Abstract
A functional transcription elongation complex can be formed without passing through a promoter by adding a complementary RNA primer and core Escherichia coli RNA polymerase in trans to an RNA-primed synthetic bubble-duplex DNA framework. This framework consists of a double-stranded DNA sequence with an internal noncomplementary DNA “bubble” containing a hybridized RNA primer. On addition of core polymerase and the requisite NTPs, the RNA primer is extended in a process that manifests most of the properties of in vitro transcription elongation. This synthetic elongation complex can also be assembled by using holo rather than core RNA polymerase, and in this study we examine the interactions and fate of the σ70 specificity subunit of the holopolymerase in the assembly process. We show that the addition of holopolymerase to the bubble-duplex construct triggers the dissociation of the sigma factor from some complexes, whereas in others the RNA oligomer is released into solution instead. These results are consistent with an allosteric competition between σ70 and the nascent RNA strand within the elongation complex and suggest that both cannot be bound to the core polymerase simultaneously. However, the dissociation of σ70 from the complex can also be stimulated by binding of the holopolymerase to the DNA bubble duplex in the absence of a hybridized RNA primer, suggesting that the binding of the core polymerase to the bubble-duplex construct also triggers a conformational change that additionally weakens the sigma–core interaction.
Keywords: Escherichia coli RNA polymerase, core and holo RNA polymerase, sigma factor, synthetic bubble-duplex elongation complexes
The σ70 subunit of the Escherichia coli RNA polymerase serves as a specificity factor to direct the binding of the polymerase to its target promoters (1). After the formation of initiation complexes between the holo RNA polymerase and the promoter region, and during the initial stages of de novo synthesis of nascent RNA chains, the sigma subunit is separated from the complex, leaving the core RNA polymerase to engage in processive template-directed RNA synthesis within a mature elongation complex. The release of the σ70 subunit marks the point at which RNA polymerase becomes committed to processive RNA synthesis and represents an important regulatory event in the overall control of gene expression.
It is known that the sigma subunit interacts with elements of both the −35 and the −10 regions of the E. coli promoter (2, 3). In particular, the interaction at the −10 region seems to be between σ70 and the nontemplate strand (4, 5). This interaction implicates σ70 not only in promoter recognition but also in DNA melting and open promoter complex formation (6). It is also known that abortive initiation can occur, resulting in the release of the initial RNA transcript and the subsequent reformation of the open promoter complex, within which sigma is still present and fully functional (7). The exact template position of the transition to stable and processive elongation, which is manifested by the release of sigma and the end of abortive initiation, seems to be somewhat promoter specific and occurs at nascent transcript lengths ranging from 8 to 12 nt (1, 8–11).
The molecular events of the initiation-elongation transition of the transcription complex, which result in the release of sigma and stabilization of the interaction of the nascent RNA with the polymerase, have not been well defined, although several models for the process have been proposed. These models fall into three general classes. The first class suggests that, perhaps for steric reasons, the transcription complex cannot accommodate both the sigma subunit and a nascent RNA chain of significant length. In this view, the sigma subunit must be ejected from the complex once an RNA chain longer than 8 to 12 nt residues has been synthesized, with the length of the nascent RNA determining the release point of the sigma subunit.
A second class of models posits that the sigma subunit does not translocate from the promoter with the rest of the transcription complex. Rather, because it is known that σ70 interacts with both the DNA of the promoter and the core polymerase, if the forces holding sigma at the promoter are stronger than those holding it to the core polymerase, then translocation of the core polymerase downstream along the template will result (after some deformation of the complex) in its eventual separation from sigma, which remains weakly bound at the promoter and then dissociates into solution. We note that such a scenario applies to most of the promoter activation and specificity factors involved in eukaryotic transcript initiation, although many of these factors bind tightly enough to the DNA genome to permit their retention at the promoter even after the transcription complex has departed (12).
Finally, a third type of model has focused on the kinetics of promoter clearance, suggesting that the transcription complex may function as a “molecular clock,” with the point of release of the sigma factor depending on the time that has elapsed after the departure of the holopolymerase from the initiation position (13), rather than on the length of nascent RNA that has been formed. Clearly these models are not mutually exclusive, and elements of all may apply under certain conditions.
We have used the synthetic RNA–DNA bubble-duplex method of direct elongation complex formation to investigate the release of sigma factor from the transcription complex. Here the RNA–DNA bubble duplex was a synthetic construct composed of two DNA strands that form a DNA bubble flanked by duplex regions, together with an oligomeric RNA transcription primer hybridized within the bubble. On the addition of core RNA polymerase, this system forms a functional elongation complex without passing through the normal processes of promoter-dependent transcript initiation (14–16).‡ Because this system uncouples the elongation steps of transcription from promoter-dependent initiation, we should be able to use it to distinguish the first class of models from the others, because the second and third types of models attribute the sigma release signal primarily to factors related to polymerase binding to the promoter, whereas the first focuses on the growing RNA chain as the critical component that interferes with the retention of sigma within the transcription complex.
To help discriminate and refine these models, we have examined the consequences of binding the holo form of E. coli RNA polymerase to the synthetic RNA–DNA bubble duplex and have followed the fates of the sigma subunit and the RNA primer on elongation complex formation. Our results argue against changes in σ70–promoter interactions as the primary determinant of sigma release and tend to support models that focus on competitive sigma–RNA interactions within the elongation complex and on interactions of the polymerase with the fully formed transcription bubble. These studies also illuminate interactions that may be involved in the regulation of abortive initiation and promoter clearance in promoter-initiated transcription complexes.
MATERIALS AND METHODS
The materials and procedures used in this study, with the exception of those described below, have been published previously (14–16).
RNA Primer Exchange Assay.
Approximately 40-nM concentrations of RNA–DNA bubble duplexes (32P-labeled at the 5′-end of the RNA and assembled as described in ref. 14) were incubated with 70-nM concentrations of either core (provided by Kevin Wilson, University of Oregon, Eugene, OR) or holo (provided by William Rees, University of Oregon, Eugene, OR) E. coli RNA polymerase in transcription buffer (1× TB), which contains 20 mM Hepes, pH 8.0, 150 mM NaOAc, 1 mM DTT, 0.5 mM EDTA, and 125 μg/ml BSA, at a final reaction volume of 10 μl. The reactions were incubated for 3 to 5 min at 30°C, after which an unlabeled RNA primer, 12 nt in length and identical in sequence to the RNA primer present in the RNA–DNA bubble duplexes, was added to a final concentration of 24 μM. After an additional 3 min of incubation, NTPs (ATP, CTP, UTP, GTP) and Mg2+ (at final concentrations of 1 mM and 10 mM, respectively) were added to some of the reactions, and all were incubated for an additional 5 min to permit transcription to proceed.
Transcription was halted by the addition of an equal volume of SDS loading buffer to each sample, resulting in a final concentration of 6% glycerol/0.1% SDS/0.025% bromophenol blue/0.025% xylene cyanole/1× TBE buffer (89 mM Tris borate and 2.5 mM EDTA, pH 8.3) in each sample. Reaction tubes were kept on ice before loading onto a 0.7-mm 10% polyacrylamide (20:1 acrylamide/bis) gel that was 10 cm in length and contained 1× TBE, 0.1% SDS, and 8 mM Mg2+. The running buffer contained 1× TBE, 8 mM Mg(OAc)2, and 0.025% SDS. The gel was run for 2.5 hr at a constant current of 20 mAmp, dried on Whatman 3M paper, and autoradiographed on x-ray film (Kodak X-Omat). The dried gel was quantitated by using a radioanalytical scanner (AMBIS).
Sigma Release Assay.
The proteins used (sigma subunit and core or holo RNA polymerase), with or without added nucleic acid, were incubated for 10 min at 25°C in 10-μl samples containing 20 mM Hepes (pH 8.0), 10 mM Mg(OAc)2, 1 mM DTT, and 0.5 mM EDTA. Here the NTPs included in the transcription reaction included only ATP, CTP, and UTP (100 μM each), resulting in the stalling of the polymerase at position +16 of the template (14). Gel loading buffer was added to a final concentration of 6% glycerol, 1× TBE, 0.025% bromophenol blue, and 0.025% xylene cyanole, and the reaction samples were loaded onto a 0.7-mm 5% polyacrylamide gel (112:1 acrylamide/bis) containing 1× TBE buffer. Gels (7 cm in length) were run in 1× TBE for 40 min at 15 mAmp and then either stained directly with silver (20) or dried on Whatman 3MM paper and subjected to autoradiography on x-ray film (Kodak X-Omat). For antibody staining, proteins within the gels were transferred to DEAE membranes according to published protocols (20, 21), and the membranes were blotted with 1HF monoclonal antibodies (kindly provided by Lam Nguyen, University of Wisconsin) that had been raised against σ70 (21).
RESULTS
RNA Primer Exchange Is Catalyzed by Holo E. coli RNA Polymerase.
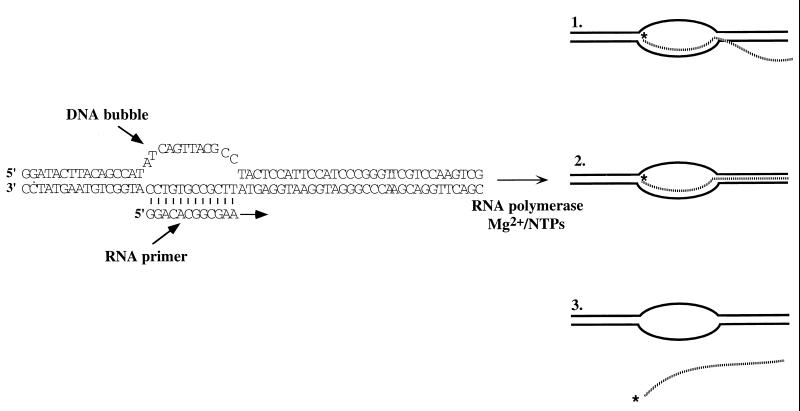
We have previously provided a detailed characterization of transcript elongation reactions performed with synthetic elongation complexes initiated at RNA–DNA bubble-duplex constructs (14–16). Fig. 1 shows the structure of the primed bubble-duplex construct used in these studies and the three types of RNA products that can be formed by adding E. coli RNA polymerase to these constructs in trans in the presence of Mg2+ and appropriate NTPs. Product 1 is a completed RNA transcript that has rehybridized to the single-stranded template sequence within the noncomplementary bubble. Product 2 is a completed transcript that remains fully hybridized to the DNA template strand. Product 3 is a completed transcript that has been properly and totally released from the bubble-duplex complex. These different hybridization states of the RNA products can be resolved by PAGE (see Fig. 2 and ref. 15). We have shown previously that large amounts of fully released RNA transcripts (Product 3) can be obtained if an excess of unlabeled RNA primer is added to the reaction mix (15). This excess RNA oligomer serves as a trap to block the rebinding of the nascent transcript to the template strand within the noncomplementary DNA bubble (i.e., to block the formation of Product 1) and shifts the distribution of RNA products so that the reaction produces primarily Product 3 (see Fig. 2, lane 5, and ref. 15).§
Figure 1.
The initial DNA bubble-duplex construct used and the RNA products obtained. The structure and sequence of the RNA primer and the DNA bubble-duplex construct used in this work are shown. The direction of synthesis is indicated by an arrow. On addition of RNA polymerase and Mg2+/NTPs, three different transcription products are obtained. Product 1 corresponds to a partially hybridized RNA transcript, Product 2 to a fully hybridized transcript, and Product 3 to a fully released transcript (see text).
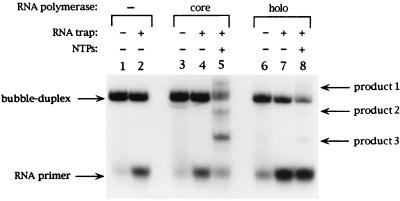
Figure 2.
RNA primer exchange is induced by holo E. coli RNA polymerase. As indicated at the top of each lane, RNA–DNA bubble- duplex constructs were incubated with buffer (lanes 1 and 2) or with either the holo (lanes 3–5) or core (lanes 6–8) forms of E. coli RNA polymerase in the presence or absence of NTPs and of an RNA trap, and the products obtained were resolved by nondenaturing gel electrophoresis (see Materials and Methods). The band positions of the RNA primer and of RNA Products 1, 2, and 3 are indicated on the right side of the gel.
This gel assay was used in the present study to compare the elongation activity of core and holo RNA polymerase on binding to the RNA–DNA bubble duplex (Fig. 2). Comparison of lane 5 (core RNA polymerase) to lane 8 (holo RNA polymerase) suggests that the substitution of holo E. coli RNA polymerase for core results in no detectable difference in the types and relative amounts of RNA products obtained, meaning that the holo and core forms of polymerase respond to the presence of the RNA oligomer trap in a qualitatively similar fashion. In addition, analysis of the transcribed RNA on a denaturing PAGE gel also revealed no difference between the types of transcripts obtained with the holo and the core enzyme (data not shown). The major difference observed in the products of the reactions catalyzed by the two forms of the polymerase is that a significantly smaller amount of total RNA primer is extended by the holoenzyme, relative to the amount extended by the core polymerase (compare lanes 8 and 5 of Fig. 2), despite the fact that equivalent initial polymerase concentrations were used in the two reactions. This activity variation could reflect a different binding affinity of the two forms of the enzyme to the RNA–DNA bubble duplex. However, a careful examination of Fig. 2 suggests an alternate explanation.
We note that incubation of the RNA “trap oligomer” with primed RNA–DNA bubble duplex in the absence of polymerase leads to exchange between the unlabeled trap oligomer and the initially bound (labeled) RNA primer, as manifested by an increase in the amount of free labeled RNA primer observed in the reaction mix in the presence of the unlabeled trap primer (compare the intensities of the free primer bands in lanes 1 and 2 of Fig. 2). The same extent of exchange was observed when core RNA polymerase was incubated with the primed bubble duplex and the RNA trap in the absence of NTPs (compare the free labeled primer bands in lanes 2 and 4 of Fig. 2). On the other hand, incubation with holoenzyme resulted in a dramatic increase in the amount of labeled RNA primer released.
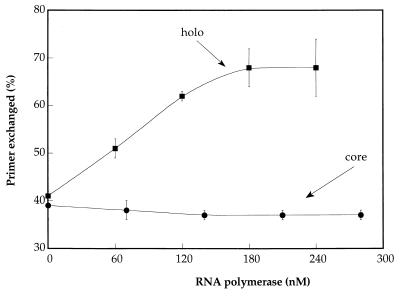
A quantitative analysis of this process is presented in Fig. 3, which shows that increasing the concentration of core RNA polymerase does not affect the basal level of RNA primer exchange, which remains at ≈40% under our standard reaction conditions. (In fact, a slight inhibition of primer release is observed with increasing core polymerase concentration.) In contrast, the fraction of labeled RNA primer that is exchanged increases significantly with increasing holoenzyme concentration, reaching saturation at an exchange level of ≈70% of the initial labeled primer under these conditions. The release of larger fractions of the labeled primer by the holoenzyme also explains the apparent reduction in labeled transcripts produced with holoenzyme in Fig. 2 (compare lanes 5 and 8), because labeled primer that has been exchanged into solution cannot be extended by the enzyme, whereas transcript formed with unlabeled (exchanged) primer is not detected in the assay.
Figure 3.
Exchange of RNA primer is catalyzed by holo E. coli RNA polymerase. The percentage of released primer (calculated from the ratio of free primer to free primer plus primer bound to the bubble duplexes) resulting from incubation with either holo (circles) or core (squares) E. coli RNA polymerase, as a function of active polymerase concentration. All incubations were performed in the presence of 24-μM concentrations of RNA trap oligomer.
Sigma Factor and RNA Primer Are Both Released on Addition of Holo RNA Polymerase to the Primed Bubble-Duplex Complex.
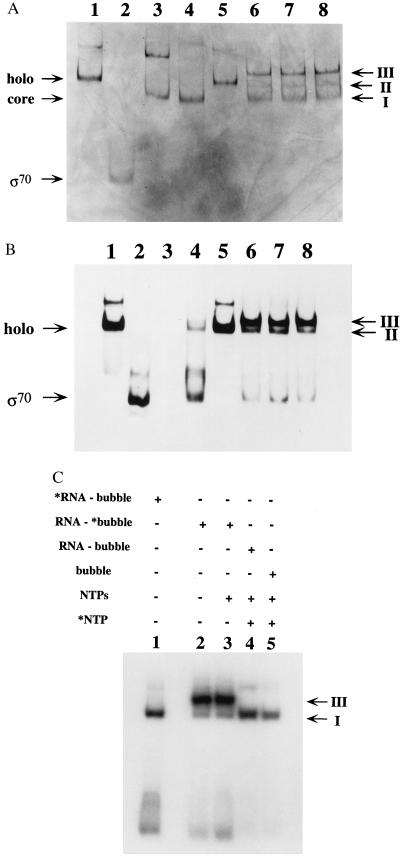
The results presented above suggest that binding of holopolymerase to the RNA-primed synthetic bubble-duplex construct drives the release of labeled RNA primer from the transcription complex, whereas core polymerase is relatively inactive in facilitating this exchange process. The sigma subunit, which is present only in the holoenzyme, must therefore be playing a role in this RNA release. To determine the state of association of the sigma subunit with the synthetic complexes, reaction mixtures similar to those used in the experiment shown in Fig. 2 were resolved on nondenaturing polyacrylamide gels, so that stable interactions between the protein and the nucleic acid components of the primed bubble duplex could be maintained. Incubation of holo RNA polymerase with the RNA-primed DNA bubble-duplex construct resulted in formation of three bands, as detected by silver staining of the gel (see Fig. 4A, lane 6). The lowest band of the three (band I in Fig. 4A) runs in a position that corresponds to core RNA polymerase (compare lanes 3 and 6). The middle band (band II in Fig. 4A) corresponds to free holo RNA polymerase (compare lanes 1 and 6), whereas the upper band (band III in Fig. 4A) migrates at a different mobility than any other component in the system.
Figure 4.
Binding of RNA primer and σ70 to synthetic transcription complexes is mutually exclusive. (A) Holo RNA polymerase (1.2 pmols), 1.4 pmols of core RNA polymerase, and 2 pmols of purified sigma subunit (provided by Lam Nguyen) were resolved on a nondenaturing gel (Materials and Methods) in the absence and presence of nucleic acids and were visualized by silver staining of the protein components. Lane 1 contains holo RNA polymerase; lane 2 contains sigma subunit; lane 3 contains core RNA polymerase; lane 4 contains holo RNA polymerase plus 2 μg of yeast tRNA; lane 5 contains holo RNA polymerase plus 35 pmols of RNA primer that is 12 nt in length; lane 6 contains holo RNA polymerase plus 1.4 pmols RNA–DNA bubble duplex; lane 7 contains holo RNA polymerase plus RNA–DNA bubble duplexes and 100 μM concentrations of ATP, CTP, and UTP; lane 8 contains holo RNA polymerase plus 1.7 pmols of DNA bubble duplex lacking the RNA primer. The gel positions of the monomeric forms of holo, core, and sigma subunit are marked on the left side of the gel, whereas the positions of gel bands I, II, and III are marked on the right. (B) Staining of a gel identical to that shown in Fig. 4A with a sigma subunit monoclonal antibody (see Materials and Methods). (C) An autoradiogram of a nondenaturing gel, similar to those shown in Fig. 4 A and B, in which 1.2 pmols of holo RNA polymerase were incubated with radiolabeled RNA–DNA bubble duplexes in 10-μl reaction volumes. Lane 1 contains 0.7 pmols of 5′-end labeled RNA primer plus unlabeled DNA bubble duplex; lane 2 contains 0.5 pmols of a DNA bubble duplex radiolabeled on the bottom strand plus a nonlabeled RNA primer; lane 3 contains the same materials as lane 2 plus 100 μM of ATP, CTP, and UTP; lane 4 contains 0.45 pmols of a nonlabeled RNA–DNA bubble duplex in the presence of 100 μM ATP, 100 μM UTP, and 1.5 μM 32P-α CTP; lane 5 contains 0.55 pmols of a nonlabeled DNA bubble duplex (lacking an RNA primer) plus 100 μM ATP, 100 μM UTP, and 1.5 μM 32P-αCTP. The radiolabeled species are marked (∗) at the top of the lanes, and the positions of bands I and III are marked on the right side of the gel.
It had previously been shown that incubation of holo RNA polymerase with yeast tRNA resulted in binding of the tRNA to the enzyme and the concomitant release of the sigma subunit into solution (22, 23). We have reproduced this behavior here under the same electrophoretic conditions used with our other reactions (Fig. 4A, lane 4). Indeed, the presence of yeast tRNA shifts the position of the E. coli RNA polymerase gel band from that of the holoenzyme (Fig. 4A, lane 1) to that of the core polymerase (Fig. 4A, lane 3). Band I in lane 6 also migrates similarly to the complex of core polymerase and tRNA, supporting the conclusion that it consists of complex that has lost sigma subunit.
In addition to tRNA, other RNA components have been shown to drive the release of σ70 from free E. coli holo RNA polymerase (24). We therefore tried incubating only the 12-nt RNA primer with the holoenzyme in the absence of the DNA bubble-duplex construct (Fig. 4A, lane 5). This treatment did not cause a mobility shift of the holoenzyme to the core band position, showing that the mobility shift induced when holoenzyme binds to the entire RNA–DNA bubble-duplex complex (Fig. 4A, lane 6, band III) does not reflect sigma release driven by any free RNA primer that might be present in the reaction mix.
To further identify the molecular components that comprise each of the three bands in lane 6 of Fig. 4A, an identical gel was run and probed with a monoclonal antibody directed against the sigma subunit (Fig. 4B). Staining with the σ70 antibody provides a direct demonstration that the sigma subunit has indeed been released from the complex, because a band migrating as free sigma is now detected (compare lane 6 to lane 2 in Fig. 4B). The same band can also be seen in the control reaction, in which sigma release from the holoenzyme was induced by tRNA binding (Fig. 4B, lane 4). Free sigma was not detected in Fig. 4A, most likely because of the much lower sensitivity of silver staining relative to staining with antibodies. In addition, comparison of the band pattern of lane 6 of Fig. 4B to that of lane 6 of Fig. 4A shows that only gel bands III and II, but not band I, contain the sigma subunit. This finding verifies that band I does indeed correspond to a polymerase species that has lost the sigma subunit and that band III contains the complete holoenzyme.
Finally, the identity of the nucleic acid components in each of the bands was explored by carrying out reactions identical to those described above, but this time by using RNA–DNA bubble duplexes in which either the RNA primer or the DNA bubble had been radioactively labeled. The reaction products were resolved by gel electrophoresis performed under the same conditions used in Fig. 4 A and B. An autoradiogram of this gel (Fig. 4C) revealed only two bands, corresponding to bands I and III of Fig. 4A; these band assignments were confirmed by careful alignment of the three gels. Both bands could be observed in reactions with the labeled DNA bubble-duplex construct, whereas only band I appeared in reactions in which the RNA primer was labeled.
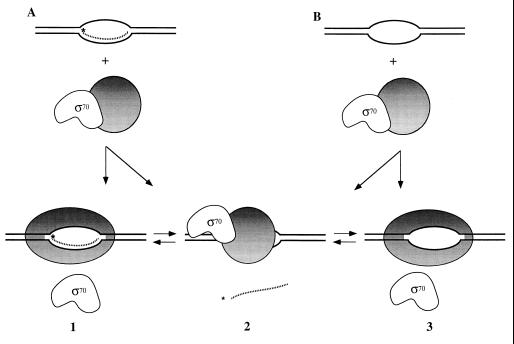
The results of the experiments shown in Figs. 4 A–C lead us to conclude that band I contains core RNA polymerase bound to either the RNA–DNA bubble duplex or the DNA bubble duplex alone. Band II contains only the holoenzyme and no nucleic acids, and band III contains holo RNA polymerase and the DNA bubble construct, but no RNA primer. This analysis suggests that two types of ternary complexes are formed on addition of E. coli holo RNA polymerase to the RNA–DNA bubble duplexes, one that contains σ70 but no RNA primer and one that contains RNA primer but no σ70 (see Discussion and Fig. 5).
Figure 5.
Release of σ70 or RNA primer on binding of E. coli RNA holopolymerase to DNA bubble-duplex constructs. This schematic depicts the structures proposed for the synthetic elongation complexes present in our assay system. Complexes 1 and 2 are formed on addition of holoenzyme to RNA–DNA bubble duplexes (corresponding to bands I and III of Fig. 4, respectively). Complexes 2 and 3 (the former in the absence of an RNA primer) are formed on addition of holoenzyme to DNA bubble duplexes (corresponding, respectively, to bands III and I of Fig. 4). Core polymerase is depicted as a shaded circle when bound to σ70 and as an ellipse (representing a different conformation) when bound to the DNA bubble. We suggest that this putative conformational change triggers the release of σ70 (complexes 1 and 3). Failure of the polymerase to undergo this conformational change on binding to the bubble-duplex construct induces the release of the RNA strand instead of σ70 (complex 2) and may resemble the events of abortive initiation in promoter-initiated transcription complexes.
We note that the presence of NTPs in the reaction mixture does not alter this pattern of bands and the addition of NTPs does not result in a qualitative change in the relative intensities of the three bands (compare Fig. 4A, lanes 6 and 7, with Fig. 4B, lanes 6 and 7). This fact suggests (in accord with the assumption that conformational and binding equilibrium is achieved within the elongation complex at each template position; see refs. 18 and 19) that the rates of release of σ70 and of the RNA primer are faster than the rate of transcript extension in these reactions. In addition, inclusion of a radioactively labeled NTP as the sole labeled species in the reaction mixture (Fig. 4C, lane 4) reveals that only band I (which has lost the sigma factor; see above) is labeled. Addition of labeled NTP to holo RNA polymerase alone (in the absence of any nucleic acids) resulted in no labeled bands (data not shown), suggesting that the band seen in lane 4 of Fig. 4C represents ternary complexes that are, at least, active in binding NTP. However, band III, which retains the sigma subunit but has lost the RNA primer, was not labeled by the radioactive NTP and therefore is not likely to correspond to a transcriptionally active complex.¶
Finally, we note that similar results are obtained when a bubble lacking an RNA primer is incubated with holo RNA polymerase (Fig. 4 A and B, lane 8, and Fig. 4C, lane 5). This observation means that binding of holopolymerase to the DNA bubble framework, even in the absence of the RNA primer, can induce σ70 release to an extent that is similar to that induced by DNA bubble-duplex constructs that do carry an annealed RNA primer. The significance of this observation will be considered below.
DISCUSSION
Previously, we established that the addition of core RNA polymerase to synthetic RNA–DNA bubble-duplex constructs in the presence of NTPs results in specific extension of the RNA primer that is characteristic of the elongation phase of a normal transcription cycle (14–16). In addition, we deduced that interactions within these synthetic elongation complexes reach conformational and binding equilibrium at most nucleotide addition steps of the transcription process (18, 19). Here we demonstrate that synthetic transcription complexes formed with holo RNA polymerase produce transcripts similar to those formed with core polymerase (Fig. 2). Thus it seems likely that the similar behaviors of the two enzymatic forms reflect the conversion of the holo to the core form of the polymerase as a consequence of the release of the sigma subunit. By using nondenaturing gel electrophoresis, we have been able to resolve the elongation complexes on the basis of molecular size and so have demonstrated directly that sigma is indeed released from at least some of these complexes on polymerase binding (Fig. 4B, lane 6, band I). In addition, we have shown that these complexes are likely to be transcriptionally active, at least as judged by their ability to bind NTPs.
In this work, we have shown an inverse correlation between the presence of the RNA primer and the sigma subunit in these synthetic transcription complexes. Figs. 2 and 3 provide direct evidence that holo RNA polymerase, but not core RNA polymerase, promotes the exchange of labeled and unlabeled primer. This result suggests that sigma participates in this exchange by inducing the release of the RNA primer. Fig. 4 provides direct evidence that no single transcription complex contains both σ70 and the RNA primer. Thus it appears that the sigma subunit and the RNA primer bind to the core RNA polymerase of the elongation complex in a mutually exclusive fashion. We note that in a recent independent study with similar synthetic elongation complexes, Sidorenkov et al. (17) have also shown that σ70 is displaced when the holoenzyme is bound to RNA-primed nucleic acid bubble-duplex constructs in which the length of the RNA–DNA hybrid exceeds ≈8 bp.
Our results, which are depicted schematically in Fig. 5, are compatible with and extend the first class of models described in the Introduction. These models argue that the sigma subunit is released during the initiation to elongation transition process, perhaps as a consequence of steric interference between the binding of the σ70 and the RNA primer to the complex. The majority of experimental evidence that is available to date supports this interpretation (11, 25–27). In addition, our results are inconsistent with the second and third class of models (see Introduction), at least to the extent that promoter binding comprises an essential part of the proposed mechanism for sigma release, because no promoter sequences are present within the synthetic bubble-duplex constructs that we have used.
This latter conclusion is strengthened by our observation (Fig. 4) that binding of polymerase to a DNA bubble duplex lacking an RNA primer can also induce σ70 release from some complexes. This fact argues that binding interactions with the fully formed DNA bubble alone may suffice to cause the markedly reduced affinity for sigma that is observed when the transcription complex enters the elongation phase of transcription (28), and that the σ70-dependent release of the RNA primer may occur largely as a consequence of a conformational change that is induced in the core polymerase on binding to the DNA bubble duplex, rather than by a direct steric displacement by the RNA primer. These results further suggest that this conformational change occurs on polymerase binding to the DNA bubble, whether or not an RNA primer is present. Such a direct demonstration of the individual contributions of the RNA–DNA hybrid and the DNA bubble duplex to the stability of the core–σ70 interaction requires an equilibrium bubble-duplex model transcription system of the sort used here (see ref. 18) and would be difficult to achieve with nonequilibrium systems (e.g., ref. 17) in which the prior formation of the RNA–DNA hybrid is required to permit polymerase binding.
By eliminating promoter effects, the use of synthetic DNA–RNA bubble-duplex constructs also opens up the possibility of learning something more about the premature RNA release process (abortive initiation) that can occur during the initiation phase of transcription. The proposed equilibrium between the two forms of core polymerase that is depicted schematically in Fig. 5 could correlate with the presence or absence of this RNA release process, suggesting that abortive initiation could reflect failure of the polymerase to lose contact with σ70 properly at the end of initiation. This failure could, in turn, inhibit the conformational change of the core polymerase that is required to enter the elongation phase of transcription and thus destabilize the binding of the nascent RNA to the initiation complex.
In summary, our results are consistent with the following picture of sigma release as a “normal” (i.e., promoter-initiated) transcription complex moves into the elongation phase of the transcription process. Initially, in the open promoter complex, sigma is stabilized by interactions both with the core polymerase and with the promoter sequence. This fact is consistent with the observation that the binding constant of the σ70–core polymerase interaction increases from ≈108 M−1 for the free holoenzyme to ≈1011 M−1 within the open promoter complex (28). As the nascent transcript becomes longer, these interactions become strained, and eventually the loss of the binding interaction of σ70 with the promoter removes this aspect of the stability of the complex. Simultaneously, the elongating RNA chain, in combination with the concommitant and progressive opening of the transcription bubble and the interactions of core polymerase with the template and nontemplate DNA sequences of the bubble that this opening permits, appears to further destabilize the interaction between sigma and the core subunits of the holopolymerase, reducing the binding constant for sigma to core within the elongation complex to its final value of ≈105.5 M−1 (28). At this level of binding affinity, the sigma subunit is lost into solution and under physiological conditions (and in vivo) is replaced within the elongation complex by the competitively bound NusA elongation and termination factor (28, 29). The incompatibility of the simultaneous binding of the nascent RNA and σ70 to the core polymerase within the transcription complex could reflect a direct steric displacement of σ70 by other components of the fully formed elongation complex, an allosteric incompatibility of these components, or, more likely, both effects.
Acknowledgments
A portion of this work was submitted to the Graduate School of the University of Oregon (by S.S.D.) in partial fulfillment of the requirements for the Ph.D. in Chemistry. We acknowledge gifts of purified σ70 protein and monoclonal antibodies from Lam Nguyen and extend special thanks to Lisa Rees for help with antibody staining procedures. These studies were supported in part by National Institutes of Health Research Grants GM-15792 and GM-29158 and by a grant from the Lucille P. Markey Charitable Trust to the Institute of Molecular Biology. S.S.D. was partially supported by the U. S. Department of Education Program for Graduate Assistance in Areas of National Need. P.H.v.H. is an American Cancer Society Research Professor of Chemistry.
Footnotes
Since its initial development, this method has been modified by others to avoid the requirement for a “permanent” noncomplementary bubble within the initial nucleic acid construct under some conditions (17) and also has been extended to demonstrate in more detail that the elongation complexes that result do indeed resemble closely those obtained by normal initiation processes (ref. 17; see also refs. 18, 19).
We have shown that this binding of the 5′-end of the nascent transcript to the bubble sequence is a kinetically controlled rebinding reaction. It does not occur at standard NTP concentrations with T7 RNA polymerase, which transcribes about 10-fold faster than the E. coli enzyme. However, as expected for such a mechanism, slowing down transcription by the T7 enzyme ≈10-fold by limiting the input NTP concentrations does result in rebinding the RNA transcript to the bubble with this polymerase as well (15).
We note that complexes retaining the sigma subunit are expected to resemble “normal” initiation complexes, which usually begin transcription by using a purine-containing rather than a pyrimidine-containing nucleotide as the initiating NTP. Because the only labeled NTP used here to detect binding was 32P-αCTP, we cannot rule out the possibility that complexes retaining σ70 are also active by this binding criterion.
References
- 1.Hansen U M, McClure W R. J Biol Chem. 1980;255:9556–9563. [PubMed] [Google Scholar]
- 2.Dombroski A J, Walter W A, Record M T, Siegele D A, Gross C A. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 3.Dombroski A J, Walter W A, Gross C A. Genes Dev. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- 4.Roberts C W, Roberts J W. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 5.DeHaseth P L, Helmann J D. Mol Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 6.Rong J C, Helmann J D. J Bacteriol. 1994;176:5218–5224. doi: 10.1128/jb.176.17.5218-5224.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munsan L M, Reznikoff W S. Biochemistry. 1981;20:2081–2085. doi: 10.1021/bi00511a003. [DOI] [PubMed] [Google Scholar]
- 8.Krummel B, Chamberlin M J. Biochemistry. 1989;28:7829–7842. doi: 10.1021/bi00445a045. [DOI] [PubMed] [Google Scholar]
- 9.Carpousis A J, Gralla J D. J Mol Biol. 1985;183:165–177. doi: 10.1016/0022-2836(85)90210-4. [DOI] [PubMed] [Google Scholar]
- 10.Straney D C, Crothers D M. Cell. 1985;43:449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- 11.Stackhouse T M, Telesnitsky A P, Meares C F. Biochemistry. 1989;28:7781–7788. doi: 10.1021/bi00445a038. [DOI] [PubMed] [Google Scholar]
- 12.Hochschild A, Dove S L. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 13.Shimamoto N, Kamigochi T, Utiyama H. J Biol Chem. 1986;261:11859–11865. [PubMed] [Google Scholar]
- 14.Daube S S, von Hippel P H. Science. 1992;258:1320–1324. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- 15.Daube S S, von Hippel P H. Biochemistry. 1994;33:340–347. doi: 10.1021/bi00167a044. [DOI] [PubMed] [Google Scholar]
- 16.Daube S S, Hart C R, von Hippel P H. Proc Natl Acad Sci USA. 1994;91:9539–9543. doi: 10.1073/pnas.91.20.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidorenkov I, Komissarova N, Kashlev M. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 18.Wilson K S, Conant C R, von Hippel P H. J Mol Biol. 1999;289:1179–1194. doi: 10.1006/jmbi.1999.2814. [DOI] [PubMed] [Google Scholar]
- 19.von Hippel P H. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsh E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 21.Strickland M S, Thompson N E, Burgess R R. Biochemistry. 1988;27:5755–5762. doi: 10.1021/bi00415a054. [DOI] [PubMed] [Google Scholar]
- 22.Krakow J S. Methods Enzymol. 1971;210:520–528. [Google Scholar]
- 23.Spassky A, Busloy S J W, Danchin A, Buc H. Eur J Biochem. 1979;99:187–201. doi: 10.1111/j.1432-1033.1979.tb13245.x. [DOI] [PubMed] [Google Scholar]
- 24.Altmann C R, Solow-Cordero D E, Chamberlin M J. Proc Natl Acad Sci USA. 1994;91:3784–3788. doi: 10.1073/pnas.91.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen U M, McClure W R. J Biol Chem. 1980;255:9564–9570. [PubMed] [Google Scholar]
- 26.Bowser C A, Hanna M M. J Mol Biol. 1991;220:227–239. doi: 10.1016/0022-2836(91)90009-u. [DOI] [PubMed] [Google Scholar]
- 27.Metzger W, Schikor P, Meier T, Werel W, Heumann H. J Mol Biol. 1993;232:35–49. doi: 10.1006/jmbi.1993.1368. [DOI] [PubMed] [Google Scholar]
- 28.Gill S C, Weitzel S E, von Hippel P H. J Mol Biol. 1991;220:307–324. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- 29.Greenblatt J, Li J. Cell. 1981;24:421–428. doi: 10.1016/0092-8674(81)90332-9. [DOI] [PubMed] [Google Scholar]