Abstract
The retrieval of DNA from fossils remains controversial. To substantiate claims of DNA recovery, one needs additional information on the preservation of other molecules within the same sample. Flash pyrolysis with GC and MS was used to assess the quality of protein preservation in 11 archaeological and paleontological remains, some of which have yielded ancient DNA sequences authenticated via a number of criteria and some of which have consistently failed to yield any meaningful DNA. Several samples, including the Neanderthal-type specimen from which DNA sequences were recently reported, yielded abundant pyrolysis products assigned to 2,5-diketopiperazines of proline-containing dipeptides. The relative amounts of these products provide a good index of the amount of peptide hydrolysis and DNA preservation. Of these samples, four stem from arctic or subarctic regions, emphasizing the importance of cooler temperatures for the preservation of macromolecules. Flash pyrolysis with GC and MS offers a rapid and effective method for assessing fossils for the possibility of DNA preservation.
Keywords: pyrolysis, GC, MS, ancient DNA, collagen, Neanderthal fossil
Most archeological and paleontological remains do not contain endogenous DNA that can be amplified via PCR but do contain DNA from exogenous sources such as bacteria and fungi, as well as contaminating DNA from contemporary humans (1). In the few cases where endogenous DNA is preserved, no simple correlation between the age of a sample and the preservation of its DNA is observed (2, 3). Instead, largely unknown aspects of burial conditions seem to be more important. One beneficial factor for long-term DNA preservation seems to be low temperature during burial (2, 3). This benefit has been illustrated by the fact that the oldest DNA sequences that have been reported—and subsequently reproduced in other laboratories—stem from remains of the woolly mammoth found in the Siberian permafrost, which are over 50,000 years old (4–7). By contrast, DNA sequences determined from fossils that are millions of years old have failed to be reproduced (8, 9) or have been shown to be derived from identifiable contaminants (10).
The modification and degradation of DNA in the archaeological and paleontological record is believed to be similar to some of the chemical processes that can be studied in the laboratory over short periods of time (11–13). It has been suggested, based on aqueous-solution studies on the DNA molecule, that no meaningful genetic information should be preserved for longer than 104–105 years in most environments (11–13). Obviously, other organic macromolecules that are present in the same specimen as the DNA will be subject to the same environmental conditions. Analyses of such molecules may therefore provide a rapid way to identify samples likely to yield DNA sequences and lend support to claims of DNA preservation. Proteins offer an especially promising possibility for such evaluation.
Flash pyrolysis with GC and MS (Py-GC/MS) is a technique that has been used successfully to characterize otherwise intractable organic molecules, such as lignocellulose (14–16), chitin (17–19), resins (20), algae (21), and plant cuticles (22). It has also been applied to the study of proteins (23–25), although its applications to DNA have been limited (26). Recently, it was used to assess plant-tissue preservation in 17- to 20-million-year-old leaves from the Clarkia deposit in Idaho (27). Py-GC/MS allows complex mixtures of organic materials to be analyzed, is relatively rapid, and requires milligram sample sizes and little sample preparation. We have analyzed 11 archaeological and paleontological samples by Py-GC/MS. Of these, six have consistently yielded endogenous DNA sequences, whereas five have consistently failed to yield any meaningful DNA sequences.
MATERIALS AND METHODS
Sample Treatment.
The samples listed in Table 1, along with a modern piece of human epidermis, were cleaned to remove exterior contamination and ground under liquid nitrogen in a bone grinder (Freezer/mill 6700, Spex Industries, Edison, NJ). The powdered samples were sonicated three times for 10 min in CH2Cl2 to remove lipids and contaminants originating from handling of the specimens and were dried under a stream of nitrogen.
Table 1.
Sample list
| Scientific name (common name) | Age, Kyr | Tissue type | Condition, location | Ref. |
|---|---|---|---|---|
| Equus caballus (horse) | 0.04 | Bone | Open desert, Mojave, USA | — |
| Papio cf. cynocephalus (baboon) | 2.3 | Bone | Mummified animal, tomb, Egypt | — |
| Equus ferus (horse) | 5.5 | Bone | ND, Reusten, Germany | — |
| Mylodon darwinii (ground sloth) | 13 | Bone | Mylodon cave, Patagonia, Chile | 6, 30 |
| Megalonyx sp. (ground sloth) | 13 | Tooth | Peat beds, Florida, USA | — |
| Ursus spelaeus (cave bear) | ND | Bone | Cave, France | — |
| Equus hemionus (onager) | 27 | Bone | Open permafrost, Fairbanks, USA | 6, 30 |
| Homo neandertalensis (Neanderthal) | 30–100 | Bone | Cave, Dusseldorf, Germany | 31 |
| Equus ferus (horse) | 42 | Bone | Permafrost, Siberia | 6, 30 |
| Mammuthus primigenius (mammoth) | 50 | Tissue | Permafrost, Kathanga, Siberia | 4–7 |
| Vertebrate | 60 | Bone | Border cave, South Africa | — |
List of samples used in this study. Where DNA sequences have been published, the relevant references are given. Kyr, 1,000 years; ND, not determined.
Py-GC/MS.
Approximately 0.15 mg of each sample was pyrolyzed under helium flow (0.65 kg/cm2) for 10 s by using a CDS 1000 pyroprobe (CDS Analytical, Oxford, PA). Pyrolysis products were separated in a Carlo Erba Gas Chromatograph (Milan, Italy) by using a 50-m CP Sil-5CB column (inner diameter of 0.32 mm; film thickness of 0.4 μm) connected to a Finnigan-MAT 4500 Mass Spectrometer (San Jose, CA). The pyrolysis interface was maintained at 260°C; the GC injector at 270°C; and the GC/MS transfer line at 310°C. The GC oven was operated as follows: 5 min at 35°C; increasing 4°C/min to 300°C; and 10 min at 300°C. The mass spectrometer was operated in full-scan mode [35–650 Da; 1 scan per s; 70-eV electron energy (1 eV = 1.602 × 10−19 J); 300-mA emission current; 170°C ionization source temperature]. Peaks were identified based on their mass spectral characteristics by comparison with GC/MS data and with the retention times and mass spectra of pyrolysates of amino acids (histidine, aspartic acid, valine, alanine, proline, hydroxyproline, tyrosine, tryptophan, cysteine, serine, glycine, lysine, arginine, and leucine), dipeptides (Pro-Ala, Pro-Val, Pro-Lys, and Pro-Gly), polypeptides (Thr-Pro-Arg-Lys and Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His-Leu), and collagen (23).
Amino Acid Racemization.
The extent of amino acid racemization was determined by using fluorescent derivatization of hydrolysates of powder from the samples. The eluate was separated on reverse-phase HPLC as described in Zhao and Bada (28). Results presented here were determined previously and have been published elsewhere (3, 29). A fresh piece of human skin was processed as described (3) for comparison with the archeological samples.
GC/MS Detection of Hydantoins.
The extent of oxidative damage in 10 of the 11 samples presented here was measured as described and has been presented by Höss et al. (2).
DNA Extraction, Amplification, and Sequencing.
DNA extraction and amplification by PCR were performed as described by Höss and Pääbo (30) and Krings et al. (29). DNA sequences from five of the six samples that have consistently yielded endogenous DNA sequences have been published previously (2–7, 29–31). In the case of the Mylodon (6, 31), Neanderthal (29), Equus ferus (6, 30), Equus hemionus (6, 30), and mammoth samples (4, 6), DNA results were reproduced in separate labs by using tissue or bone from the same specimen. In addition, several other mammoth tissues from remains that were preserved in permafrost have yielded authentic ancient DNA (5, 7). From those samples that did not yield any authentic DNA, extractions were performed, and a short amplification of a 141-bp fragment of the 16S rRNA gene of the mitochondria, sensitive to a single-target molecule, was attempted at least twice.
RESULTS
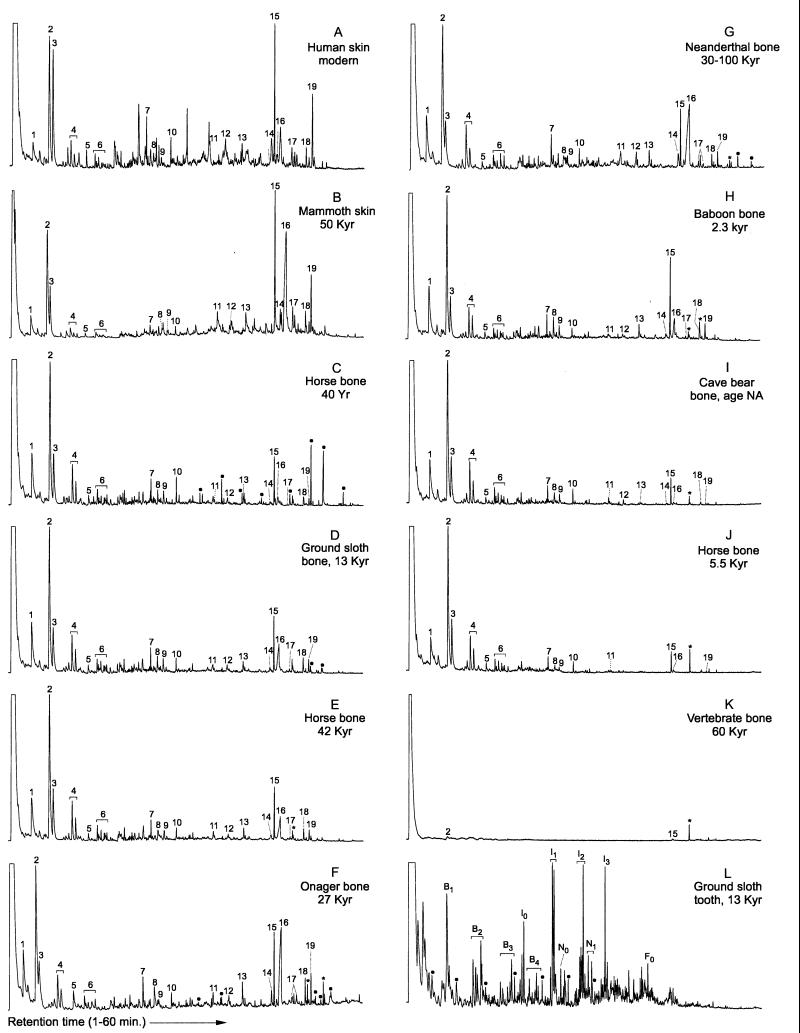
The 11 archeological samples analyzed ranged in age from 40 years to approximately 50,000 years (Table 1); 10 represent mineralized tissues (bone and tooth) from a variety of environmental conditions with one soft tissue remain from the Siberian permafrost. In addition, one sample of modern human epidermis was analyzed for comparison. Py-GC/MS was performed on all 12 samples. Fig. 1 shows the resultant chromatograms, and Tables 2 and 3 summarize the results.
Figure 1.
Total ion chromatograms of pyrolysates of the 11 samples studied. (A) Fresh skin; (B) Mammuthus primigenius; (C) Equus caballus; (D) Mylodon darwinii; (E) Equus ferus; (F) Equus hemionus; (G) Homo neandertalensis; (H) Papio cf. cynocephalus; (I) Ursus spelaeus; (J) Equus ferus; (K) vertebrate; (L) Megalonyx sp. Numbers and symbols above the peaks correspond to Table 2. ∗, contaminants; ⋅, nonextractable lipids; Bn, alkylbenzenes; In, alkylindenes; Nn, alkylnaphthalenes; Fn, fluorene; n, the extent of alkyl substitution (i.e., 0, none; 1, methyl; 2, dimethyl or ethyl; etc.).
Table 2.
Py-GC/MS data for all samples
| Scientific name (common name) | Locant (from Fig. 1) | Products of pyrolysis assigned to single amino acids (peak no.)
|
Products of pyrolysis assigned to 2,5-diketopiperazines (peak no.)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pro Hyp (1) | Pro (2) | Phe (3) | Pro (4) | Phe (5) | Pro (6) | Phe (7) | Phe (10) | Hyp (15) | Pro-Ala (14) | Pro-Gly (16) | Pro-R/V (17) | Pro-Hyp (18) | Pro-Pro (19) | ||
| Mammuthus primigenius (mammoth) | B | 13 | 73 | 32 | 7 | 3 | 3 | 7 | 6 | 100 | 9 | 70 | 19 | 16 | 41 |
| Equus caballus (horse) | C | 31 | 100 | 27 | 21 | 7 | 11 | 18 | 20 | 36 | 2 | 6 | 8 | 6 | 5 |
| Mylodon darwinii (ground sloth) | D | 30 | 100 | 24 | 18 | 5 | 9 | 16 | 9 | 39 | 2 | 19 | 2 | 10 | 9 |
| Equus ferus (horse) | E | 25 | 100 | 28 | 20 | 5 | 10 | 13 | 7 | 36 | 3 | 15 | 2 | 8 | 7 |
| Equus hemionus (onager) | F | 35 | 100 | 23 | 12 | 12 | 9 | 21 | 11 | 54 | 10 | 56 | 6 | 9 | 23 |
| Homo neandertalensis (Neanderthal) | G | 33 | 100 | 23 | 26 | 5 | 9 | 22 | 13 | 42 | 8 | 44 | 2 | 9 | 11 |
| Papio cf. cynocephalus (baboon) | H | 31 | 100 | 24 | 17 | 5 | 7 | 17 | 6 | 59 | 3 | 13 | 2 | 13 | 11 |
| Ursus spelaeus (cave bear) | I | 25 | 100 | 23 | 24 | 4 | 11 | 12 | 10 | 18 | 1 | 2 | ND | 2 | 1 |
| Equus ferus (horse) | J | 19 | 100 | 26 | 17 | 6 | 8 | 10 | 8 | 15 | 1 | 2 | ND | 2 | 1 |
| Vertebrate | K | ND | 37 | 100 | ND | 12 | ND | ND | ND | 8 | ND | 27 | ND | ND | 8 |
| Megalonyx sp. (ground sloth) | L | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
The relative amounts of single amino acid products and 2,5-diketopiperazines as indicated by the Py-GC/MS analyses are given. Numbers in parentheses indicate peaks of pyrolysis products marked in Fig. 1. Amounts of pyrolysis products are given relative to the highest peak (100) on all chromatograms. ND, not detected; Pro-R/V, Pro-Arg and/or Pro-Val.
Table 3.
Biochemical data for all samples
| Scientific name (common name) | Age, Kyr | Ratio AA/DkP | Racemization, D/L ratios (Asp, Ala) | Hydantoins, nmol/mg
|
DNA, bp | |
|---|---|---|---|---|---|---|
| 5-OH-5Me | 5-OH | |||||
| Homo sapiens (modern human) | 0 | 7 | 0.05, 0.00* | 0.42† | 0.016 | kbp‡ |
| Mammuthus primigenius (mammoth) | 50 | 2 | 0.06, 0.01 | 1.25 | 0.70 | 200 |
| Equus caballus (horse) | 0.04 | 10 | 0.05, 0.01 | 1.82 | 1.20 | 340 |
| Mylodon darwinii (ground sloth) | 13 | 7 | 0.05, 0.00 | 3.09 | 1.72 | 140 |
| Equus ferus (horse) | 42 | 8 | 0.06, 0.01 | 0.78 | 0.66 | 140 |
| Equus hemionus (onager) | 27 | 3 | 0.07, 0.01 | 2.08 | 1.19 | 140 |
| Homo neandertalensis (Neanderthal) | 30–100 | 4 | 0.12, 0.01 | NA | NA | 170 |
| Papio cf. cynocephalus (baboon) | 2.3 | 7 | 0.18, 0.02 | 8.22 | 9.07 | 0 |
| Ursus spelaeus (cave bear) | ND | 40 | NA | 14.69 | 19.20 | 0 |
| Equus ferus (horse) | 5.5 | 41 | 0.15, 0.01 | 15.81 | 17.09 | 0 |
| Vertebrate (ND) | 60 | ND | 0.99, ND | 11.17 | 10.53 | 0 |
| Megalonxy sp. (ground sloth) | 13 | ND | 0.33, 0.44 | 12.73 | 9.86 | 0 |
Molecular preservation of the samples analyzed. Ratio AA/DKP indicates the sum of peak heights (data not shown) for amino acids divided by the sum of the peak heights for 2,5-diketopiperazines. D/L ratios indicate the extent of racemization of aspartic acid and alanine (3). Also shown are the amounts (nmol/mg) of oxidative DNA lesions for 5-hydroxy-5-methylhydantoin (5-OH-5Me) and 5-hydroxyhydantoin (5-OH) (2) and the maximum number of base pairs (bp) of mitochondrial DNA amplified. NA, not attempted; ND, not determined.
The extent of racemization measured on a piece of human epidermis.
The amounts of oxidative DNA lesions from a DNA control sample from modern tissue.
The assumed length of DNA amplifiable from modern human tissue.
Two samples yielded few or no pyrolysis products that could be assigned to biological molecules. One (Table 2, Megalonyx sp.) was a 13,000-year-old tooth from Florida that showed a series of alkylated aromatic compounds such as benzenes, indenes, naphthalenes, and fluorenes (Fig. 1L). Previously, similar products have been seen in pyrolysates of fossil arthropod cuticles (17). The origin of these compounds may be either the result of extensive diagenetic alteration of the endogenous macromolecules or an artifact of the pyrolysis procedure (32). Alternatively, such compounds may stem from contamination of the sample; such contamination would agree with the racemization results, as a D/L ala ratio greater than the D/L asp ratio is known to be a marker for contamination (33). The second sample (Table 2, Vertebrate) was a 60,000-year-old bone from South Africa that showed only traces of two pyrolysis products related to proline (Fig. 1K). The other nine samples contained a complex mixture of compounds. The large majority of these pyrolysis products can be assigned to amino acids and protein debris either by virtue of their molecular structures or by comparative interpretation of pyrolysates of polypeptides, collagen, and fresh skin. For example, pyrroline (no. 1; numbers refer to those in Fig. 1), pyrrole (no. 2), C1 (no. 4), and C2 (no. 6) alkylpyrroles are characteristic products of proline; diketodipyrrole (no. 15) and pyrroline (no. 1) are derived from hydroxyproline, whereas toluene (no. 3), styrene (no. 5), ethylcyanobenzene (no. 7), and propylcyanobenzene (no. 10) are produced during the pyrolysis of phenylalanine (23, 24). Furthermore, several 2,5-diketopiperazines are seen. These are formed via a so-called “McLafferty rearrangement” (23, 34, 35) during pyrolysis of dipeptide sequences in which proline is one of the amino acids. In most of the samples studied, the following products can be assigned to 2,5-diketopiperazines, which can be recognized in terms of their dipeptide precursors: Pro-Ala (no. 14), Pro-Gly (no. 15), Pro-Val and/or Pro-Arg (no. 17), Pro-Hyp (no. 18), and Pro-Pro (no. 19).
The fact that derivatives of proline, glycine, alanine, and hydroxyproline are prominent in the pyrolysates suggests that collagen may be present in the samples. Collagen, the most abundant protein in mammalian skin and bone, consists of three polypeptide chains in which glycine (≈33%), alanine (≈10%), proline (≈12%), and hydroxyproline (≈10%) are the major constituents. To investigate whether other products seen in the pyrolysates may be derived from collagen, a sample of collagen and fresh skin (Fig. 1A) was pyrolyzed in parallel with the ancient tissues. Besides many minor pyrolysis products, compounds no. 8 and no. 9 (tentatively identified as a piperidinone derivative), alkylpyrimidones (no. 11 and no. 12), and a pyrazine derivative (no. 13) were all prominent in the collagen, fresh skin, and some of the ancient samples. Thus, a majority of the organic compounds detected in the pyrolysates seem to be assignable to collagen-type debris. Indeed, in agreement, relatively large amounts of products assignable to hydroxyproline are also detected in the samples.
Four of the samples also contained C16 and C18 fatty acids, n-alk-1-enes and n-alkanes, alkyl-nitriles, and amides, presumably derived from bound lipid components (Fig. 1 C, D, F, and G, indicated by ⋅ symbols). The specimen of the bone from a 40-year-old horse showed the highest relative abundance of the latter components (Fig. 1C). These lipids may be bound to tissue structures or encapsulated in micropores such that they cannot be removed completely by organic extraction before pyrolysis.
Table 2 summarizes the relative amounts of pyrolysis products (highest peak assigned as 100) related to single amino acids and 2,5-diketopiperazines on all samples. Table 3 gives the ratio of amino acid peak height to 2,5-diketopiperazine peak height, the extent of racemization of two amino acids (3, 29), the extent of oxidative DNA damage to the pyrimidine bases (5-hydroxy-5-methylhydantoin and 5-hydroxyhydantoin) assessed by GC/MS (2), and the length of DNA fragment amplifiable from the extract. It has been possible to retrieve DNA sequences from six of the samples. These positive results have been verified through reproduction within a single laboratory and, with the sole exception of the 40-year-old sample, by workers in other laboratories (2–7, 29–31). It has proven impossible to amplify DNA sequences from the remaining five samples, despite repeated attempts with various extraction protocols all performed in the same laboratory.
DISCUSSION
Of the 11 samples analyzed, samples B–H (as identified in Fig. 1 and Table 2) show good levels of protein debris as indicated by the relative amounts of pyrolysis products compared with those of collagen and fresh skin, especially those products assigned to diketopiperazines. Samples I and J had fewer relative amounts of diketopiperazines, whereas samples K and L lacked any indication of protein preservation.
There is no clear correlation between the relative abundance of most pyrolysis products assigned to single amino acids (e.g., peaks 1–7) and the preservation of endogenous DNA fragments. For example, the relative abundance of these products is similar in samples E and I, but the latter has not yielded genuine ancient DNA sequences. However, the relative abundance of the products assigned to 2,5-diketopiperazines in the pyrolysate of samples B–H is greater than those of samples I–L. Additionally, one peak, no. 17 (Pro-Arg and/or Pro-Val), was not detectable in samples I–L. The preservation of peptide linkages involving these amino acids does show a better correlation with DNA preservation. In fact, by looking at the ratio of the summed peak heights of single amino acid products and the summed peak heights of the 2,5-diketopiperazine products (Table 3, AA/DKP), we observed a distinct hierarchy of preservation. For samples B–H, the ratio of AA/DKP peak heights ranged from 2–10, with fresh skin tissue at 5; however, samples I and J both had ratios of 40 and 41. We interpret this ratio as suggesting the extent of hydrolysis of the intact proteins within the sample. In cases in which preservation has been good and little hydrolysis has taken place, the peptides are longer in length, and, therefore, more diketopiperazines are formed due to the pyrolysis procedure. On the other hand, when peptides are cleaved into shorter fragments because of a more complete hydrolysis, the pyrolysis of these tissues yields fewer diketopiperazines. Therefore, this ratio may be one way to estimate the relative amounts of peptide hydrolysis in the samples.
Proteins hydrolyze under geochemical conditions through cleavage of an internal peptide bond, an internal amino lysis reaction at the N terminal position, and hydrolysis at the C terminal position (36). In bones, the hydrolysis of collagen is rapid, and little intact collagen is found after 10,000–30,000 years in temperate environments (36).
Of the seven samples (B–H) that show good protein debris, the first six have yielded authentic DNA sequences (Table 3). In addition, five of these samples have D/L ratios for aspartic acid between 0.05 and 0.07, whereas the Neanderthal-type specimen had a D/L ratio for aspartic acid of 0.12. The remaining samples that failed to yield DNA sequences had D/L ratios of 0.15 or higher and have between 2.7–20.2 and 5.3–29.2 times more oxidized pyrimidine bases than those samples that yielded DNA sequences. Thus, in cases where protein preservation is good and little racemization and base oxidation has taken place, amplifiable DNA is present. The Py-GC/MS results of the Neanderthal-type specimen indicate good protein preservation with relatively high levels of diketopiperazines. These results agree with the amino acid racemization and DNA results published for this specimen (29). Nevertheless, the baboon bone (sample H) shows that these different parameters of preservation do not always correlate well with each other. This sample yielded relative amounts of all pyrolysis products comparable to those of samples C and D but failed to yield any authentic ancient DNA. Additionally, the ratio of amino acids to diketopiperazines was 7 and thus well within the range (2–10) of “good” samples. Thus, the preservation of proteinaceous debris in bone alone does not guarantee the presence of amplifiable DNA. Despite these results, the sample did have relatively high amounts of oxidatively damaged bases and a moderately high level of racemization (D/L Asp of 0.18).
It is generally believed that the degradation of organic matter in fossils is a function of limited microbial activity (37, 38). Environments where bacterial growth is restricted, such as permafrost, hypersaline pools, acidic moorlands, and arid regions, may therefore provide good long-term preservation. However, although such environments often ensure excellent morphological preservation, they are not always conducive to macromolecular preservation. A case in point may be the Egyptian baboon bone; it is well preserved macroscopically, and protein debris is relatively well preserved as assessed by Py-GC/MS. The bone seems to have been protected from extensive microbial degradation, a situation typical with many remains from Egypt because of the arid climate (39). In spite of this preservation, the sample does not yield amplifiable DNA, which is in fact typical of most naturally and artificially mummified remains of humans and animals from Egypt (40). Therefore, the recognition of Py-GC/MS products compatible with a collagen origin does not guarantee DNA survival, a result which is in agreement with previous results (3). It is possible that samples preserved under these circumstances have undergone condensation or Maillard-type reactions, which may have cross-linked the proteins with the DNA, and thus the samples may contain endogenous DNA that is inaccessible to current methods of DNA extraction (as has been shown recently in coprolites from the Southwestern United States; ref. 41). Further work is needed to elucidate this possibility. Additionally, samples deposited in warm environments may be preserved from extensive hydrolysis through rapid drying but are still subject to oxidative damage, which would block the extension of the polymerase in the PCR. Samples like these are lost sources of ancient DNA unless the DNA can be repaired before DNA amplification.
The results presented here are restricted to relatively few samples, because the number of ancient specimens from which the authenticity of ancient DNA sequences have been rigorously established is limited. Nevertheless, the results indicate that Py-GC/MS may provide a fast and sensitive way to differentiate samples that are unlikely to yield any DNA from those that may contain DNA. At present, both Py-GC/MS and amino acid analyses that allow the extent of racemization to be determined seem suited for the purpose of identifying samples suitable for DNA extraction (3, 42). Additionally, Py-GC/MS has the advantage of being able to determine the extent of diagenetic alteration of a sample more easily than other methods of analysis on fossil material. On the other hand, Py-GC/MS is a less sensitive method than HPLC coupled with fluorescence and can yield products that are often the result of rearrangements of the original molecules occurring during the pyrolysis procedure. Recent analysis of amber-entombed fossils by Py-GC/MS (43) has shown extensive diagenetic alteration contrary to the results obtained from an amino acid analysis by HPLC (44), which indicated amino acid preservation and low racemization. It may be that these particular amino acids were in low concentration and thus not detectable by Py-GC/MS; in addition, they may have been “masked” by the resin markers in the total pyrolysate. Based on the diagenetic transformation of more decay-resistant molecules such as lignin and chitin, it was postulated that more decay-prone molecules such as DNA would not survive (43). In any event, fossils that are millions of years old will require even more discriminatory methods of analysis before the exact nature of preservation can be elucidated. As the number of ancient samples from which verified DNA sequences have been published increases, it will be possible to evaluate more precisely which of these methods of analysis best correlates with DNA preservation. In any event, because the amplification of DNA by the PCR is notoriously prone to contamination, it is extremely valuable to analyze protein preservation from archaeological and paleontological remains before DNA analysis.
Our knowledge of the influence of particular environmental conditions on the preservation of biomacromolecules is still incomplete. In spite of this fact, most investigations stress that a favorable burial environment will limit biodegradation and thus enhance preservation. However, preservation is often selective, and in some cases, the presence of one molecule will not necessarily guarantee the persistence of another. In general, preservation seems to follow a predicted hierarchy of molecules dictated by the relative strength of their bonds (45). This model, however, does not take into consideration the fact that molecules may be altered once they are cross-linked, adsorbed to minerals, and/or trapped within glycosylation end products, resulting in preferential preservation. Although permafrost may provide an excellent burial condition for biomacromolecules, it is still not clear which environmental factors may enhance preservation of some macromolecules, while, at the same time, leading to the degradation of others. In any event, time is not the crucial factor when the specimens are deposited/buried in a favorable environment where the initial rapid degradation can be inhibited.
Acknowledgments
We thank J. Bada, H. Bland, V. Börner, D. Poinar, G. Eglinton, M. Hofreiter, M. Höss, and M. Krings for assistance; R. Schmitz and H. Krainitzki for the Neanderthal samples; J. F. Carter and A. Gledhill for technical assistance with the mass spectrometry; and Svante Pääbo for constructive criticism and excellent working facilities, as well as financial support of the Deutsche Forschungsgemeinschaft to Svante Pääbo. Py-GC/MS analyses were performed while B.A.S. was in the Department of Chemistry, University of Bristol, United Kingdom, and supported by Natural Environment Research Council (London) Ancient Biomolecules Initiative Grant GST/02/1027 to D. E. G. Briggs and R. P. Evershed.
ABBREVIATIONS
- Py-GC/MS
flash pyrolysis with gas chromatography and mass spectrometry
- Kyr
1,000 years
References
- 1.Handt O, Höss M, Krings M, Pääbo S. Experientia. 1994;50:524–529. doi: 10.1007/BF01921720. [DOI] [PubMed] [Google Scholar]
- 2.Höss M, Jaruga P, Zastawny T H, Dizdaroglu M, Pääbo S. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poinar H N, Höss M, Bada J L, Pääbo S. Science. 1996;272:864–866. doi: 10.1126/science.272.5263.864. [DOI] [PubMed] [Google Scholar]
- 4.Höss M, Vereshchagin M K, Pääbo S. Nature (London) 1994;370:333. doi: 10.1038/370333a0. [DOI] [PubMed] [Google Scholar]
- 5.Hagelberg E, Thomas M H, Cook C E, Jr, Sher A V, Baryshnikov G F, Lister A M. Nature (London) 1994;370:333–334. doi: 10.1038/370333b0. [DOI] [PubMed] [Google Scholar]
- 6.Taylor P G. Mol Biol Evol. 1996;13:283–285. doi: 10.1093/oxfordjournals.molbev.a025566. [DOI] [PubMed] [Google Scholar]
- 7.Noro M, Masuda R, Dubrovo I, Yoshida M, Kato M. J Mol Evol. 1998;46:314–326. doi: 10.1007/pl00006308. [DOI] [PubMed] [Google Scholar]
- 8.Austin J J, Ross A, Smith A, Fortey R, Thomas R. Proc R Soc London Ser B. 1997;264:467–474. doi: 10.1098/rspb.1997.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidow A, Wilson A C, Pääbo S. In: Molecules Through Time. Eglington G, Curry G B, editors. Vol. 333. London: R. Soc. London; 1991. pp. 429–433. [Google Scholar]
- 10.Zischler H, Höss M, Handt O, van der Kuyl A C, Goudsmit J, Pääbo S. Science. 1995;268:1192–1193. doi: 10.1126/science.7605504. [DOI] [PubMed] [Google Scholar]
- 11.Pääbo S, Wilson A C. Curr Biol. 1991;1:45–46. doi: 10.1016/0960-9822(91)90125-g. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl T. Nature (London) 1993;365:700. doi: 10.1038/365700a0. [DOI] [PubMed] [Google Scholar]
- 13.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 14.Boon J J, Stout S A, Genuit W, Spackman W. Acta Bot Neerl. 1989;38:391–404. [Google Scholar]
- 15.Hatcher P G, Lerch H E, III, Kotra R K, Verheyen T V. Fuel. 1988;67:1069–1075. [Google Scholar]
- 16.Van Bergen P F, Goni M, Collinson M E, Barrie P J, Sinninghe Damste J S, de Leeuw J W. Geochim Cosmochim Acta. 1994;58:3823–3844. [Google Scholar]
- 17.Stankiewicz B A, Briggs D E G, Evershed R P, Flannery M B, Wuttke M. Science. 1997;276:1541–1543. [Google Scholar]
- 18.Stankiewicz B A, Mastalerz M, Kruge M A, van Bergen P F, Sadowska A. New Phytol. 1997;135:375–393. [Google Scholar]
- 19.van der Kaaden A, Boon J J, de Leeuw J W, de Lange F, Schuyl P J W, Schulten H-R, Bahr U. Anal Chem. 1984;56:2160–2165. [Google Scholar]
- 20.van Aarssen B G K, de Leeuw J W, Collinson M, Boon J J, Goth K. Geochim Cosmochim Acta. 1994;58:223–229. [Google Scholar]
- 21.Derrene S, Largeau C, Casadevall E. Org Geochem. 1991;17:597–602. [Google Scholar]
- 22.Tegelaar E W, Visscher H, Schenk P A, de Leeuw J W. Paleobiology. 1991;17:133–144. [Google Scholar]
- 23.Stankiewicz B A, Hutchins J C, Thomson R, Briggs D E G, Evershed R P. Rapid Commun Mass Spectrom. 1997;11:1884–1890. doi: 10.1002/(SICI)1097-0231(199711)11:17<1884::AID-RCM62>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Tsuge S, Matsubara H. J Anal Appl Pyrol. 1985;8:49–64. [Google Scholar]
- 25.Munson T O, Vick J. J Anal Appl Pyrol. 1985;8:493–501. [Google Scholar]
- 26.Jarman M. J Anal Appl Pyrol. 1980;2:217–223. [Google Scholar]
- 27.Logan G A, Boon J J, Eglinton G. Proc Natl Acad Sci USA. 1993;90:2246–2250. doi: 10.1073/pnas.90.6.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao M, Bada J L. J Chromatogr A. 1995;690:55–60. doi: 10.1016/0021-9673(94)00927-2. [DOI] [PubMed] [Google Scholar]
- 29.Krings M, Stone A, Schmitz R, Krainitzki H, Stoneking M, Pääbo S. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 30.Höss M, Pääbo S. Nucleic Acids Res. 1993;21:3913–3914. doi: 10.1093/nar/21.16.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Höss M, Dilling A, Currant A, Pääbo S. Proc Natl Acad Sci USA. 1996;93:181–185. doi: 10.1073/pnas.93.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stankiewicz B A, Briggs D E G, Evershed R P. Energy Fuels. 1997;11:515–521. [Google Scholar]
- 33.Bada J L, Kvenvolden K A, Peterson E. Nature (London) 1973;245:308–310. [Google Scholar]
- 34.Smith G G, Sudhakar Reddy G, Boon J J. J. Chem. Soc. Perkin Trans. 2. 1988. , 203. [Google Scholar]
- 35.Ratcliff J M A, Medley E E, Simmonds P G. J Org Chem. 1974;39:1482. doi: 10.1021/jo00924a007. [DOI] [PubMed] [Google Scholar]
- 36.Bada J L. Philos Trans R Soc London B. 1991;333:349–358. [Google Scholar]
- 37.Eglinton G, Logan G. Philos Trans R Soc London B. 1991;333:315–328. doi: 10.1098/rstb.1991.0081. [DOI] [PubMed] [Google Scholar]
- 38.Briggs D E, Eglinton G. Chem Br. 1994;11:907–912. [Google Scholar]
- 39.van Bergen P F, Bland H A, Evershed R P. Geochim Cosmochim Acta. 1997;61:1919–1930. [Google Scholar]
- 40.Krings M. Dissertation. Munich: University of Munich; 1998. [Google Scholar]
- 41.Poinar H N, Hofreiter M, Spaulding G S, Martin P S, Stankiewicz A B, Bland H, Evershed R P, Possnert G, Pääbo S. Science. 1998;281:402–406. doi: 10.1126/science.281.5375.402. [DOI] [PubMed] [Google Scholar]
- 42.Cooper A, Poinar H N, Pääbo S, Radovcic J, Debenath A, Caparros M, Barroso-Ruiz C, Bertranpetit J, Nielsen-Marsh C, Hedges R E, et al. Science. 1997;277:1021–1023. doi: 10.1126/science.277.5329.1021b. [DOI] [PubMed] [Google Scholar]
- 43.Stankiewicz B A, Poinar H N, Briggs D E G, Evershed R P, Poinar G O., Jr Proc R Soc London Ser B. 1998;265:641–647. [Google Scholar]
- 44.Bada J L, Wang S X, Poinar H N, Pääbo S, Poinar G O., Jr Geochim Cosmochim Acta. 1994;58:3131–3135. doi: 10.1016/0016-7037(94)90185-6. [DOI] [PubMed] [Google Scholar]
- 45.Tegelaar E W, de Leeuw J W, Derenne S, Largeau C. Geochim Cosmochim Acta. 1989;53:3103–3106. [Google Scholar]