatives, e.g. (9,10)]. The second goal is to organize the interactions into biological network models—i.e. collections of interactions hypothesized to work together towards a particular cellular function or within a common pathway (11–13).
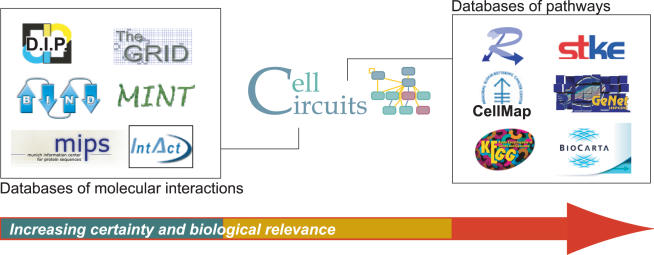
Interaction analysis is currently supported by two types of available databases (Figure 1). First, the raw material for analysis is provided by databases of molecular interactions including the Database of Interacting Proteins (14), the Munich Center for Information on Protein Sequences (15), the Biomolecular Interaction Network Database (16), the BioGRID (17) and IntAct (18). Many of these databases provide confidence scores with each measured and predicted interaction. Second, there are a growing number of so-called pathway databases, in which canonical diagrams of metabolic, signaling or regulatory pathways have been hand-curated from review articles and textbooks. Metabolic pathways are the focus of Reactome (19), MetaCyc (20) and the Kyoto Encyclopedia of Genes and Genomes (21), while databases such as BioCarta (http://www.biocarta.com/genes), CellMap (http://cellmap.org), the Signal Transduction Knowledge Environment (22), GeNet (23) and TransPATH (24) are primarily concerned with signaling and transcription. All of these pathway databases are relevant to the second and perhaps ultimate goal of interaction analysis—models of well-defined and well-validated functional relationships among genes, proteins and/or metabolites.
Figure 1.
The need for a new type of database. The CellCircuits database is positioned between raw molecular interaction databases (left) and databases of rigorously validated cellular pathways (right). Interaction database icons represent (clockwise from top left) the Database of Interacting Proteins [DIP (14)]; the General Repository of Interaction Datasets [GRID (17)]; Molecular INTeractions Database [MINT (48)]; the IntAct molecular interactions database (18); the interaction database at the Munich Information Center for Protein Sequences [MIPS (15)]; and Biomolecular Interaction Network Database [BIND (16)]. Pathway database icons represent Reactome (19); Signal Transduction Knowledge Environment [STKE (22)]; Gene Networks database [GeNet (23)]; BioCarta (http://www.biocarta.com/genes); Kyoto Encyclopedia of Genes and Genomes [KEGG (21)]; and CellMap (http://cellmap.org).
Automatic inference of accurate and detailed molecular pathways, however, is well beyond the capability of current interaction analyses and integrative modeling approaches. Although current approaches attempt to place interactions into subnetworks according to their putative function (11–13), such subnetworks are hypothetical in nature and thus inappropriate for entry into any of the existing databases of canonical pathways. Rather, the subnetwork models produced by automated approaches are typically embedded in figures, tables or supplementary information in the primary published literature. Although it is certainly possible to read about the models, there are several problems with this traditional method of dissemination. First, the size and number of models from even a single publication can be overwhelming, making models relevant to a particular gene or function difficult to locate. Second, in many cases, network modeling papers target bioinformatic, rather than biological or medical, audiences. As a result, the models remain largely inaccessible to those who have the most knowledge to interpret them and the most to gain from their successful interpretation.
Recent opinion articles (25,26) have recognized a related problem for the case of protein functional predictions, calling for a clearinghouse of hypotheses generated by bioinformatics analyses and searchable by experimental biologists. In the same vein, the BioModels Database (27) has recently been adopted as a working repository for simulations of kinetic quantitative systems based on ordinary differential equations. Subnetworks inferred from genome-scale data, however, do not fall into this category.
Motivated by these considerations, we have designed CellCircuits as an open-access general repository of models distilled from protein networks. By aggregating models derived from many separate studies into a single resource, CellCircuits bridges the gap between databases of individual pairwise interactions and fully curated, biologically validated pathway models. The CellCircuits database enables experimentalists to readily access and cross-reference models across multiple publications. It also enables the meta-analysis of the entire set of models to reveal inter-model relationships and to answer global questions; for instance, which models overlap in terms of the genes and/or cellular processes represented? How novel is a new result given the models that are already present in the database?
MATERIALS AND METHODS
Data processing
A data processing pipeline was used to extract information from the textual representation of a model and store that information in a MySQL (http://www.mysql.org) relational database. The data processing pipeline required a digital image of each model and a text file containing the genes, proteins, metabolites, other small molecules and interconnections represented in the model. In cases when a network model was published in graphical form only, the text file was manually transcribed (see Supplementary Table S1).
To ensure that the CellCircuits database used a consistent set of gene identifiers, we mapped each gene name found in the text file for a model to a Gene Ontology (GO) gene id using tables from the GO database. Gene names found in a model but not in the GO database were automatically inserted into the appropriate database tables and flagged as being externally added. Future curation efforts could be directed towards handling these genes missing from the GO database. After models were entered into the database, they were scored using the hypergeometric test for GO annotation enrichment.
Web interface
We used Perl CGI scripts (http://www.perl.org) in conjunction with the Apache web server (http://httpd.apache.org), mod_perl (http://perl.apache.org) and Perl DBI (http://dbi.perl.org) to serve HTML content, handle user input and query the MySQL database. Script.aculo.us version 1.61 (http://script.aculo.us), an open source JavaScript library, was used to generate visual effects on the web pages that display search results.
Scoring models for Gene Ontology annotation
Using the latest release of the GO database, models were scored for a statistically significant number of genes in the model that were annotated with a particular GO term. We first identified the complete set of genes associated with each GO term. This set included the genes directly annotated with that term as well as those annotated with any of the term's descendents in the GO hierarchy. Next, we used the hypergeometric distribution (28,29) to test the genes in each model against the genes annotated with each of the GO terms. The resulting P-values were stored in the database.
Scoring similarity between publications
For each pair of publications we compared all models in one publication to all of the models in the other. To capture model similarity as sensitively as possible, we defined two models to be similar if they shared at least one interaction. The similarity score of a pair of publications was defined to be the number of distinct models that participated in any overlap divided by the total number of models in the pair. For example, consider publication A containing two models and publication B containing six models. If model 1 in A overlaps with models 1–5 in B, and model 2 in A only overlaps with model 1 in B, then the total number of distinct overlapping models is 7, and the similarity score between publications is 7/8.
RESULTS
A spectrum of network models
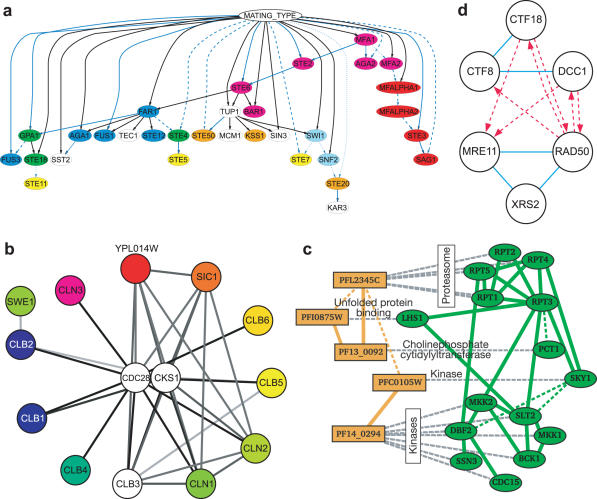
To date, interactions have been organized by searching for essentially three types of subnetworks: linear paths of interactions, interaction clusters or parallel clusters. Representative models of each type are shown in Figure 2. Linear (or branching) paths of interactions have been used to represent biological pathways such as metabolic processes or regulatory cascades (Figure 2a) (30–32). Clusters in an interaction network are regions of dense interconnections and are suggestive of functional protein complexes (Figure 2b) (33–37). Parallel clusters are two (or more) similar network clusters in which the proteins in one cluster are, in some way, associated with the proteins in the other cluster. Parallel clusters have been used to represent protein complexes conserved across species (Figure 2c) (38–40), in which pairs of proteins spanning the two clusters are orthologs associated by sequence-similarity relationships. They have also been used to identify the physical basis for genetic interactions (Figure 2d) (41), in which two protein interaction clusters are linked by many genetic interactions if the clusters perform redundant or synergistic cellular functions.
Figure 2.
Representative network models stored in CellCircuits. (a) A collection of linear regulatory pathways downstream of mating-type locus in yeast (31) (b) An interaction cluster of co-expressed proteins suggestive of a functional complex (34) (c) Parallel clusters conserved between P.falciparum and yeast (40). (d) Parallel clusters that are highly connected by genetic interactions (41).
Finally, integrating the interaction network with external data, such as gene expression profiles and other molecular states, has also been a key methodology used to identify significant subnetworks. For instance, these approaches have been used to find protein interaction clusters that exhibit coherent expression changes in response to panels of perturbations (33,35,36) or as a function of the cell cycle (34). Other works (42) have reported network ‘motifs’, defined as patterns of interactions that occur more often in the network than expected by chance. However, these approaches (by design) focus on general patterns rather than subnetworks of particular proteins. Therefore, they are not considered here.
Database coverage and assembly
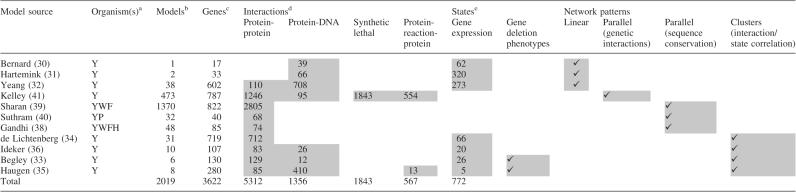
This CellCircuits initial release (version 1.0) was designed as proof-of-principle of the value of a searchable database of network models. We focused on providing a clear database interface and representative, albeit incomplete, coverage of the types of network models possible. For version 1.0, the database includes models from 11 publications, spanning linear, clustered or parallel subnetworks, with priority given to publications with models available in both graphical representations and machine-readable formats (Table 1). Graphical representations of network models are a particularly valuable method of disseminating interactions and/or pathways, in much the same way that DNA sequence logos (43) are used to visualize position-specific score matrices of DNA-binding motifs. Conversely, machine-readable formats, such as SBML (44), BioPAX (45) or the Cytoscape SIF format (46), greatly facilitate database entry, model curation and subsequent computational analysis. Four publications provided models in both graphical and machine-readable formats (32,39–41). For the remaining seven, models were manually curated from published figures (30,31,33–36,38).
Table 1.
Sources of data
aY = Yeast; W = Worm; F = Fly; H = Human; P = P.falciparum.
bCounts refer to total number of models across all organisms modeled.
cCounts refer to number of distinct genes in yeast only across all models.
dCounts refer to number of distinct interactions in yeast only across all models.
eFor gene expression, counts refer to number of profiles used.
Shading indicates which publications utilize particular types of Interaction data, State data, or Network patterns.
Manual curation involved downloading figures containing each network model, and then transcribing the genes and interactions in the models into a machine-readable format. For most publications, one figure, or each subpanel in a figure, contained a single network model. However, in three publications (31,34,38) the figures contained multiple, unconnected networks that were not divided by the authors into separate subpanels. In these cases, each unconnected component was entered as one model in CellCircuits, and in one case, networks were further subdivided into smaller models if they contained several sparsely connected, but functionally annotated, clusters of proteins (see Supplementary Table S1).
These curation activities resulted in a total of 2019 protein network models in the database. Models in the database include protein interactions from five organisms: yeast (Saccharomyces cerevisiae; 91% of all models), fly (Drosophila melanogaster; 58%), nematode worm (Caenorhabditis elegans; 27%), a malarial parasite (Plasmodium falciparum; 2%) and human (2%; these percentages total >100% due to cross-species comparisons covering multiple species in a single model). The models include up to four types of interactions (protein–protein, protein–DNA, genetic and metabolic) as well as two types of external data (gene expression and gene deletion phenotypes).
Network model query
Models in the CellCircuits database are queried through a web-based interface. In the simplest use case, entering a standard gene name (e.g. RAD9) into the search field will return all models containing that gene. Wild-card searches are permitted (e.g. RAD* will search for models containing any gene with a name that begins with the letters RAD, see Figure 3). All gene queries are also checked against a list of gene name synonyms, which are drawn from the latest release of the GO database (47). In addition, searches can be limited to models from specific publications or to models containing genes from specific organisms.
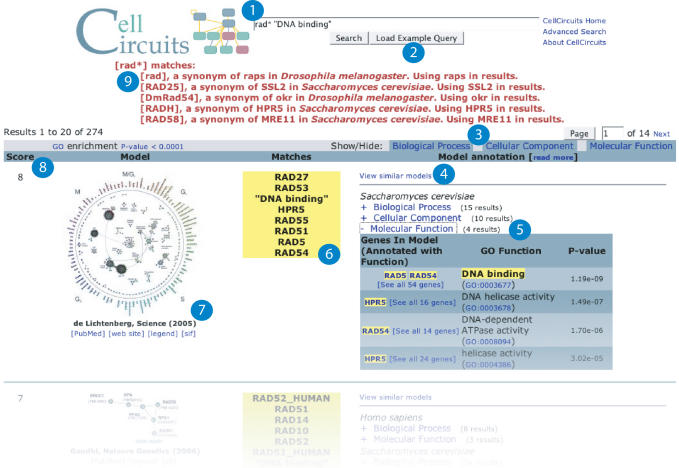
Figure 3.
Web interface (www.cellcircuits.org). Results using RAD* and ‘DNA binding’ as the search query (circle 1). A total of 274 subnetwork models are returned. The search output includes a graphical representation of the model (circle 7), the genes and GO terms from the model that match the query (circle 6), alternative gene names or synonyms matching the query (circle 9), the total number of matches (circle 8), enriched GO terms (circle 5 and 3), a link to view similar models (circle 4) and a link to example search queries (circle 2).
Searches based on gene function are also supported. The CellCircuits database automatically scores all models for GO functional enrichment using the hypergeometric test (see Materials and Methods). Such tests had been originally applied in only 3 out of the 11 curated publications. The enrichment results are stored with each model in the database as meta-data, allowing users to search for models that are enriched for genes having a particular annotation. For example, some of the same models retrieved by searching for RAD9 can also be retrieved by searching for GO annotations associated with this gene. Queries may include exact GO ID numbers (e.g. GO:0006974) or partial or complete GO term names (e.g. ‘DNA damage’ or ‘integrity checkpoint’; these must be enclosed in double quotes).
More than one gene, GO annotation or wild-card may be included in a query. If a model matches multiple search terms, it will be ranked higher in the results. All search results include graphical representations of the models, links to the original publication, the organism(s) modeled, the genes or GO annotations from the search query that were found in each model and the hypergeometric P-value of enrichment for any GO annotations (Figure 3).
Meta-analysis of models
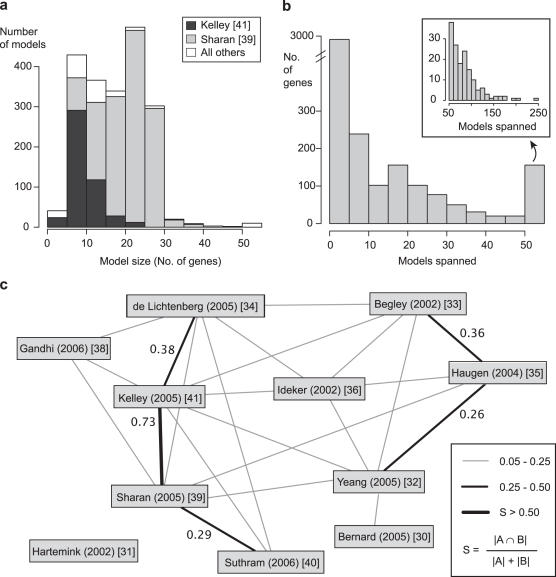
Collecting published network models within a single database allowed us to survey the state of computational analysis of large interaction datasets. Scoring all models for GO functional enrichment (described in the previous section) is an example of such analyses. Another example, the observed sizes of models from all 11 publications, is shown in Figure 4a. On average, the 2019 models in the database contained ∼18 proteins and 36 interactions with 95% of models containing between 5 and 30 proteins. However, this distribution was heavily influenced by two publications (39,41) which together contributed over 90% of the models in the database.
Figure 4.
Meta-analysis of models. (a) Histogram of the number of genes or proteins per model. (b) Histogram of the number of genes (y-axis) that are contained in a given number of models (x-axis). The inset is an expanded view of the genes that span over 50 models. (c) Overlap between network modeling publications. Thicker lines represent greater similarity between the sets of models published in two publications (see legend). Similarity is measured by the number of distinct models that share one or more interactions (yeast interactions only) divided by the total number of models in both publications. Interactions are shared between almost every pair of publications, but for clarity, similarity scores <0.05 are not shown.
To assess the overlap between models, we examined the extent to which the same proteins appeared in multiple models (Figure 4b). Although a protein was shared by approximately nine models on average, the majority were found in only one or two models. Thirty-five proteins appeared in over 100 models (<5% of all models in the database). Interestingly, among these were all six of the yeast ATPases in the 26S proteasome (RPT1–6), components of the yeast and worm 20S proteasome, and several yeast, worm and fly protein kinases. The pervasiveness of these proteins in models may reflect their broad evolutionary conservation across species, a high degree of connectivity in the protein network, their popularity in the biological literature or their functional roles in many distinct biological processes (i.e. pleiotropy).
The results of our model overlap analyses are accessible through the web interface. Each model is annotated in the CellCircuits database with a list of similar models, defined as those that contain at least three of the same genes. Clicking the ‘View similar models’ link in the search results will display these models (Figure 3, circle 4). Currently, only the number of shared genes is used to assess similarity between models. However, more complex measures could be envisioned, potentially making CellCircuits, itself, a resource for comparing several similar models (perhaps corresponding to the same biological process) and showing the differences between them.
On a broader scale, we also assessed the extent to which publications covered overlapping regions of the protein interactome using a pairwise similarity score (see Materials and Methods). Results are shown in Figure 4c. Although our similarity score was permissive such that some overlap was expected between every pair of publications, only 5 out of the 55 possible pairs showed over 25% similarity. Thus, it appears that the different modeling publications are, to some degree, capturing different regions of the protein interaction network [excluding (39,41), see Figure 4c]. Furthermore, in the future, this kind of meta-analysis could be used to determine how the results from new publications differ from existing models.
DISCUSSION
In summary, CellCircuits version 1.0 provides a clearinghouse in which hypothetical pathway models derived from large-scale protein networks may be easily accessed, queried and exported for further study. The 11 publications included in this initial release were chosen to cover a broad range of network model types with a bias towards publications that provided models in both graphical and machine-readable format. Beyond this proof-of-principle, a significant question is whether, or to what extent, all past and future network models might be incorporated.
On one hand, the field of network biology is still young such that the number of relevant previous publications is probably <50. On the other hand, the rapid adoption of systems and network approaches will make capturing information from all future works a daunting prospect if the models are not readily accessible. CellCircuits complements existing efforts that have begun to address this challenge, such as markup languages for describing models [BioPAX (45) and SBML (44)] and the BioModels Database of quantitative, kinetic models (27). Similar to biological sequence and microarray databases, we envision CellCircuits as a valuable resource for storing, accessing and updating network models across the wider biological research community.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We acknowledge funding from the National Science Foundation (NSF 0425926) and thank the members of the Ideker lab for testing and suggesting improvements to the web interface. Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation (NSF 0425926).
Conflict of interest statement. None declared.
REFERENCES
- 1.Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cusick M.E., Klitgord N., Vidal M., Hill D.E. Interactome: gateway into systems biology. Hum. Mol. Genet. 2005;14:R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 3.Reguly T., Breitkreutz A., Boucher L., Breitkreutz B.-J., Hon G., Myers C., Parsons A., Friesen H., Oughtred R., Tong A., et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ooi S.L., Pan X., Peyser B.D., Ye P., Meluh P.B., Yuan D.S., Irizarry R.A., Bader J.S., Spencer F.A., Boeke J.D. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 2006;22:56–63. doi: 10.1016/j.tig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Stuart J.M., Segal E., Koller D., Kim S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 6.Krallinger M., Valencia A. Text-mining and information-retrieval services for molecular biology. Genome Biol. 2005;6:224. doi: 10.1186/gb-2005-6-7-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bader J.S., Chaudhuri A., Rothberg J.M., Chant J. Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- 8.von Mering C., Krause R., Snel B., Cornell M., Oliver S.G., Fields S., Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 9.Jansen R., Yu H., Greenbaum D., Kluger Y., Krogan N.J., Chung S., Emili A., Snyder M., Greenblatt J.F., Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 10.Lee I., Date S.V., Adai A.T., Marcotte E.M. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- 11.Joyce A.R., Palsson B.O. The model organism as a system: integrating ‘omics’ data sets. Nature Rev. Mol. Cell Biol. 2006;7:198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- 12.Sharan R., Ideker T. Modeling cellular machinery through biological network comparison. Nat. Biotechnol. 2006;24:427–433. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- 13.Vidal M. Interactome modeling. FEBS Lett. 2005;579:1834–1838. doi: 10.1016/j.febslet.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 14.Xenarios I., Salwinski L., Duan X.J., Higney P., Kim S.M., Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mewes H.W., Frishman D., Mayer K.F., Munsterkotter M., Noubibou O., Pagel P., Rattei T., Oesterheld M., Ruepp A., Stumpflen V. MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res. 2006;34:D169–D172. doi: 10.1093/nar/gkj148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bader G.D., Betel D., Hogue C.W. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stark C., Breitkreutz B.-J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A., et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caspi R., Foerster H., Fulcher C.A., Hopkinson R., Ingraham J., Kaipa P., Krummenacker M., Paley S., Pick J., Rhee S.Y., et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006;34:D511–D516. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gough N.R., Ray L.B. Mapping cellular signaling. Sci. STKE. 2002;2002:EG8. doi: 10.1126/stke.2002.135.eg8. [DOI] [PubMed] [Google Scholar]
- 23.Ananko E.A., Podkolodny N.L., Stepanenko I.L., Podkolodnaya O.A., Rasskazov D.A., Miginsky D.S., Likhoshvai V.A., Ratushny A.V., Podkolodnaya N.N., Kolchanov N.A. GeneNet in 2005. Nucleic Acids Res. 2005;33:D425–D427. doi: 10.1093/nar/gki077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krull M., Pistor S., Voss N., Kel A., Reuter I., Kronenberg D., Michael H., Schwarzer K., Potapov A., Choi C., et al. TRANSPATH(R): an information resource for storing and visualizing signaling pathways and their pathological aberrations. Nucleic Acids Res. 2006;34:D546–D551. doi: 10.1093/nar/gkj107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karp P.D. Call for an enzyme genomics initiative. Genome Biol. 2004;5:401. doi: 10.1186/gb-2004-5-8-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts R.J. Identifying protein function—a call for community action. PLoS Biol. 2004;2:E42. doi: 10.1371/journal.pbio.0020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Novere N., Bornstein B., Broicher A., Courtot M., Donizelli M., Dharuri H., Li L., Sauro H., Schilstra M., Shapiro B., et al. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34:D689–D691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feller W. An Introduction to Probability Theory and Its Application, 3rd edn. NY: John Wiley & Sons, Inc.; 1968. [Google Scholar]
- 29.Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernard A., Hartemink A.J. Informative structure priors: joint learning of dynamic regulatory networks from multiple types of data. Pac. Symp. Biocomput. 2005;2005:459–470. [PubMed] [Google Scholar]
- 31.Hartemink A.J., Gifford D.K., Jaakkola T.S., Young R.A. Combining location and expression data for principled discovery of genetic regulatory network models. Pac. Symp. Biocomput. 2002;2002:437–449. [PubMed] [Google Scholar]
- 32.Yeang C.H., Mak H.C., McCuine S., Workman C., Jaakkola T., Ideker T. Validation and refinement of gene-regulatory pathways on a network of physical interactions. Genome Biol. 2005;6:R62. doi: 10.1186/gb-2005-6-7-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Begley T.J., Rosenbach A.S., Ideker T., Samson L.D. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol. Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- 34.de Lichtenberg U., Jensen L.J., Brunak S., Bork P. Dynamic complex formation during the yeast cell cycle. Science. 2005;307:724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
- 35.Haugen A.C., Kelley R., Collins J.B., Tucker C.J., Deng C., Afshari C.A., Brown J.M., Ideker T., Van Houten B. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 2004;5:R95. doi: 10.1186/gb-2004-5-12-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ideker T., Ozier O., Schwikowski B., Siegel A.F. Discovering regulatory and signaling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl. 1):S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 37.Maciag K., Altschuler S.J., Slack M.D., Krogan N.J., Emili A., Greenblatt J.F., Maniatis T., Wu L.F. Systems-level analyses identify extensive coupling among gene expression machines. Mol. Syst. Biol. 2006;2:E1–E14. doi: 10.1038/msb4100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gandhi T.K., Zhong J., Mathivanan S., Karthick L., Chandrika K.N., Mohan S.S., Sharma S., Pinkert S., Nagaraju S., Periaswamy B., et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nature Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 39.Sharan R., Suthram S., Kelley R.M., Kuhn T., McCuine S., Uetz P., Sittler T., Karp R.M., Ideker T. Conserved patterns of protein interaction in multiple species. Proc. Natl Acad. Sci. USA. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suthram S., Sittler T., Ideker T. The Plasmodium protein network diverges from those of other eukaryotes. Nature. 2005;438:108–112. doi: 10.1038/nature04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelley R., Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat. Biotechnol. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milo R., Shen-Orr S., Itzkovitz S., Kashtan N., Chklovskii D., Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 43.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hucka M., Finney A., Sauro H.M., Bolouri H., Doyle J.C., Kitano H., Arkin A.P., Bornstein B.J., Bray D., Cornish-Bowden A., et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 45.BioPAX working group. 2004. BioPAX—biological pathways exchange language. Level 1, Version 1.0 Documentation. [Google Scholar]
- 46.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]