Abstract
ERA forms a unique family of GTPase. It is widely conserved and essential in bacteria. ERA functions in cell cycle control by coupling cell division with growth rate. ERA homologues also are found in eukaryotes. Here we report the crystal structure of ERA from Escherichia coli. The structure has been determined at 2.4-Å resolution. It reveals a two-domain arrangement of the molecule: an N-terminal domain that resembles p21 Ras and a C-terminal domain that is unique. Structure-based topological search of the C domain fails to reveal any meaningful match, although sequence analysis suggests that it contains a KH domain. KH domains are RNA binding motifs that usually occur in tandem repeats and exhibit low sequence similarity except for the well-conserved segment VIGxxGxxIK. We have identified a βααβ fold that contains the VIGxxGxxIK sequence and is shared by the C domain of ERA and the KH domain. We propose that this βααβ fold is the RNA binding motif, the minimum structural requirement for RNA binding. ERA dimerizes in crystal. The dimer formation involves a significantly distorted switch II region, which may shed light on how ERA protein regulates downstream events.
ERA is an essential GTPase found in every bacterium sequenced to date (1–4). In bacteria, ERA has a regulatory role in cell cycle control by coupling cell growth rate with cytokinesis (5, 6). Cell division is signaled when a threshold of ERA GTPase activity is reached. Artificially reducing the expression or impairing the GTPase activity of ERA results in bacterial cell cycle arrest at a predivisional two-cell stage (5, 6). The arrest lasts until ERA activity accumulates to the threshold level, allowing another cell cycle to start (5). Because the synthesis of ERA itself is positively correlated with growth rate, the cell division rate is thus coordinately maintained (5). It has been shown that era genes from other bacteria are capable of complementing Escherichia coli mutants defective in ERA production (3, 4), suggesting that the cell cycle regulation function of ERA is conserved in bacteria. Highly conserved ERA homologues also are found in eukaryotes, such as Antirrhinum (7), Caenorhabditis elegans, mouse, and human (5). The Antirrhinum homologue has been shown to be essential for embryogenesis (7).
The N-terminal GTPase domain of ERA is closely related to Ras p21 (8, 9) and other GTPases, whereas the C-terminal domain is unique. Recently, sequence studies have shown that a C-terminal region of ERA contains an RNA binding KH domain (I. S. Mian, Lawrence Berkeley National Laboratory, University of California, personal communication), consistent with the findings that ERA binds RNA in vitro (R. Simons, University of California, Los Angeles, personal communication) and in vivo (H. Peters, N. Costantino, and D.L.C., unpublished results). Four lines of evidence suggest that ERA interacts with the cell’s translational machinery. First, ERA copurifies with ribosome free 16S rRNA (H. Peters, N. Costantino, and D.L.C., unpublished results; G. Zhao, Eli Lilly and Company, personal communications). Second, a cold-sensitive mutant of ERA affects ribosomal maturation (10). Third, the 16S rRNA dimethyltransferase, when expressed from a multicopy plasmid, suppresses a cold-sensitive mutation of ERA (11). Fourth, the E. coli ERA coexpresses with RNaseIII, an endonuclease involved in rRNA maturation (12). Both the N and C domains of ERA are critical to its function. Lethal mutations in both domains have been identified (13–15), but no functional coordination between the N-terminal GTPase domain and the C-terminal domain has been revealed.
GTPase proteins are used as molecular switches in a wide variety of cellular processes, exploiting conformational changes that occur on GTP hydrolysis and information relay to effector molecules (16–18). Only a few bacterial GTPase proteins have been characterized, including cell division protein FtsZ (19), signal recognition particle components Fth and FtzY (20, 21), and translation factors IF-2, EF-Tu, EF-G, and RF-3 (22–25). Most of these GTPase proteins are conserved throughout the prokaryotic and eukaryotic kingdoms. ERA, having an essential function in cell cycle regulation, appears to be a new member of such conserved GTPases. In contrast to extensively characterized GTPase-coupled signaling in eukaryotic Ras and Gα proteins (17, 18), little is known about ancient GTPase proteins. The three-dimensional structure of ERA should shed light not only on its structure and function, but also on the mechanism of GTPase-dependent signal transduction in both bacteria and eukaryotes. As an essential protein in bacteria, including numerous pathogens, ERA is an ideal target for antimicrobial drug design. Here we report the crystal structure of ERA from E. coli (Fig. 1). The structure was solved by using a combination of the multiple isomorphous replacement (MIR) and the multiwavelength anomalous diffraction (MAD) techniques at 2.4-Å resolution.
Figure 1.
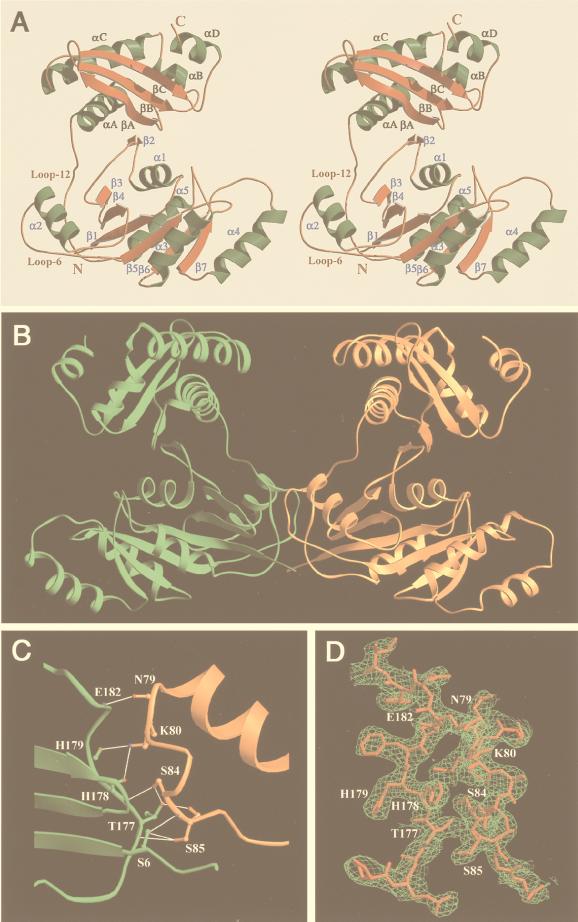
Overall structure of ERA. (A) Stereoview of the ERA monomer. (B) The ERA dimer in the crystal. A pseudo 2-fold axis is in the dimer interface. Half of the dimer interface is shown in C; the other half is not shown for clarity. The interactions consist of one backbone-to-backbone, six backbone-to-side chain, and two side chain-to-side chain hydrogen bonds (illustrated as thin white lines). (D) Characteristic electron density of MIR phasing is superimposed on final structure. The electron density from MAD phasing has comparable quality. A and D were prepared by using molscript (47) and raster3d (48), and B and C were prepared by using ribbons (49).
EXPERIMENTAL PROCEDURES
Crystallization of the Protein.
E. coli ERA (301 residues) was expressed and purified as described (26). Crystals of ERA were grown by the hanging-drop vapor diffusion method. The typical starting equilibrium contained 50 mM Tris⋅HCL (pH 8.0), 0.8 M NaCl, and 8 mg/ml protein. Crystallization of ERA in the presence of 5–20 mM GDP or GTP analogues also was attempted, but no ligand was identified in the crystal structure. Instead, a sulfate ion was found in the P loop.
X-Ray Data Collection and Processing.
The crystal containing 65% solvent is in space group P21 with unit cell dimensions a = 86.8, b = 67.6, c = 87.3 Å, α = γ = 90, and β = 115.8°. Each data set was collected from a frozen crystal at 100 K by using a MAR-Research (Hamburg, Germany) imaging-plate detector. Two types of x-ray sources were used. Single wavelength x-ray diffraction data were collected by using a Rigaku (Tokyo) rotating anode x-ray generator. The MAD data were collected by using the National Synchrotron Light Source at the Brookhaven National Laboratory at beamline X9B. All data sets were processed by using the hkl2000 suite (27).
Crystal Structure Solution and Refinement.
Heavy-atom sites of one mercury derivative [Table 1, ethylmercury phosphate (EMP), Tris buffer] were derived from the difference Patterson maps calculated with phases (28). Heavy-atom sites for other derivatives were located from cross-difference Fourier syntheses using heavy-atom phases of the first mercury derivative. Calculation, refinement, and solvent flattening for both MIR and MAD phases were carried out by using sharp (29). Tracing and model building were done with o (30). The electron density maps calculated by using either the MIR or the MAD phases were used in a manner of cross-validation and cross-reference. The two experimental electron density maps had comparable quality (Fig. 1D). The structure was refined by using x-plor (31) with 5% reflections for Rfree calculations. Bulk solvent correction was used at the final stage of the refinement. A weak restraint between the two independent molecules was applied during the initial stage of refinement and was released at the final stage. For both molecules, residues 224–228 had very poor electron density and were not included in the refinement. The average B factor for the structure is 49.5 Å2, consistent with that estimated from the Wilson plot (53.4 Å2). The Ramachandran plot showed that more than 85% of the main-chain torsion angles are in the most favored region and there is no residue in the generously allowed or disallowed regions. The MIR and MAD phasing and the refinement statistics are summarized in Table 1.
Table 1.
Crystallographic analysis (MIR and MAD phasing) and refinement statistics of ERA protein
| Resolution, Å | Redundancy,* Å | Completeness (last shell), (%) | Rsym† | # Sites | Rscale‡ | Phasing power§
|
||
|---|---|---|---|---|---|---|---|---|
| Centric, iso | Acentric, iso/ano | |||||||
| Native data | 2.4 | 6.4 | 99.7 (99.1) | 0.056 | ||||
| MIR phasing (mean figure of merit = 0.71) | ||||||||
| EMP (Tris buffer) | 3.5 | 3.2 | 96.3 (82.9) | 0.086 | 4 | 0.216 | 1.4 | 1.9/1.2 |
| EMP (Hepes buffer) | 2.95 | 3.5 | 98.3 (97.1) | 0.075 | 8 | 0.130 | 1.3 | 1.7/0.94 |
| APMA | 4.0 | 2.8 | 97.5 (97.9) | 0.098 | 4 | 0.201 | 0.98 | 1.3/0.90 |
| (UO2)(NO3)2 | 3.2 | 3.2 | 98.8 (98.8) | 0.084 | 2 | 0.080 | 0.34 | 0.28/0.26 |
| MAD phasing (EMP, mean figure of merit = 0.81) | ||||||||
| λ1 = 1.00870 Å | 2.6 | 5.4 | 99.4 (99.1) | 0.064 | 4 | 0.023 | 1.7 | 2.1/2.6 |
| λ2 = 1.00764 Å | 2.6 | 5.5 | 99.5 (99.1) | 0.070 | 4 | 0.022 | 1.3 | 1.6/2.3 |
| λ3 = 0.99184 Å | 2.6 | 5.6 | 99.5 (99.2) | 0.076 | 4 | 0.034 | n.a. | n.a./2.6 |
| λ4 = 1.00903 Å | 2.6 | 5.7 | 99.5 (99.1) | 0.063 | 4 | 0.034 | 1.1 | 1.5/2.0 |
| Structure refinement | ||||||||
| Resolution (Å), 8-2.4 | ||||||||
| Reflections (I/σ(I) > 2), 28, 848 | ||||||||
| Completeness (last shell), 82.9 (56.9) | ||||||||
| R/Rfree, 0.243/0.298 | ||||||||
| rms deviations, bond/angle, 0.009 Å/1.55° | ||||||||
| Protein atoms, 4,618 | ||||||||
| Sulfate ions, 5 | ||||||||
| Water molecules, 150 | ||||||||
APMA, 4-aminophenylmercuric acetate. n.a., not applicable.
Redundancy is the average number of observations for each unique reflection.
Rsym = Σ|In − 〈In〉|/ΣIn, where 〈In〉 is the average intensity over symmetric equivalents.
Rscale = Σ|IPH − IP|/ΣIP.
Phasing power 〈fh〉/〈E〉, where 〈fh〉 and 〈E〉 are the rms deviation of the heavy-atom structure factor and the lack of closure error, respectively.
RESULTS AND DISCUSSION
Overall Structure and Domain Interactions.
There are two independent molecules in the crystallographic asymmetric unit (residues 4–295 for molecule A and residues 4–296 for molecule B). The two ERA monomers have almost identical conformation and display a novel two-domain arrangement (Fig. 1A). The N-terminal GTPase domain resembles p21 Ras (8, 9), the G domain of transducin α (32), and domain I of EF-Tu (33). It consists of a central six-stranded β-sheet flanked by five helices. Uniquely, the loop before the G2 region in other GTPases (16) is a β-strand (β2) in ERA formed from a parallel continuation of a β-sheet in the C-terminal domain (Fig. 1A). This region (33ISITSR38) has been reported to undergo autophosphorylation in vitro in the presence of GTP (34). In the current structure, however, the γ-phosphate binding site of GTP appears to be quite far (>8 Å) from this region. Although the importance of this cross-domain β-sheet has not been elucidated, alterations of T36 and S37 to alanine (34) or S34 to proline (13) have been shown to be lethal. A sulfate ion from the crystallization buffer is bound in the cleft between the N-terminal domain (to T36, K27, and T23) and the C-terminal domain (to T215 and K282).
The C-terminal domain folds tightly into a structure comprised of a three-stranded β-sheet on one side and three helices on the other (Fig. 1A). It is connected to the N-terminal domain by a 15-residue linker (loop 12 in Fig. 1A). The C-terminal residues 286–295 form α-helix αD, which is solvent exposed and probably functionally dispensable (2). Compared with other multidomain GTPases, the disposition of the C-terminal domain of ERA facing the nucleotide binding site remotely resembles the helical domain of Gα of the heterotrimeric G proteins (32). However, the helical domain of Gα interacts strongly with the nucleotide binding site and has been proposed to have an internal GTPase-activating function for the bound GTP (17, 18), whereas the C domain of ERA extends out from the N domain and does not interact with the nucleotide binding site. Whether the C domain of ERA is able to approach and affect the bound nucleotide in vivo is not clear.
Crystal Packing and Dimerization.
The two independent ERA monomers in the crystal are related by a pseudo 2-fold axis and interact extensively at their interface (Fig. 1B) between loop 6 of one molecule and loop 12 of the other (Fig. 1A). Eighteen hydrogen bonds are present in the two contact regions related by the pseudo 2-fold axis. One of the two regions is illustrated in Fig. 1C. The dimer formation appears to be unique and specific to ERA for two reasons. First, the switch II region of ERA, the loop–helix–loop motif between β4 and β5, has a seven-residue insertion as compared with the same region in H-Ras or Gαs. Five of the seven inserted residues are within loop 6 that is involved in ERA dimerization. Second, loop 12 of ERA, in the vicinity of loop 6, is not seen in other multidomain G proteins. Whether ERA dimerizes in vivo is not clear (see below). The protein behaves as a monomer during gel filtration chromatography (26).
The Unique C-Terminal Domain and RNA Binding Motif.
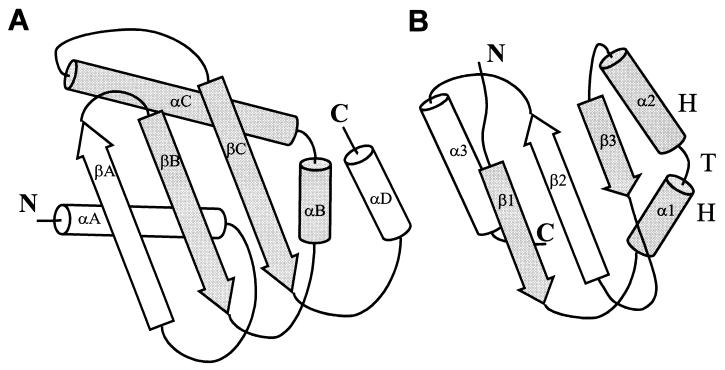
Although a structure-based topological search (35) for the fold of ERA C domain did not reveal any meaningful match, sequence studies indicated that it might contain a KH domain. A segment of ≈70 residues within the C-terminal domain of ERA (≈120 residues) can be aligned with KH sequences that contain ≈70 aa (I. S. Mian, personal communication). As widely conserved RNA binding motifs (36, 37), the KH domains usually occur in tandem repeats and exhibit low sequence similarity except for a conserved segment, VIGxxGxxIK. This segment also is found in the C-terminal domain of ERA. The NMR structures of isolated KH domains have revealed a globular fold consisting of a three-stranded β-sheet packed against three helices with a topology βααββα (36, 37) (Fig. 2B). In the C domain of ERA, however, corresponding secondary structural elements are arranged as αββααβ (Fig. 2A, excluding the C-terminal αD helix). We notice that a common βααβ fold, containing the VIGxxGxxIK sequence, is shared between the two domains. The RNA binding site in the KH domains has not been identified. We propose that this βααβ fold, with two central helices mediating RNA binding and the flanking β-strands as structural support, is the minimal RNA binding motif. The two α-helices in the βααβ motif form a helix–turn–helix (HTH) structure with a sharp turn where the consensus sequence VIGxxGxxIK (Fig. 2) is located. Mutations in this region reduce RNA binding ability of ERA in vitro and are lethal to the cell (R. Simons, personal communication). The first helix and the following loop, which are not well defined in the NMR structures, have been suggested to bind RNA through an induced-fit mechanism (36, 37). The HTH motif also has been implicated in RNA binding in a few ribosomal proteins (38–40). The clustering of positively charged residues (R239, K243, K244, K250, K253, K255, R262, and K263) along one face of the HTH motif in ERA, especially along the αC-helix, could favor RNA interactions. The mutation of R262 to methionine significantly affects the growth of E. coli (14, 15). The secondary structural elements of the C domain of ERA (αA-C and βA-C) are held together by a hydrophobic core formed by aliphatic side chains. At the edge of the hydrophobic core is I254, the cognate residue of I304 in the KH domain of the fragile X mental retardation gene product. The fragile X syndrome is the result of I304N mutation (41, 42). In the C domain of ERA, I254 holds the N terminus of the αC-helix, playing a critical role in maintaining the conformational integrity of the HTH motif. In addition to RNA binding, the KH domain has been shown to mediate RNA-dependent protein–protein interactions (43). A KH domain-containing protein, Sam68, has been shown to be essential for mammalian cell cycle progression (44).
Figure 2.
Topological diagrams showing the similarities and differences between (A) the C-terminal domain of ERA (this work) and (B) the KH domain of FMR1, the protein responsible for the fragile X syndrome (37). Although in general the two domains are topologically different, they share a βααβ fold (highlighted), of which the two central helices (αB and αC in ERA, α1 and α2 in FMR1) form a HTH structure with a sharp turn containing a sequence segment VIGxxGxxIK that is highly conserved among RNA binding KH domains.
The N-Terminal GTPase Domain and ERA-Nucleotide Interactions.
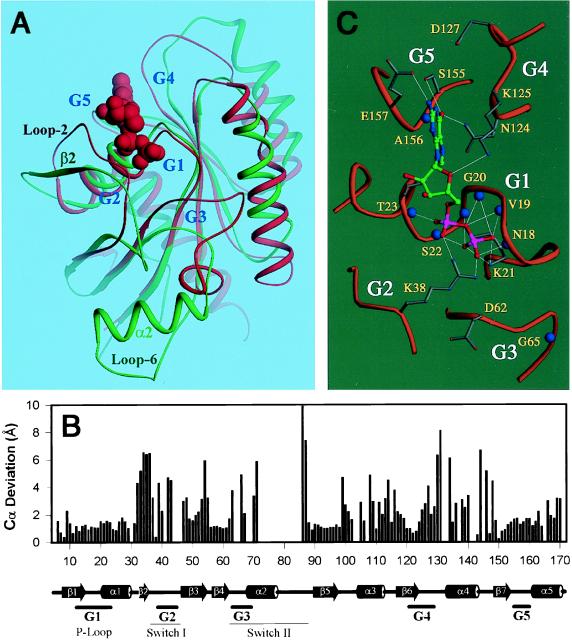
The ERA GTPase domain appears to be the most divergent when the GTPase domains of ERA, Gαs, EF-Tu, and Ras proteins are aligned based on the three-dimensional structures (data not shown). The ERA GTPase domain can be superimposed with the H-Ras⋅GDP complex (9) with an rms deviation of 2.8 Å for 131 Cα pairs (Fig. 3 A and B). The G1-G5 regions (17, 18) in ERA, responsible for nucleotide binding, appear to move outward when the protein is nucleotide free. Therefore, the present structure is equivalent to the apo form of ERA. In the ERA/Ras alignment, segments around the G2 and G3 regions show the largest deviations, and the segment immediately after G4 region shows notable differences. The central β-sheet and the G1 and G5 regions superimpose well (Fig. 3 A and B). A GDP molecule can be fit into the nucleotide binding site of ERA without significant steric conflicts based on the alignment of ERA GTPase domain with the H-Ras⋅GDP complex (9). A preliminary energy minimization using x-plor (31) eliminated a few clashes between the amino acid side chains and the GDP molecule, suggesting that the ligand–protein interaction in ERA⋅GDP complex is similar to that observed in the H-Ras⋅GDP complex (Fig. 3 A and C).
Figure 3.
Comparion of the ERA GTPase domain with H-Ras⋅GDP complex (9). (A) Superposition of the GTPase domain of ERA (in green) and the H-Ras⋅GDP complex (in red). GDP is shown as space-filling model. The G regions for nucleotide binding and the switch regions are labeled, as are the β2-strand, α2-helix, and loop 6 of the switch II region in ERA and loop 2 in H-Ras. (B) Cα deviations between ERA and Ras from the alignment in A. Empty space in the histogram indicates the loss of register between corresponding Cα atoms. Secondary structure assignment of ERA is indicated at the bottom. (C) A GDP molecule is modeled into the nucleotide binding pocket of ERA based on the superposition in A followed by a preliminary energy minimization (see text). The GDP molecule is shown in ball-and-stick model with atom-specific colors (carbon in green, oxygen in red, nitrogen in blue and phosphorus in magenta). Hydrogen bonds are illustrated as thin white lines and backbone amide nitrogen atoms as large blue spheres. The G1–G5 regions involved in nucleotide binding are labeled. A and C were prepared by using ribbons (49).
In the model of ERA⋅GDP complex, the GDP molecule binds tightly to a highly conserved ERA sequence (15GRPNxGKSTL24) in the G1 region (P loop), mostly through phosphate-amide hydrogen bonds (Fig. 3C). The side chain of K21 interacts with the β-phosphate in a manner similar to that of K16 in H-Ras (9). In addition, the side chain of T23 is hydrogen-bonded to N7 of the guanine base. Bacterial viability is extremely sensitive to the mutations in this area (H. Peters, B. Powell, N. Costantino, and D.L.C., unpublished results; refs. 13–15). The G5 region of ERA interacts with the guanine base with the conserved residues 155SA156 (145SA146 in H-Ras). E157 in ERA also recognizes N2 of the guanine base. The G4 region of ERA resembles that of H-Ras with a consensus sequence 124NKxD127 (116NKxD119 in H-Ras), where N124 and K125 interact with the guanine base in a way similar to that in H-Ras. However, unlike D119 in H-Ras that interacts with the nucleotide, D127 in ERA is 6.5 Å away from the base. D127 is absolutely conserved in ERA proteins. The binding of the nucleotide might bring this residue closer to the base.
The switch I region between β2 and β3 (Fig. 3 A and B) in ERA corresponds to the regulatory segment that has been shown to mediate effector binding in Ras GTPases (16–18). However, the β2-strand in ERA is a loop (loop 2) in Ras (Fig. 3A). In ERA, the unique β2-strand interacts with the C-terminal domain by forming a cross-domain β-sheet β2-βA-βB-βC, which alters the relative position of β2 and the following switch I when compared with H-Ras (9). Switch I includes G2 that interacts, through T35 in H-Ras, with the γ-phosphate of the bound GTP and the coordinated Mg2+ ion, and therefore, plays an important role in the conformational change (switch) on GTP hydrolysis (8). In ERA, however, the cognate threonine residue, T42 or T43, is far from the γ-phosphate. The requirement of this threonine residue in ERA function is supported by the strict conservation of a four-residue stretch (41QTTR44) and by the finding that the mutation of 42TT43 to alanines severely impairs the GTPase activity and inhibits cell growth without affecting GTP binding (45). In the present ERA structure, residues 38–41 exhibit poor electron density, suggesting the intrinsic flexibility of this region in apo-ERA. We speculate that it is either T42 or T43 of ERA that has the identical function of T35 in H-Ras. Thus, a positional shift of this threonine in ERA must take place through either the disruption of the β2-βA interactions or the movement of the C domain, which pulls β2 and the following G2 off the center of the GTPase domain. Exactly how this may happen is not clear. The occurrence of the threonine repeat in the G2 region of ERA (38RKAQTTR44) resembles that in Gα (RxxTTG) (32). The interaction of R38 of ERA with the α- and β-phosphates in the model of ERA⋅GDP complex (Fig. 3C) resembles that of a conserved arginine in Gαs (32), which has been implicated in maintaining constitutive GTP hydrolysis (16). Nevertheless, ERA exhibits a much poorer GTPase activity when compared with Gα (26).
The switch II region of G proteins undergoes striking conformational changes on GTP hydrolysis (16–18). The conformation of the switch II region in ERA is such that a big segment of the α2-helix and the following loop are out of register when ERA is aligned to H-Ras (Fig. 3B). The G3 region of ERA is far away from the nucleotide binding site (Fig. 3 A and B) and the α2-helix is almost perpendicular to the central β-sheet (Fig. 3A). In other G proteins, however, the α2-helix and the central β-sheet are nearly parallel in both GDP- and GTP-bound states. The insertion of the seven residues in the switch II region of ERA, with respect to H-Ras and Gαs, evidently augments the protruding conformation of this region. The unusual conformation of the switch II region in ERA is most likely one of the consequences of the dimerization that involves loop 6 (Fig. 1 A and B). The corresponding loop in Gsα is known to be involved in effector activation in the GTP-bound state (46). In the G3 region of ERA proteins, sequence 62DTPG65 is conserved. This sequence corresponds to 57DTAG60 in H-Ras where D57 and G60 interact with the γ-phosphate of GTP (8). It therefore is expected that ERA interacts with the γ-phosphate of GTP in a similar manner. If this is true, the switch II region of ERA can exist only in one of the two conformations: the γ-phosphate-accepting conformation as seen in H-Ras, or the conformation in the present structure on dimerization. Assuming the dimerization of ERA is biologically relevant, we propose the following regulatory mechanism. On GTP binding, the G2 region interacts with the γ-phosphate of GTP and switch II is retained to the core of the GTPase domain. After GTP hydrolysis, switch II is released and mediates dimerization. The dimerization of ERA may be required for functional signaling through interactions with RNA. To complete the GTPase cycle, the dimer should dissociate when GDP is replaced by GTP. Therefore, the dimer form cannot be too stable, which may account for the fact that we failed to detect ERA dimer by using gel filtration chromatography (26).
Acknowledgments
We thank Dr. Z. Dauter for his assistance during x-ray data collection and Drs. H. Ellis, D. Morrison, and D. Waugh for critical reading of the manuscript. This work was supported by the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories.
ABBREVIATIONS
- MIR
multiple isomorphous replacement
- MAD
multiwavelength anomalous diffraction
- EMP
ethylmercury phosphate
- HTH
helix-turn-helix
Footnotes
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1ega).
References
- 1.March P E, Lerner C G, Ahnn J, Cui X, Inouye M. Oncogene. 1988;2:539–544. [PubMed] [Google Scholar]
- 2.Inada T, Kawakami K, Chen S M, Takiff H E, Court D L, Nakamura Y. J Bacteriol. 1989;171:5017–5024. doi: 10.1128/jb.171.9.5017-5024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuber M, Hoover T A, Dertzbaugh M T, Court D L. Gene. 1997;189:31–34. doi: 10.1016/s0378-1119(96)00813-x. [DOI] [PubMed] [Google Scholar]
- 4.Zuber M, Hoover T A, Powell B S, Court D L. Mol Microbiol. 1994;14:291–300. doi: 10.1111/j.1365-2958.1994.tb01290.x. [DOI] [PubMed] [Google Scholar]
- 5.Britton R A, Powell B S, Dasgupta S, Sun Q, Margolin W, Lupski J R, Court D L. Mol Microbiol. 1998;27:739–750. doi: 10.1046/j.1365-2958.1998.00719.x. [DOI] [PubMed] [Google Scholar]
- 6.Gollop N, March P E. J Bacteriol. 1991;173:2265–2270. doi: 10.1128/jb.173.7.2265-2270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingram G C, Simon R, Carpenter R, Coen E S. Curr Biol. 1998;8:1079–1082. doi: 10.1016/s0960-9822(98)70445-2. [DOI] [PubMed] [Google Scholar]
- 8.Pai E F, Krengel U, Petsko G A, Goody R S, Kabsch W, Wittinghofer A. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong L A, de Vos A M, Milburn M V, Kim S H. J Mol Biol. 1991;217:503–516. doi: 10.1016/0022-2836(91)90753-s. [DOI] [PubMed] [Google Scholar]
- 10.Nashimoto H. In: The Translational Apparatus. Nierhaus K H, editor. New York: Plenum; 1993. pp. 185–195. [Google Scholar]
- 11.Lu Q, Inouye M. J Bacteriol. 1998;180:5243–5246. doi: 10.1128/jb.180.19.5243-5246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Court D L. In: Control of Messenger RNA Stability. Belasco J, Brawerman G, editors. San Diego: Academic; 1993. pp. 77–116. [Google Scholar]
- 13.Pillutla R C, Sharer J D, Gulati P S, Wu E, Yamashita Y, Lerner C G, Inouye M, March P E. J Bacteriol. 1995;177:2194–2196. doi: 10.1128/jb.177.8.2194-2196.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lerner C G, Gulati P S, Inouye M. FEMS Microbiol Lett. 1995;126:291–298. doi: 10.1111/j.1574-6968.1995.tb07432.x. [DOI] [PubMed] [Google Scholar]
- 15.Lerner C G, Sood P, Ahnn J, Inouye M. FEMS Microbiol Lett. 1992;74:137–142. doi: 10.1016/0378-1097(92)90419-o. [DOI] [PubMed] [Google Scholar]
- 16.Milburn M V, Tong L, deVos A M, Brunger A, Yamaizumi Z, Nishimura S, Kim S H. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 17.Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 18.Sprang S R. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 19.Lowe J, Amos L A. Nature (London) 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 20.Freymann D M, Keenan R J, Stroud R M, Walter P. Nature (London) 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 21.Montoya G, Svensson C, Luirink J, Sinning I. Nature (London) 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- 22.Brock S, Szkaradkiewicz K, Sprinzl M. Mol Microbiol. 1998;29:409–417. doi: 10.1046/j.1365-2958.1998.00893.x. [DOI] [PubMed] [Google Scholar]
- 23.Nyborg J, Liljas A. FEBS Lett. 1998;430:95–99. doi: 10.1016/s0014-5793(98)00624-3. [DOI] [PubMed] [Google Scholar]
- 24.Laalami S, Grentzmann G, Bremaud L, Cenatiempo Y. Biochimie. 1996;78:577–589. doi: 10.1016/s0300-9084(96)80004-6. [DOI] [PubMed] [Google Scholar]
- 25.Tuite M F, Stansfield I. Mol Biol Rep. 1994;19:171–181. doi: 10.1007/BF00986959. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Chen S-M, Powell B, Court D L, Ji X. FEBS Lett. 1999;445:425–430. doi: 10.1016/s0014-5793(99)00178-7. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Furey W, Swaminathan S. Methods Enzymol. 1997;277:590–620. doi: 10.1016/s0076-6879(97)77033-2. [DOI] [PubMed] [Google Scholar]
- 29.de La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 30.Jones T A, Kjeldgaard M. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 31.Brünger A T, Rice L M. Methods Enzymol. 1997;277:243–269. doi: 10.1016/s0076-6879(97)77015-0. [DOI] [PubMed] [Google Scholar]
- 32.Noel J P, Hamm H E, Sigler P B. Nature (London) 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 33.Jurnak F. Science. 1985;230:32–36. doi: 10.1126/science.3898365. [DOI] [PubMed] [Google Scholar]
- 34.Sood P, Lerner C G, Shimamoto T, Lu Q, Inouye M. Mol Microbiol. 1994;12:201–208. doi: 10.1111/j.1365-2958.1994.tb01009.x. [DOI] [PubMed] [Google Scholar]
- 35.Holm L, Sander C. Proteins Struct Funct Genet. 1994;19:165–173. doi: 10.1002/prot.340190302. [DOI] [PubMed] [Google Scholar]
- 36.Musco G, Stier G, Joseph C, Castiglione Morelli M A, Nilges M, Gibson T J, Pastore A. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 37.Musco G, Kharrat A, Stier G, Faternali F, Gibson T J, Nilges M, Pastore A. Nat Struct Biol. 1997;4:712–716. doi: 10.1038/nsb0997-712. [DOI] [PubMed] [Google Scholar]
- 38.Xing Y, Guha Thakurta D, Draper D E. Nat Struct Biol. 1997;4:24–27. doi: 10.1038/nsb0197-24. [DOI] [PubMed] [Google Scholar]
- 39.Berglund H, Rak A, Serganov A, Garber M, Hard T. Nat Struct Biol. 1997;4:20–23. doi: 10.1038/nsb0197-20. [DOI] [PubMed] [Google Scholar]
- 40.Hosaka H, Nakagawa A, Tanaka I, Harada N, Sano K, Kimura M, Yao M, Wakatsuki S. Structure. 1997;5:1199–1208. doi: 10.1016/s0969-2126(97)00270-0. [DOI] [PubMed] [Google Scholar]
- 41.De Boulle K, Verkerk A J, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra B A, Willems P J. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 42.Siomi H, Siomi M C, Nussbaum R L, Dreyfuss G. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 43.Chen T, Damaj B B, Herrera C, Lasko P, Richard S. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barlat I, Maurier F, Duchesne M, Guitard E, Tocque B, Schweighoffer F. J Biol Chem. 1997;272:3129–3132. doi: 10.1074/jbc.272.6.3129. [DOI] [PubMed] [Google Scholar]
- 45.Shimamoto T, Inouye M. FEMS Microbiol Lett. 1996;136:57–62. doi: 10.1111/j.1574-6968.1996.tb08025.x. [DOI] [PubMed] [Google Scholar]
- 46.Berlot C H, Bourne H R. Cell. 1992;68:911–922. doi: 10.1016/0092-8674(92)90034-a. [DOI] [PubMed] [Google Scholar]
- 47.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 48.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 49.Carson M. J Mol Graphics. 1987;5:103–106. [Google Scholar]