Abstract
Transgenic plants producing environmentally benign Bacillus thuringiensis (Bt) toxins are deployed increasingly for insect control, but their efficacy will be short-lived if pests adapt quickly. The diamondback moth (Plutella xylostella), a worldwide pest of vegetables, is the first insect to evolve resistance to Bt toxins in open-field populations. A recessive autosomal gene confers resistance to at least four Bt toxins and enables survival without adverse effects on transgenic plants. Allelic variants of this gene confer resistance in strains from Hawaii, Pennsylvania, and the Philippines. Here we exploited the biphasic nature of Lepidopteran genetic linkage to map this gene in diamondback moth with 207 amplified fragment length polymorphisms as DNA markers. We also cloned and sequenced an amplified fragment length polymorphism marker for the chromosome containing the Bt resistance gene. The results provide a powerful tool for facilitating progress in understanding, monitoring, and managing resistance to Bt.
The common soil bacterium Bacillus thuringiensis (Bt) produces crystals containing proteins that are toxic to certain insects, but are harmless to most other organisms including people, wildlife, and most beneficial insects (1). Genes encoding Bt toxins have been incorporated into and expressed by crop plants, thus providing environmentally benign control of insect pests (2). Lepidopteran larvae, which include some of the world’s most damaging crop pests, are the primary targets of more than 99% of the acreage of currently deployed, Bt-producing transgenic plants (3).
Evolution of resistance by pests is the most serious threat to the continued efficacy of Bt toxins. Although the diamondback moth (Plutella xylostella) is the only insect with resistance to Bt toxins in open-field populations, laboratory selection has produced resistance in several other species of Lepidoptera (4). With millions of hectares of Bt toxin-producing transgenic plants grown yearly (3), other pests are likely to evolve resistance quickly unless effective countermeasures are designed and implemented soon (5, 6). Better understanding of the genetic basis of resistance is essential for developing such countermeasures.
Despite their enormous economic and ecological importance, surprisingly little is known about the genetics of most Lepidoptera. Because of their large number of chromosomes, linkage is difficult to detect and a great many markers are required. The cultivated silkworm Bombyx mori has received the most study; the classical linkage map based on approximately 200 morphological markers (7) is now being augmented by molecular maps based on restriction fragment length polymorphisms (8) and randomly amplified polymorphic DNA fragments (RAPDs) (9, 10). The recent dense coverage of the silkworm genome with RAPD markers (10) illustrates the feasibility of the intensive mapping approach and also the large amount of effort required. Still to be achieved are the integration of the various molecular maps with the classical map and their application in identifying genes underlying silk production and other interesting traits—undoubtedly a long-term effort.
However, most agronomically important Lepidoptera are crop pests, and there is an urgent need to rapidly identify genes causing Bt resistance, before saturation of the entire genome with genetic markers is achieved. An efficient way to overcome this problem is to exploit the biphasic nature of genetic linkage found in Lepidoptera (11–13). Any two loci on the same chromosome are absolutely linked in females, because no crossing-over occurs during oogenesis and the loci cannot recombine. Linkage in males, however, depends on the recombinational distance separating the two loci, because crossing-over does occur during spermatogenesis. This biphasic nature of linkage facilitates a sequential approach to mapping resistance genes. First, absolute linkage in offspring of female-informative crosses is used to identify the linkage groups, each of which corresponds to a chromosome, contributing to resistance. Second, the location of resistance genes within those groups is narrowed down further by analyzing recombinant offspring of male-informative matings.
We used this approach to map the major gene conferring resistance to Bt in diamondback moth. In this species, a recessive autosomal gene confers resistance to at least four Bt toxins (14) and enables survival without adverse effects on transgenic plants (15). Allelic variants of this gene confer resistance in strains from Hawaii, Pennsylvania, and the Philippines (16). Mapping of this gene along with cloning and sequencing of a sequence-tagged site (STS), as described here, can accelerate global efforts to elucidate the molecular mechanism of Bt resistance and to combat it in the field.
MATERIALS AND METHODS
Insect Strains, Crosses, and Bioassays.
We used two strains of diamondback moth from Hawaii: resistant strain NO-QA and susceptible strain LAB-PS. NO-QA was derived from a population in Pearl City, Oahu, that had evolved moderate resistance in response to repeated sprays of Bt subsp. kurstaki in the field (17) and subsequently was selected for extremely high levels of resistance to Bt subsp. kurstaki in the laboratory (18). NO-QA is extremely resistant to the three Cry1A toxins in Bt subsp. kurstaki (Cry1Aa, Cry1Ab, and Cry1Ac) and cross-resistant to Cry1F and Cry1J (14). LAB-PS is a 100% susceptible strain derived via an isofemale line from strain LAB-P (19), which originally was collected from near Pulehu, Maui (20), and reared in the laboratory without exposure to any insecticide for >200 generations.
To generate informative families of diamondback moth for mapping, we crossed the susceptible LAB-PS strain with the Bt-resistant NO-QA strain and separately backcrossed male and female F1 progeny to NO-QA (Fig. 1). Because the Bt resistance is recessive, the F1 should be susceptible, and backcross progeny are expected to segregate 1:1 for resistant vs. susceptible phenotypes. All marker loci heterozygous in the F1 should also segregate 1:1 in the backcrosses (barring segregation distortion or viability effects). Larvae from the parental strains and from F1 and backcross families were tested for resistance by allowing them to feed on cabbage leaf disks that had been dipped in a solution containing 0.1 mg/ml Bt toxin Cry1Ac (14). Mortality was recorded after 5 days. Treated survivors were reared to adulthood and frozen at −70°C. Untreated control larvae from all families also were reared to adulthood without exposure to Cry1Ac and frozen.
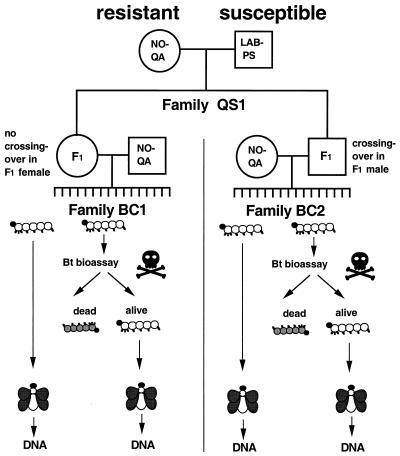
Figure 1.
Experimental design for crosses and bioassay to map Bt resistance. Single-pair crosses between LAB-PS and NO-QA virgin adults produced families of hybrid F1 offspring. A female F1 from hybrid family QS1 was backcrossed to a male NO-QA to produce a family BC1 of offspring; one of her F1 brothers was backcrossed to a female NO-QA to produce family BC2. Approximately 80 backcross offspring from BC1 and BC2 were tested at 0.1 mg/ml Cry1Ac. Survivors were reared to adulthood and frozen for DNA analysis. About 30 additional backcross offspring from each family were reared to adulthood without exposure to Bt and frozen for DNA analysis.
Genetic Markers.
DNA was isolated from adults as described previously (21), but with a sample volume of 20 μl. To obtain amplified fragment length polymorphism (AFLP) markers (22), we used the AFLP Analysis System 1 (Life Technologies, Gaithersburg, MD) according to the manufacturer’s instructions, except that additional primers were synthesized with two instead of three selective bases on the Eco primer. We used all 10 combinations of 1 Eco primer (E-AT or E-AA) and 1 Mse primer (M-CAC, M-CAT, M-CTA, M-CTC, or M-CTG). 33P end-labeled fragments were run on a 6% acrylamide-denaturing sequencing gel and exposed to Kodak X-Omat film for a week.
Phase One: Identification of Linkage Groups and Tests of Their Contribution to Resistance.
We identified which linkage groups segregating in backcross (BC) family BC1 originated from NO-QA and which came from LAB-PS by comparing AFLPs of offspring, parents, and grandparents. This comparison requires two steps: (i) determining which AFLPs are informative for linkage in the female F1 parent and (ii) determining the source (LAB-PS or NO-QA) of each informative band. Only bands that were present in the F1 mother and absent in the NO-QA father of the backcross were informative. All informative AFLP bands were expected to segregate 1:1 in the untreated backcross progeny. Because AFLPs are dominant, any bands present in the father would confound this pattern in the offspring.
Informative AFLPs then were classified as L bands (derived from LAB-PS) or N bands (derived from NO-QA). L bands were present in the susceptible LAB-PS grandparent and absent from the resistant NO-QA grandparent. L bands thus had the same linkage phase as the susceptible allele at resistance loci in backcross BC1, and linkage to resistance was detected by underrepresentation of L bands among treated survivors. Conversely, N bands were absent from the LAB-PS grandparent and present in the NO-QA grandparent. N bands thus had the same linkage phase as the resistant allele at resistance loci in BC1, with linkage to resistance detected by the overrepresentation of N bands among treated survivors.
As expected, a majority of informative AFLPs were L bands. N bands were less common because they had to be present in the NO-QA grandparent and absent in the NO-QA parent of the backcross. This requirement tends to exclude bands that were either very common or very rare in NO-QA. Note that by definition, N bands are not fixed in the NO-QA colony, because the NO-QA grandparent has them and the NO-QA parent of the backcross lacks them. Because AFLPs are dominant, any bands that are fixed in NO-QA and absent in LAB-PS would not be informative in a backcross to NO-QA; such bands were not used here.
A Pascal program written by D.G.H. (DBM3Lnk.p) was used to identify groups of cosegregating AFLPs in the female-informative family BC1. Two L bands cosegregate if they show exactly the same pattern of presence and absence across the entire set of offspring; likewise for two N bands. An L band and an N band cosegregate if they show exactly the opposite pattern of presence and absence across the entire set of offspring (because they are of opposite phase). The program looks for groups of cosegregating, nonrecombinant loci while allowing for a small number of genotyping errors, to ensure that such errors do not artificially inflate the number of independently segregating groups.
After linkage groups were identified, we determined the source (NO-QA or LAB-PS) of the chromosome passed on from the F1 mother to her progeny for each AFLP linkage group and for each offspring. For example, if an individual backcross offspring lacked all the L bands from a particular linkage group, we inferred that the F1 mother had contributed the NO-QA-derived chromosome corresponding to that linkage group. Such an offspring thus carried two NO-QA-derived homologues (the other from its NO-QA father). Conversely, if the offspring had all the L bands for the linkage group, the LAB-PS chromosome must have been transmitted. That offspring would carry one NO-QA-derived and one LAB-PS-derived homologue of that chromosome. N bands showed an inverse pattern that permitted the same types of deductions.
To test whether each linkage group contributed to resistance in BC1, the interaction χ2 statistic was calculated from 2×2 tables comparing counts of Bt survivors and untreated controls with chromosomal origin (NO-QA or LAB-PS).
Phase Two: Mapping Resistance Within a Linkage Group.
Because crossing-over does occur in males, we could map resistance within a linkage group by using the frequencies of recombinants from backcross BC2, in which an F1 male was backcrossed to an NO-QA female (Fig. 1). For markers on AFLP group 7 that were also informative in BC2, mapmaker/exp 3.0 (23) was used to determine the most likely order of markers and to estimate recombination rates. Bailey’s method of examining a 2×2 contingency table for evidence of a deficiency of double crossovers (ref. 23, p. 145) was used to test for interference.
Cloning and Sequencing an AFLP Marker for Resistance.
The AFLP band corresponding to marker EaaMcta-04.70 was excised from five lanes of the dried acrylamide gel with a razor blade and soaked in 100 μl of distilled water overnight. The product was reamplified from 10 μl of supernatant by using the same conditions as the first (nonselective) PCR amplification in the AFLP procedure, with the nonselective Eco and Mse primers. The product was isolated from an agarose gel and cloned into the plasmid vector pGEM-T Easy by following the supplier’s instructions (Promega). Sequencing was performed on an ABI 373A sequencer (Applied Biosystems). The sequence obtained was used to search GenBank, using the National Center for Biotechnology Information server at http://www.ncbi.nlm.nih.gov and blast and tblastn searches of the nucleotide databases. The sequence also was translated in all six reading frames and used for blastp searches of the protein databases.
Specific primers designed from this sequence were c39–451p1 (5′-CCG TGC TGA GCA TTG GAC AGT GAG-3′) and c39–451p2 (5′-TTA ACT ATA TTT GTT GGT GAC GAT AAG GTG-3′). These primers were used to amplify a fragment from genomic DNA as follows: PCR was carried out in a 25-μl reaction mixture consisting of 10 mM Tris (pH 8.5), 50 mM KCl, 1.5 mM MgCl2, 200 μM each of dATP, dTTP, dGTP, and dCTP, 50 ng of each primer, 10 ng of genomic DNA, and 0.5 unit of AmpliTaq DNA Polymerase (Perkin–Elmer). Cycling conditions on the Perkin–Elmer DNA Thermal Cycler 480 were 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 2 min, and a final 5 min at 72°C.
RESULTS
Phase One: Identification of the Linkage Group Contributing to Resistance.
Feeding on cabbage leaves treated with 0.1 mg/ml Bt toxin Cry1Ac killed 100% of 30 larvae from the susceptible LAB-PS strain, but only 5% of 60 larvae from the resistant NO-QA strain. In each of six F1 families produced by crossing LAB-PS with NO-QA, mortality was 100%, as expected for recessive resistance. Mean mortality in six backcross families (F1 × NO-QA) was 47.5% (range = 37.5–57.5%, n = 79–82 larvae per family), which is consistent with the expected 1:1 segregation of homozygous-resistant and heterozygous-susceptible individuals.
To obtain marker loci, we scored AFLPs (22) by using DNA isolated from 24 offspring of backcross family BC1 (female F1 × male NO-QA) that had survived the leaf dip bioassay and from 26 of their sibs that had been reared to adulthood without being treated (Fig. 1). Because the F1 parent of BC1 was female, we could use nonrecombining sets of AFLPs to identify linkage groups, each of which corresponds to a chromosome.
The analysis identified 29 linkage groups that each had 2–15 markers. We also included 2 additional linkage groups that had only single markers, because cytogenetic evidence shows 31 chromosome pairs for diamondback moth (24). The net result was a total of 207 AFLPs clustered into 31 linkage groups (Fig. 2). The sex chromosome (Z) was labeled group 1. The autosomal linkage groups were labeled from 2 to 31, with group 2 having the most AFLP markers and groups 30 and 31 having the fewest.
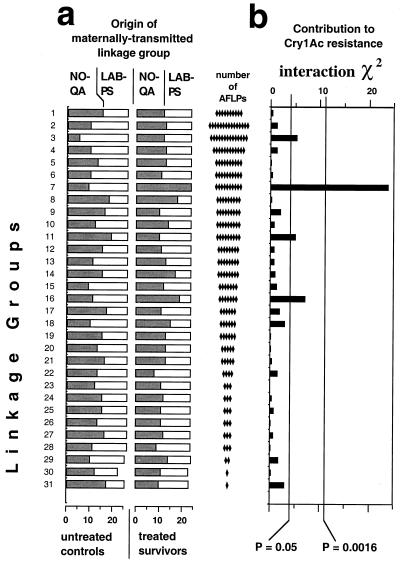
Figure 2.
Scanning the linkage groups for their contribution to Bt resistance in family BC1. (a) Distribution of the homologues transmitted by the F1 parent, derived from NO-QA and LAB-PS for each linkage group among Bt survivors and untreated controls. (b) χ2 tests of independence between treatments (Bt survivors vs. untreated controls) and chromosomal origin (NO-QA vs. LAB-PS), conducted for each AFLP linkage group. The Bonferroni correction for 31 simultaneous statistical tests at the nominal level of α = 0.05 was used to evaluate the results, producing a corrected probability value of P = 0.0016 (0.05/31). Of the 31 linkage groups, only AFLP group 7 had a χ2 value associated with a probability below this threshold (χ2 = 21.8, df = 1, P < 0.0001), thus making a significant contribution to resistance.
We identified which linkage groups segregating in the backcross originated from NO-QA and which came from LAB-PS by comparing AFLPs of offspring, parents, and grandparents (see Materials and Methods for details). Among the 26 untreated progeny, the expected 1:1 segregation was observed for each NO-QA-derived linkage group and its LAB-PS homologue (Fig. 2), except for AFLP group 3, with the NO-QA homologue overrepresented, and AFLP group 11, with it underrepresented.
To determine which linkage groups contributed to Bt resistance, we examined AFLPs of the 24 resistant survivors of the bioassay. Any chromosomes derived from NO-QA carrying one or more resistance genes should be overrepresented in this group, relative to the untreated controls, leading to a significant interaction between chromosomal origin (NO-QA vs. LAB-P) and treatment (treated survivors vs. controls).
Only one linkage group, AFLP group 7, was significantly associated with resistance by this criterion (Fig. 2). Moreover, all treated survivors from BC1 had both of their AFLP group 7 chromosomes derived from NO-QA (one from the F1 mother and the other from the NO-QA father), consistent with the recessive nature of resistance. We conclude that group 7 contains the major gene or genes responsible for the Cry1Ac resistance in NO-QA.
Three other AFLP groups (3, 11, and 16) showed suggestive responses that merit further consideration (Fig. 2). The probability levels associated with the interactions for these three groups would have been considered significant if they were from single tests of a priori hypotheses, but do not attain statistical significance given that we performed multiple a posteriori comparisons. Further, the deviations seen in groups 3 and 11 are not due to overrepresentation of NO-QA-derived chromosomes among survivors, as would be expected if these groups contained resistance genes. Instead, the deviations occurred in the untreated controls, as noted above, and cannot be caused by resistance because these individuals were not exposed to toxin. AFLP group 16, on the other hand, does show the pattern expected of a minor resistance gene—slight overrepresentation of the NO-QA homologue among treated survivors compared with a more equal representation among untreated controls. Thus, we revisit this suggestive but nonsignificant group in more detail below.
Phase Two: Mapping the Resistance Gene Within a Linkage Group.
To map the group 7 genes in the second phase of the analysis, we examined 26 bioassay survivors and 26 untreated controls from the reciprocal backcross family BC2 (female NO-QA × male F1), whose F1 father was a brother of the F1 mother of family BC1 (Fig. 1). Seven of the 10 group 7 AFLPs were examined in BC2 offspring, and 5 of these were informative in that family. The pattern of recombinants among treated and control offspring enabled construction of a linkage map 47 cM in length (Fig. 3). The large number of recombinants among BC2 offspring of the F1 male contrasts with the absolute linkage of these same loci observed in BC1 offspring of the F1 female, consistent with the biphasic nature of linkage.
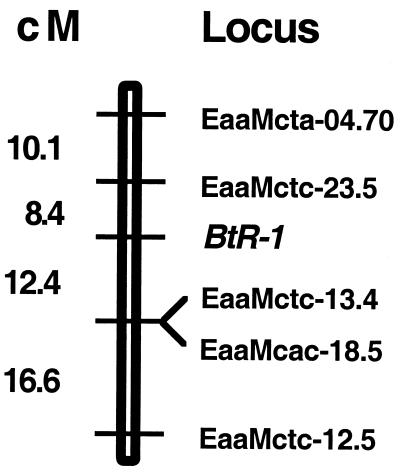
Figure 3.
Mapping the resistance locus within AFLP group 7 in family BC2. Standard likelihood methods (16) were used to determine the order and spacing of the five group 7 AFLPs informative in family BC2 and to position the BtR-1 resistance locus among them. Positive interference (a deficiency of double crossovers) was detected among the AFLP loci (χ2 = 4.48, df = 1, P < 0.05), so the Kosambi mapping function was used. The locus order shown is more than 100 times more likely than any other possible order of the loci.
The resistance trait segregating in these backcrosses maps to a single location between the AFLP markers EaaMctc-13.4 (12.4 ± 6 cM) and EaaMctc-23.5 (8.4 ± 6 cM). All survivors of exposure to Cry1Ac were homozygous for the NO-QA-derived sequence in this chromosomal region. No other region of group 7 shows this behavior. Thus, the gene or genes responsible for Cry1Ac resistance in NO-QA must reside here. We name this locus BtR-1 because it is the first Bt resistance factor mapped in diamondback moth.
Two of the group 16 markers (EatMcta-13.8 and EaaMcac-25.0) were informative in the 52 offspring of the BC2 family, giving us the opportunity to determine whether their nonsignificant but suggestive association with resistance in BC1 held up in an independent test with a larger sample size. Segregation of both markers was nearly 1:1 in both treated survivors and untreated controls, with no evidence of the pattern seen in BC1. Thus, neither marker showed a significant interaction with resistance in BC2 (interaction χ2 = 0.018 for EatMcta-13.8 and 0.04 for EaaMcac-25.0; P > 0.80 for each marker).
Cloning and Sequencing of an AFLP Marker for Resistance.
We cloned and sequenced the AFLP band for marker EaaMcta-04.70, which was linked to BtR-1 at 18.5 ± 6 cM. The 776-bp sequence showed no significant similarity to other sequences in GenBank. Thus, this new sequence represents an STS for the chromosome containing BtR-1. This STS (GenBank accession no. AF149306) can be used in linkage tests with other resistant strains, whether or not they show the same AFLP polymorphisms as the NO-QA and LAB-PS strains. To verify this, we constructed PCR primers to amplify an internal fragment of the AFLP band. Genomic DNA from LAB-PS produced a PCR product upon amplification, which was expected because the original AFLP band was LAB-PS-derived. DNA from NO-QA also produced a PCR product, showing that the failure of NO-QA to produce the LAB-PS-derived EaaMcta-04.70 AFLP band was not a result of a deletion of the entire region, but of changes in the AFLP restriction site or primer region. Moreover, the NO-QA PCR product was about 20 bp smaller than the LAB-PS PCR product, a size difference easily visualized by 6% PAGE (not shown). As expected, the size polymorphism showed the same segregation pattern in backcross offspring as the presence/absence pattern of the original AFLP.
DISCUSSION
The simplest explanation of our results is that the BtR-1 locus consists of a single gene. Evidence from earlier tests of allelism (16) strongly suggests that a single gene is responsible for the recessive resistance to Cry1Ac of NO-QA. Moreover, cross-resistance tests have shown that the gene in NO-QA conferring resistance to Cry1Ac also confers resistance to Cry1Aa, Cry1Ab, and Cry1F (14). Thus, BtR-1 likely corresponds to the multitoxin Bt resistance gene of diamondback moth (14).
The mapping of a major Bt resistance gene from diamondback moth immediately opens several important avenues of investigation. First, mapping BtR-1 greatly facilitates linkage tests of candidate resistance genes in diamondback moth. The leading hypothesis is that resistance is conferred by altered Bt toxin-binding sites in the larval midgut epithelium (25–27); however, elucidation of the role of various putative Bt receptor proteins in resistance has proved elusive (28–30). Linkage tests with the STS generated from EaaMcta-04.70 now can reject or help to confirm the involvement of individual candidate genes in NO-QA resistance. Second, mapping BtR-1 in diamondback moth enables comparison with the only other insect in which Bt resistance genes have been mapped (31), Heliothis virescens. Third, the biphasic approach can be used to analyze Bt-resistant strains of other Lepidoptera such as Indianmeal moth, beet armyworm, European corn borer, and pink bollworm, enabling characterization of other resistance genes that may be homologous to BtR-1 of diamondback moth.
Moreover, the biphasic approach can be used to efficiently map the major loci underlying any inherited trait in Lepidoptera, which should greatly facilitate future studies in genetics and comparative genomics of this important insect order. Although the biphasic approach has been applied for some time to higher Diptera such as Drosophila, Musca, and Lucilia, it is especially advantageous with members of the Lepidoptera because of their large number of chromosomes. Despite the relative simplicity of the biphasic method with two backcrosses (one with crossing-over during meiosis and one without), the most recent mapping efforts in the most intensively studied lepidopteran, Bombyx mori, have not exploited this approach. Instead, Bombyx geneticists have used F2 mapping populations in which the products of the two types of meiosis are confounded in the offspring (8–10). This necessitates ad hoc methods of analysis that do not make full use of the genetic information in the cross (e.g., ref. 10).
Although some Bt resistance mechanisms might be species-specific, a general pattern suggestive of a mechanism common to many species is beginning to emerge. Several strains of diamondback moth, the YHD2 strain of H. virescens and the 343R strain of Indianmeal moth, are characterized by extremely high resistance to at least one Cry1A toxin, little or no cross-resistance to Cry1C, recessive inheritance, and reduced binding of at least one Cry1A toxin (32). If the genomics approach reveals that this mode of Bt resistance has a common genetic basis in different species of insects, as has been observed for resistance to some synthetic insecticides (33–35), efforts to monitor and manage resistance to Bt in many pests could be greatly enhanced.
Acknowledgments
We thank Frantisek Marec and Quang Hoa Nguyen Thi for information on diamondback moth cytogenetics, Susan Meyer for technical assistance, and Yves Carriere, Daphne Pham, Yoonseong Park, George Roderick, and three anonymous reviewers for comments on the manuscript. This work was supported by U.S. Department of Agriculture National Research Initiative Competitive Grant 96–35302-3470, U.S. Department of Agriculture Western Regional Pesticide Impact Assessment Program Grant 97RA1810/WR337, and Cooperative State Research Service–Tropical and Subtropical Agricultural Research Program Grant 95–34135-1771. This is Technical Contribution No. 4499 of the South Carolina Agriculture and Forestry Research System.
ABBREVIATIONS
- AFLP
amplified fragment length polymorphism
- BC
backcross
- Bt
Bacillus thuringiensis
- STS
sequence-tagged site
Footnotes
Data deposition: The sequence described in this paper has been deposited in the GenBank database (accession no. AF149306).
References
- 1.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler D R, Dean D H. Microbiol Mol Biol Rev. 1998;62:755–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuler T H, Poppy G M, Kerry B R, Denholm I. Trends Biotechnol. 1998;16:169–175. doi: 10.1016/S0167-7799(98)01298-0. [DOI] [PubMed] [Google Scholar]
- 3.James C. Int Serv Acquisit Agri-Biotech Appl Briefs. 1998;8:1–43. [Google Scholar]
- 4.Tabashnik B E. Annu Rev Entomol. 1994;39:47–79. [Google Scholar]
- 5.McGaughey W H, Whalon M E. Science. 1992;258:1451–1455. doi: 10.1126/science.258.5087.1451. [DOI] [PubMed] [Google Scholar]
- 6.Mellon M, Rissler J, editors. Now or Never: Serious New Plans to Save a Natural Pest Control. Cambridge, MA: Union of Concerned Scientists; 1998. [Google Scholar]
- 7.Goldsmith M R. In: Molecular Model Systems in the Lepidoptera. Goldsmith M R, Wilkins A S, editors. Cambridge, U.K.: Cambridge Univ. Press; 1995. pp. 21–76. [Google Scholar]
- 8.Shi J, Heckel D G, Goldsmith M R. Genet Res. 1995;66:109–126. [Google Scholar]
- 9.Promboon A, Shimada T, Fujiwara H, Kobayashi M. Genet Res. 1995;66:1–7. [Google Scholar]
- 10.Yasukochi Y. Genetics. 1998;150:1513–1525. doi: 10.1093/genetics/150.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson R. Lepidoptera Genetics. Oxford: Pergamon; 1971. [Google Scholar]
- 12.Suomalainen E, Cook L M, Turner J R G. Hereditas. 1973;74:302–304. [Google Scholar]
- 13.Tazima T. Genetics of the Silkworm. London: Logos; 1964. [Google Scholar]
- 14.Tabashnik B E, Liu Y-B, Finson N, Masson L, Heckel D G. Proc Natl Acad Sci USA. 1997;94:1640–1644. doi: 10.1073/pnas.94.5.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramachandran S, Buntin G D, All J N, Tabashnik B E, Raymer P L, Adang M J, Pulliam D A, Stewart C N. J Econ Entomol. 1998;91:1239–1244. [Google Scholar]
- 16.Tabashnik B E, Liu Y-B, Malvar T, Heckel D G, Masson L, Ballester V, Granero F, Ménsua J L, Ferré J. Proc Natl Acad Sci USA. 1997;94:12780–12785. doi: 10.1073/pnas.94.24.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabashnik B E, Cushing N L, Finson N, Johnson M W. J Econ Entomol. 1990;83:1671–1676. [Google Scholar]
- 18.Tabashnik B E, Finson N, Groeters F R, Moar W J, Johnson M W, Luo K, Adang M A. Proc Natl Acad Sci USA. 1994;91:4120–4124. doi: 10.1073/pnas.91.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y-B, Tabashnik B E. J Econ Entomol. 1998;91:1032–1037. [Google Scholar]
- 20.Tabashnik B E, Cushing N L, Johnson M W. J Econ Entomol. 1987;80:1091–1099. [Google Scholar]
- 21.Zraket C A, Barth J L, Heckel D G, Abbott A G. In: Molecular Insect Science. Hagedorn H H, Hildebrand J G, Kidwell M G, Law J H, editors. New York: Plenum; 1990. pp. 13–20. [Google Scholar]
- 22.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Fritjers A, Pot J, Peleman J, Kuiper M, Zabeau M. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey N T J. Introduction to the Mathematical Theory of Genetic Linkage. Oxford: Clarendon; 1961. [Google Scholar]
- 24.Nguyen Thi Q H. Evaluation of Population Suppression by Irradiated Lepidoptera and their Progeny (IAEA-D4-RC-561) Vienna: International Atomic Energy Agency; 1995. pp. 123–148. [Google Scholar]
- 25.Gill S S, Cowles E A, Pietrantonio P V. Annu Rev Entomol. 1992;37:615–636. doi: 10.1146/annurev.en.37.010192.003151. [DOI] [PubMed] [Google Scholar]
- 26.Ferré J, Real M D, van Rie J, Jansens S, Peferoen M. Proc Natl Acad Sci USA. 1991;88:5119–5123. doi: 10.1073/pnas.88.12.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Rie J, McGaughey W H, Johnson D E, Barnett B D, van Mellaert H. Science. 1990;247:72–74. doi: 10.1126/science.2294593. [DOI] [PubMed] [Google Scholar]
- 28.Lee M K, Rajamohan F, Gould F, Dean D H. Appl Environ Microbiol. 1995;61:3836–3842. doi: 10.1128/aem.61.11.3836-3842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo K, Tabashnik B, Adang M. Appl Environ Microbiol. 1997;63:1024–1027. doi: 10.1128/aem.63.3.1024-1027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohammed S I, Johnson D E, Aronson A I. Appl Environ Microbiol. 1996;62:4168–4173. doi: 10.1128/aem.62.11.4168-4173.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heckel D G, Gahan L J, Gould F, Anderson A. J Econ Entomol. 1997;90:75–86. [Google Scholar]
- 32.Tabashnik B E, Liu Y-B, Malvar T, Heckel D G, Masson L, Ferré J. Philos Trans R Soc London B. 1998;353:1751–1756. [Google Scholar]
- 33.Martinez-Torres D, Devonshire A L, Williams M S. Pestic Sci. 1997;51:265–270. [Google Scholar]
- 34.Brown T M. In: Managing Resistance to Agrochemicals. Green M B, LeBaron H M, Moberg W K, editors. Washington, DC: Am. Chem. Soc.; 1990. pp. 61–76. [Google Scholar]
- 35.ffrench-Constant R H, Pittendrigh B, Vaughan A, Anthony N. Philos Trans R Soc London B. 1998;353:1685–1693. doi: 10.1098/rstb.1998.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]