Abstract
The bacterial metabolism of short-chain aliphatic alkenes occurs via oxidation to epoxyalkanes followed by carboxylation to β-ketoacids. Epoxyalkane carboxylation requires four enzymes (components I–IV), NADPH, NAD+, and a previously unidentified nucleophilic thiol. In the present work, coenzyme M (2-mercaptoethanesulfonic acid), a compound previously found only in the methanogenic Archaea where it serves as a methyl group carrier and activator, has been identified as the thiol and central cofactor of aliphatic epoxide carboxylation in the Gram-negative bacterium Xanthobacter strain Py2. Component I catalyzed the addition of coenzyme M to epoxypropane to form a β-hydroxythioether, 2-(2-hydroxypropylthio)ethanesulfonate. Components III and IV catalyzed the NAD+-dependent stereoselective dehydrogenation of R- and S-enantiomers of 2-(2-hydroxypropylthio)ethanesulfonate to form 2-(2-ketopropylthio)ethanesulfonate. Component II catalyzed the NADPH-dependent cleavage and carboxylation of the β-ketothioether to form acetoacetate and coenzyme M. These findings evince a newfound versatility for coenzyme M as a carrier and activator of alkyl groups longer in chain-length than methane, a function for coenzyme M in a catabolic pathway of hydrocarbon oxidation, and the presence of coenzyme M in the bacterial domain of the phylogenetic tree. These results serve to unify bacterial and Archaeal metabolism further and showcase diverse biological functions for an elegantly simple organic molecule.
Over 200 million tons of gaseous alkenes are produced annually from biogenic and anthropogenic processes (1, 2). Bacteria that are able to grow by using short-chain aliphatic alkenes as sources of carbon and energy are believed to play an important role in remineralization of this carbon in the global carbon cycle (3). Although the bacterial consumption of these compounds has been well documented, only recently have the biochemical pathways and enzymes involved in alkene catabolism in bacteria been investigated (4, 5). For two propylene-using bacteria, Xanthobacter strain Py2, a Gram-negative rod, and Rhodococcus rhodochrous strain B276, a Gram-positive nocardioform, the pathway for propylene metabolism involves a monooxygenase-catalyzed epoxidation to form epoxypropane (6, 7), which is metabolized further by carboxylation to acetoacetate (5, 8). Acetoacetate apparently undergoes thiolysis, producing two molecules of acetyl–coenzyme A that feed into the glyoxylate and tricarboxylic acid cycles of the bacteria (9). This strategy represents a clever means for taking a toxic C3 molecule and combining it with CO2 to form a central C4 metabolite.
The carboxylation of epoxypropane is catalyzed by a four-component enzyme system that also requires NAD+ and NADPH, which are reduced and oxidized in the course of the reaction according to the balanced equation: epoxypropane + CO2 + NADPH + NAD+ → acetoacetate + H+ + NADP+ + NADH (10). Based on the biochemical properties of the four epoxide carboxylase components (designated components I–IV) and mechanistic studies of the system, component I, a hexameric zinc-containing protein, has been proposed to catalyze the reaction of epoxypropane with a nucleophilic thiolate to form a β-hydroxythioether conjugate (10). Components III and IV are NAD+-dependent short-chain dehydrogenases believed to oxidize stereoselectively the R- and S-β-hydroxythioethers formed from R- and S-epoxypropane enantiomers to form a common β-ketothioether intermediate (11). Component II is an NADPH:disulfide oxidoreductase believed to catalyze the reductive cleavage of the β-ketothioether to form the free thiolate and the enolate of acetone, which then undergoes carboxylation to form acetoacetate (10, 12, 13).
One of the unanswered questions of this unprecedented catalytic cycle is the nature of the nucleophilic thiolate that serves as the carrier of the activated epoxide during this three-step reaction. Herein, we report the identification of this thiolate as coenzyme M (CoM; 2-mercaptoethanesulfonic acid), a cofactor previously believed to be restricted to the methanogenic Archaea, where it plays a central role in the reductive formation of methane (14–17).
MATERIALS AND METHODS
Radiolabeling of Thiol Cofactor.
Purified component I (2 mg) was incubated with 500 nmol [14C]epoxypropane (250 μCi/mmol; Wizard Laboratories, Sacramento, CA) for 30 min at 30°C in 50 mM Tris⋅HCl (pH 8.2). The sample was then applied to a 1.5 × 5-cm column of Sephadex G-25 equilibrated in 50 mM Tris⋅HCl (pH 8.2) and eluted with 9 ml of 50 mM Tris⋅HCl (pH 8.2); 1-ml fractions were collected and added to scintillation vials containing 10 ml of scintillation fluid (ScintiSafe Econo 1; Fisher), and the radioactivity was analyzed by scintillation counting as described (4).
Isolation of CoM Derivative.
Component I (350 mg; >90% purity) was divided into seven 1.5-ml aliquots, and each reacted with 5 μmol epoxypropane for 10 min at 30°C in 50 mM ammonium bicarbonate (pH 8.2). Each reaction mix was then applied to a 1.5 × 5-cm column of Sephadex G-25 equilibrated in 50 mM ammonium bicarbonate (pH 8.2). The salt fraction from each reaction mix was combined and lyophilized (freeze dried) to remove unreacted epoxypropane, dissolved in 3 ml of water, and applied to a 1 × 2.5-cm column of Q-Sepharose that was washed with 3 ml each of 50 mM and 100 mM ammonium acetate (pH 8.2). The epoxypropane–cofactor adduct was recovered in the 100 mM ammonium acetate fraction, which was lyophilized and dissolved in 2H2O for spectral characterization.
Synthesis and Spectral Analysis of Epoxypropane–CoM Conjugates.
The 2-(2-hydroxypropylthio)ethanesulfonate was prepared enzymatically by incubating 125 μmol epoxypropane, 100 μmol 2-mercaptoethanesulfonic acid, and 0.5 mg component I in 0.5 ml of 50 mM ammonium bicarbonate (pH 8.2) for 30 min at 30°C. The reaction mix was then applied to a 1.5 × 5-cm column of Sephadex G-25 equilibrated in 50 mM ammonium bicarbonate (pH 8.2). The salt fraction (i.e., included volume) from the column was then lyophilized (freeze dried) overnight. The 2-(R-2-hydroxypropylthio)ethanesulfonate and 2-(S-2-hydroxypropylthio)ethanesulfonate were prepared enzymatically by using the procedure described above, except that R- and S-epoxypropane, respectively, were substituted for racemic epoxypropane. Chemically synthesized 2-(2-ketopropylthio)ethanesulfonate was prepared by dissolving 0.2 g of 2-mercaptoethanesulfonic acid (sodium salt) in 208 μl of aqueous 6 M NaOH (molar ratio of 1:1). The mixture was allowed to shake at 30°C for 5 min; then 1.9 ml of aqueous 1 M chloroacetone was added (0.5 molar excess), and the mixture was allowed to shake an additional 90 min. The sample was then lyophilized in a speed-vac concentrator. Chemically synthesized 2-(2-hydroxypropylthio)ethanesulfonate was prepared by reducing 0.5 ml of 100 mM 2-(2-ketopropylthio)ethanesulfonate with excess NaBH4. Samples used in NMR analysis were dissolved in 2H2O.
The 2-(2-ketopropylthio)ethanesulfonate was prepared enzymatically by incubating 20 μmol 2-hydroxypropyl–CoM, 40 μmol NAD+, 100 μg of component III, and 100 μg of component IV in 0.5 ml of 50 mM ammonium bicarbonate (pH 8.2) for 20 min at 30°C. The assay reaction mixture was then centrifuged for 20 min at 2,000 × g in a Centricon 30 centrifuge concentrator, and the filtrate was lyophilized in a speed-vac concentrator. The dried sample was dissolved in 100 μl of water, and CoM conjugates were separated from NAD+ and NADH by using HPLC. The sample was applied to a Supelcosil LC-18 column (25 cm × 4.6 mm) with a mobile phase gradient from 25 mM potassium phosphate (pH 6.0) to 50 mM potassium phosphate (pH 3.5) containing 10% (vol/vol) methanol, and the eluent was monitored at 210 nm. By using these conditions, 2-hydroxypropyl–CoM and 2-ketopropyl–CoM eluted together. Fractions containing CoM conjugates were lyophilized overnight with a speed-vac concentrator and then dissolved in 350 μl of 2H2O and analyzed by 1H NMR.
All 1H NMR spectra were recorded in 5-mm NMR tubes with a Bruker ARX-400 NMR spectrometer at 25°C.
Enzyme Activity Assays.
Epoxide carboxylase components I–IV were purified from Xanthobacter strain Py2 (10, 13). Complete epoxide carboxylase activity was assayed by using reagents and reaction conditions as described (5, 10), except that acetoacetate was quantified by the amount of 14C radiolabel incorporated from 14CO2/NaH14CO3 (specific activity of 59 μCi/mmol of CO2–NaHCO3). Samples (20 μl) were periodically removed from assay vials and added to 20 μl of 1 M NaBH4 in 0.5-ml microcentrifuge tubes. After a 1-min incubation, 110 μl of ethanol/formic acid (50:50, vol/vol) was added. The samples were then applied to 25-mm Whatman GF/B fiber filters, allowed to dry, placed in scintillation vials, and analyzed by scintillation counting as described (4).
Assays of individual epoxide carboxylase components were performed in triplicate in 50 mM Tris⋅HCl (pH 8.2) at 30°C in a total volume of 1 ml. Component I activity was measured by monitoring the time-dependent depletion of 2 mM racemic R- or S-epoxypropane by gas chromatography in sealed (9-ml) vials containing 3 mM CoM. Component III and IV assays were performed in 1-ml (1-cm path length) quartz cuvettes containing 1 mM NAD+ and 2 mM of 2-R-hydroxypropyl–CoM or 2-S-hydroxypropyl–CoM. The reactions were monitored photometrically by measuring the increase in absorbance at 340 nm (ɛ340 of 6.22 mM−1⋅cm−1 for NADH) over time. Component II activity was monitored by the time-dependent accumulation of acetoacetate (quantified as described above) in assays containing 4 mM 2-ketopropyl–CoM, 2 mM NADPH, and 40 mM 14CO2/NaH14CO3.
Alkene monooxygenase components were purified and assayed as described (6). The formation of R- or S-epoxypropane from propylene degradation was monitored and quantified by using gas chromatography (Shimadzu model GC-8A) equipped with a β-DEX 225 (Supelco) chiral capillary column (30 m × 0.53 mm). The gas chromatograph was operated with an injector-detector temperature of 200°C, and the column temperature was increased from 40°C to 70°C at 1°C per min.
RESULTS AND DISCUSSION
Identification of a Dissociable Thiol Cofactor Involved in Epoxide Activation.
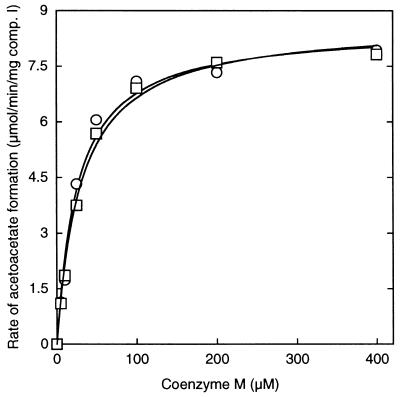
Based on the biochemical properties of the four epoxide carboxylase components and mechanistic studies of the system, the proposed reaction mechanism of epoxide carboxylation is believed to proceed by nucleophilic attack of a thiolate on the C1 atom of a terminal epoxide, leading to ring opening and the formation of a β-hydroxythioether conjugate (10). To investigate this step of the proposed mechanism, identification of the nucleophilic thiolate was pursued as follows. Because component I is believed to catalyze the initial reaction of thiolate and epoxypropane, the purified protein from Xanthobacter strain Py2 was incubated with [14C]epoxypropane followed by gel filtration chromatography, with the purpose of determining whether the putative β-hydroxythioether conjugate would remain tightly bound or dissociate from the protein. The component I thus treated was catalytically inactive in the epoxide carboxylase assay and did not contain bound radiolabel. The addition of the salt fraction (i.e., included volume) to the desalted component I restored its activity, suggesting the presence of a dissociable epoxide–cofactor conjugate. A variety of commercially available thiol compounds were tested for their ability to substitute for the salt fraction in restoring component I activity. DTT, β-mercaptoethanol, thioglycolytic acid, cysteine, lipoic acid, homocysteine, and glutathione were each unable to substitute. However, CoM not only restored epoxide carboxylase activity with inactive component I but stimulated the activity of active or inactive component I by orders of magnitude in a concentration-dependent manner (Fig. 1). At fixed concentrations of the four epoxide carboxylase components, the stimulation of activity by CoM followed saturation kinetics with an apparent Km of 26.3 ± 4.2 μM (Fig. 1). The stimulatory effect of CoM was highly specific: the related compounds 3-mercapto-1-propanesulfonate, 3-mercaptopropionate, 2-mercaptoethylamine, 2-hydroxyethanesulfonate, and 2-aminoethanesulfonate were unable to substitute for CoM in the complete epoxide carboxylase assay.
Figure 1.
Stimulation of epoxide carboxylase activity by commercially obtained CoM. Assays were performed in duplicate in sealed 9-ml vials containing purified components (5 μg of component I, 250 μg of component II, 40 μg of component III, and 25 μg of component IV) in 50 mM Tris⋅HCl (pH 8.2). □, assays containing as-isolated component I; ○, assays containing component I preincubated with epoxypropane followed by gel filtration chromatography.
Isolation of a CoM Derivative from Xanthobacter Strain Py2.
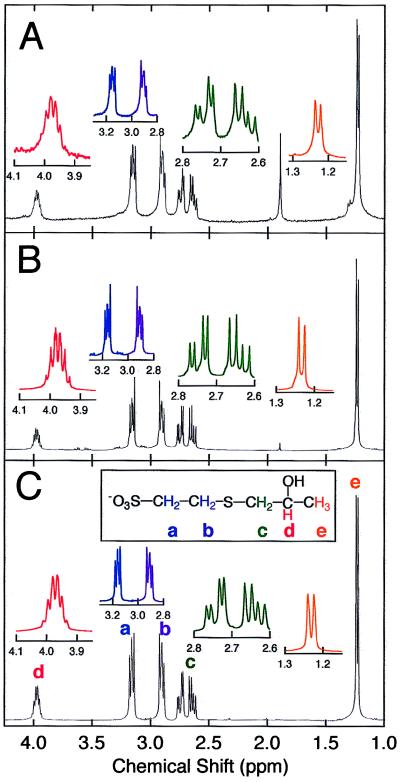
To determine whether the physiological nucleophile is CoM, the thiol-epoxypropane conjugate was isolated from component I and analyzed by 1H NMR spectroscopy. The 1H NMR spectrum of the epoxypropane–cofactor adduct is shown in Fig. 2A. This spectrum is identical to the spectrum for the product of the component I-catalyzed reaction of commercially obtained CoM with epoxypropane (Fig. 2B) and also to the spectrum of chemically synthesized 2-(2-hydroxypropylthio)ethanesulfonate (Fig. 2C). The signal assignments shown in Fig. 2C are consistent with the chemical shifts, spin integrations, and splitting patterns expected for the individual proton environments (see ref. 18 and legend to Fig. 2). Negative ion electrospray mass spectrometry of the extracted epoxypropane–cofactor adduct had a signal at 199 m/z, the value of which is identical to the expected molecular mass of the negatively charged 2-hydroxypropyl–CoM (199 g/mol) and also identical to the spectrum of enzymatically or chemically synthesized 2-hydroxypropyl–CoM (spectra not shown). To verify the presence of free CoM in as-isolated component I, a sample of the protein (25 mg) was heat-denatured followed by centrifugation and analysis of the supernatant by negative ion electrospray mass spectrometry. The sample had a strong signal that was identical to that of commercially obtained CoM and at the m/z value (140.9) expected for free CoM (spectra not shown).
Figure 2.
Spectral identification of epoxypropane-thiol adduct. (A) 1H NMR spectrum of the epoxypropane–cofactor adduct isolated from component I. (B) 1H NMR spectrum of the product of component I-catalyzed reaction of CoM with epoxypropane. (C) 1H NMR spectrum of chemically synthesized 2-hydroxypropyl–CoM. There are five signals (a–e) that correspond to protons on the carbon atoms as indicated in C. The triplet resonances at 2.91 and 3.16 ppm correspond to methylene groups (a) and (b), each integrating to two protons. The protons of methylene group (c) are not chemically equivalent and are therefore split into two quartets with resonances centered at 2.64 and 2.75 ppm, each multiplet integrating to one proton. The sextet at 3.98 ppm corresponds to the proton on carbon (d) and integrates to one proton. The protons of methyl group (e) are split to a doublet at 1.23 ppm that integrates to three protons. The resonance at 1.89 ppm in the isolated epoxypropane–cofactor adduct spectrum (A) is caused by acetate remaining in the sample.
CoM Is the C3 Carrier Throughout the Epoxide Carboxylation Pathway.
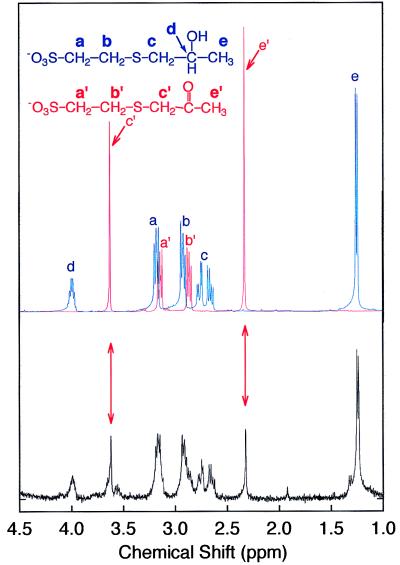
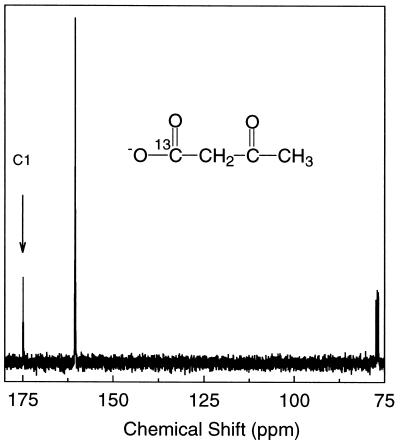
As shown in Fig. 3, the addition of NAD+ and components III and IV to 2-hydroxypropyl–CoM resulted in a change in the NMR spectrum that is consistent with the conversion of 2-hydroxypropyl–CoM to 2-ketopropyl–CoM, the expected product of dehydrogenation. Component II alone was found to catalyze the NADPH- and CO2-dependent transformation of 2-ketopropyl–CoM to acetoacetate and free CoM. This transformation is illustrated by the 13C NMR spectrum presented in Fig. 4, which shows the spectrum of the product formed on reaction of 13C-enriched CO2 with 2-ketopropyl–CoM. As expected, the acetoacetate product is specifically enriched with 13C label at the C1 position.
Figure 3.
1H NMR spectrum of the product of component III- and IV-catalyzed reaction of NAD+ and 2-hydroxypropyl–CoM. The spectra of chemically synthesized 2-hydroxypropyl–CoM (in blue) and 2-ketopropyl–CoM (in red) are overlaid and shown directly above the product spectrum. Resonances at 1.23 and 3.98 ppm correspond to methyl group (e) and proton (d) of 2-hydroxypropyl–CoM, respectively. Resonances at 2.63 and 2.74 ppm correspond to methylene group (c) of 2-hydroxypropyl–CoM. The multiplets at 2.89 and 3.15 ppm are the overlapping resonances of methylene groups (b, b′) and (a, a′) of 2-hydroxypropyl–CoM and 2-ketopropyl–CoM, respectively. The singlet at 3.63 ppm corresponds to the (c′) methylene group of 2-ketopropyl–CoM, which integrates to two protons. The signal at 2.33 ppm corresponds to methyl group (e′) of 2-ketopropyl–CoM, which integrates to three protons.
Figure 4.
13C NMR spectrum of the product of component II-catalyzed reaction of 2-ketopropyl–CoM, NaH13CO3, and NADPH. The resonance peak at 174.6 ppm corresponds to the chemical shift for the C1 (carboxyl) atom of acetoacetate as reported (8). The resonance at 160.3 ppm is caused by the NaH13CO3 present in the sample. The resonance at 77.0 ppm is caused by chloroform, which was used as the reference.
The specific activities for the individual reactions of the epoxide carboxylation pathway are summarized in Table 1. In reporting these activities, attention should be drawn to the stereoselectivity of the components for their respective reactions. Both epoxypropane and 2-hydroxypropyl–CoM contain a single chiral center. The alkene monooxygenase of Xanthobacter Py2 is highly stereoselective, forming R- and S-epoxypropane from propylene epoxidation in a ratio of about 95:5 (mol/mol). As shown in Table 1, component I also had a degree of stereoselectivity, catalyzing the reaction of CoM with R-epoxypropane at a rate approximately twice of that with S-epoxypropane. This stereospecificity is perhaps a reflection of the higher abundance of R-epoxypropane formed in vivo from propylene oxidation by the alkene monooxygenase. As shown in Table 1, component III was highly selective for dehydrogenation of 2-R-hydroxypropyl–CoM, whereas component IV was highly selective for 2-S-hydroxypropyl–CoM. Chirality is no longer a concern after the dehydrogenation step, as the oxidation of both hydroxythioether enantiomers forms a common product. The component II-catalyzed step was the slowest of the three and is probably rate-limiting in vivo. In this context, it should be noted that component II is the most highly expressed of the four proteins, accounting for up to 20% of soluble proteins in propylene-grown cells (13, 19).
Table 1.
Reactions and enzyme activities of epoxide carboxylase components
| Reaction | Component | Specific activity, unit* |
|---|---|---|
| Racemic-epoxypropane + CoM → 2-hydroxypropyl–CoM | I | 11.8 ± 0.9 |
| R-epoxypropane + CoM → 2-R-hydroxypropyl–CoM | I | 15.0 ± 0.6 |
| S-epoxypropane + CoM → 2-S-hydroxypropyl–CoM | I | 8.0 ± 0.5 |
| 2-R-hydroxypropyl–CoM + NAD+ → 2-ketopropyl–CoM + NADH + H+ | III | 45.7 ± 3.3 |
| 2-S-hydroxypropyl–CoM + NAD+ → 2-ketopropyl–CoM + NADH + H+ | III | 0.5 ± .04 |
| 2-S-hydroxypropyl–CoM + NAD+ → 2-ketopropyl–CoM + NADH + H+ | IV | 38.1 ± 1.0 |
| 2-R-hydroxypropyl–CoM + NAD+ → 2-ketopropyl–CoM + NADH + H+ | IV | 0.2 ± .04 |
| 2-Ketopropyl–CoM + CO2 + NADPH + H+ → acetoacetate + NADP+ | II | 2.5 ± 0.2 |
Micromoles of substrate consumed or product formed⋅min−1⋅mg−1 protein.
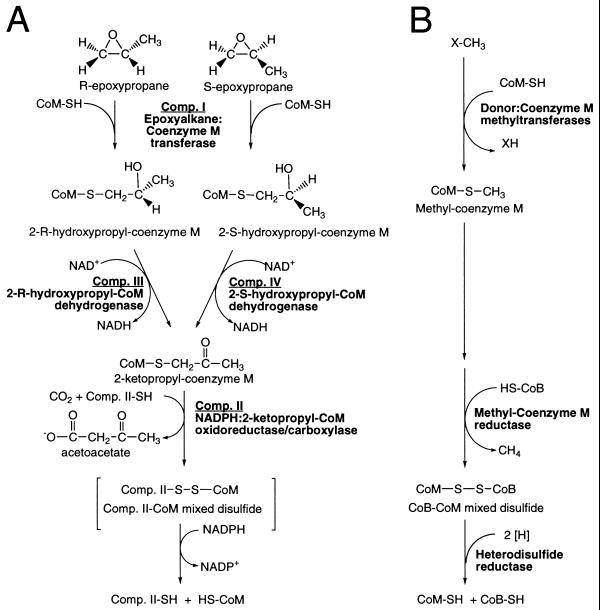
The studies described above have allowed the assignment of specific reactions for the four epoxide carboxylase proteins, which had been referred to collectively as a “multiprotein enzyme system.” It should be noted that there were no observed synergistic effects of combining components, indicating that each of the enzymes functions independently of the others. The four components can now be referenced more descriptively according to the reactions they catalyze: component I is an epoxyalkane:CoM transferase; components III and IV are stereoselective 2-hydroxypropyl–CoM dehydrogenases, and component II is a dual function NADPH:2-ketopropyl–CoM oxidoreductase/carboxylase. As shown in Fig. 5A, epoxide carboxylation is more properly viewed as a metabolic pathway involving the concerted action of four individual enzymes rather than as a multiprotein-catalyzed event. Although CoM is a substrate and forms covalent complexes with intermediates therein, it is regenerated in its original form at the conclusion of the linear pathway. The recycling of CoM explains why, in previous studies, low rates of epoxide carboxylation could be measured in the absence of exogenously added CoM. Presumably, the previous assays relied on the recycling of the limiting amount of CoM that had copurified with component I. The stimulation of the overall rate of epoxide carboxylation by orders of magnitude on addition of exogenous CoM to the four epoxide carboxylase components (Fig. 1) shows that CoM was highly rate-limiting in the previous in vitro studies (10, 11).
Figure 5.
Roles of CoM in epoxide carboxylation (A) and methanogenesis (B).
Significance of Our Findings.
Of broader interest and significance than the elucidation of the reactions of aliphatic epoxide carboxylation is the surprising discovery that CoM is the central cofactor of this process. Previously, CoM, the smallest known organic cofactor, was believed to be restricted to the methanogenic Archaea, where it plays a central role in the reductive formation of methane (14–17). Balch and Wolfe (20) screened a variety of microbes, plants, and animal tissues for CoM by using a sensitive bioassay but were unable to find the cofactor in any nonmethanogenic organism. Our discovery of CoM as a cofactor of a very specialized hydrocarbon oxidation pathway suggests that it may be expressed and used by bacteria only under specific conditions. It is intriguing that this role for CoM also involves a pathway of gaseous hydrocarbon metabolism—perhaps the pathway for oxidation of short-chain aliphatic hydrocarbons diverged at some point from archaeal C1 metabolic pathway(s). Of possible relevance to this role, Lidstrom, Thauer, and coworkers (21, 22) recently established the presence of the methanogenic coenzyme tetrahydromethanopterin (a tetrahydrofolate analog) and tetrahydromethanopterin-dependent enzymes in a methylotrophic bacterium. These findings provide exciting unifying links between the archaeal and bacterial domains and reductive and oxidative microbial C1 metabolism. Interestingly, Xanthobacter Py2 is a facultative methylotroph (23); methylotrophy may be an evolutionarily significant feature, because the only other microbes in which archaeal cofactors have been found to date are methylotrophs.
Fig. 5 A and B highlights some interesting similarities in the processes of epoxide carboxylation and methanogenesis. In both cases, the initial group transfer involves S-alkylation of CoM to form a thioether intermediate. Although a variety of compounds are capable of serving as methyl group donors in methanogenesis, in each instance, CoM must be activated as a nucleophile by deprotonation of the sulfhydryl to generate the thiolate anion. Zinc has been proposed to play an integral role in the activation of diverse thiol groups as nucleophiles in a variety of enzymes (24), including methanogenic methyl transferases (25, 26). By analogy, the zinc in epoxide carboxylase component I probably functions in the activation of CoM as well, in this instance for attack on the electrophilic epoxide substrate.
In both methanogenesis and epoxide carboxylation, a reductive dealkylation of a CoM thioether occurs, in the former system to generate methane, in the latter to form an enolate that undergoes carboxylation (Fig. 5). However, as highlighted in Fig. 5, there are notable differences in the proposed mechanisms of the respective dealkylations. Reductive methane formation requires two additional uniquely methanogenic cofactors: coenzyme B (7-mercaptoheptanoylthreonine phosphate), which forms a mixed disulfide with CoM (27–29), and coenzyme F430, a nickel tetrapyrrole to which the methyl group is transferred and reduced to methane (30). We have no evidence that either of these cofactors is involved in epoxide carboxylation. Rather, we believe at present that the reductive dealkylation of 2-ketopropyl–CoM is promoted by formation of a mixed disulfide between a cysteine residue on component II, a member of the NADPH:disulfide oxidoreductase family of enzymes, and CoM (Fig. 5A). The mixed disulfide is then reduced by using NADPH as the reductant (Fig. 5A).
It is intriguing that CoM was selected as the cofactor for epoxide carboxylation rather than a more conventional thiol such as glutathione, lipoic acid, cysteine, or homocysteine. It would be interesting to determine whether phylogenetically distinct aliphatic alkene-using bacteria (e.g., R. rhodochrous, an actinomycete) use CoM in the pathway of aliphatic epoxide metabolism as well. The identification of this cofactor in diverse nonmethylotrophic bacteria would unify the archaeal and bacterial domains further and showcase the newfound versatility of CoM.
Acknowledgments
This work was supported by National Institutes of Health Grant GM51805.
ABBREVIATION
- CoM
coenzyme M (2-mercaptoethanesulfonic acid)
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Sawada S, Totsuka T. Atmos Environ. 1986;20:821–832. [Google Scholar]
- 2.Cleveland C C, Yavitt J B. Appl Environ Microbiol. 1998;57:228–235. doi: 10.1128/aem.64.1.172-177.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartmans S, de Bont J A M, Harder W. FEMS Microbiol Rev. 1989;63:235–264. doi: 10.1016/0168-6445(89)90034-x. [DOI] [PubMed] [Google Scholar]
- 4.Small F J, Ensign S A. J Bacteriol. 1995;177:6170–6175. doi: 10.1128/jb.177.21.6170-6175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen J R, Ensign S A. J Bacteriol. 1998;180:2072–2078. doi: 10.1128/jb.180.8.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Small F J, Ensign S A. J Biol Chem. 1997;272:24913–24920. doi: 10.1074/jbc.272.40.24913. [DOI] [PubMed] [Google Scholar]
- 7.Miura A, Dalton H. Biosci Biotechnol Biochem. 1995;59:853–859. [Google Scholar]
- 8.Allen J R, Ensign S A. J Bacteriol. 1996;178:1469–1472. doi: 10.1128/jb.178.5.1469-1472.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ensign S A, Small F J, Allen J R, Sluis M K. Arch Microbiol. 1998;169:179–187. doi: 10.1007/s002030050558. [DOI] [PubMed] [Google Scholar]
- 10.Allen J R, Ensign S A. J Biol Chem. 1997;272:32121–32128. doi: 10.1074/jbc.272.51.32121. [DOI] [PubMed] [Google Scholar]
- 11.Allen J R, Ensign S A. Biochemistry. 1999;38:247–256. doi: 10.1021/bi982114h. [DOI] [PubMed] [Google Scholar]
- 12.Swaving J, Debont J A M, Westphal A, Dekok A. J Bacteriol. 1996;178:6644–6646. doi: 10.1128/jb.178.22.6644-6646.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen J R, Ensign S A. J Bacteriol. 1997;179:3110–3115. doi: 10.1128/jb.179.10.3110-3115.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfe R S. Annu Rev Microbiol. 1991;45:1–35. doi: 10.1146/annurev.mi.45.100191.000245. [DOI] [PubMed] [Google Scholar]
- 15.Thauer R K. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 16.DiMarco A A, Bobik T A, Wolfe R S. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- 17.Ferry J G. Methanogenesis: Ecology, Physiology, Biochemistry and Genetics. New York: Chapman & Hall; 1993. [Google Scholar]
- 18.Taylor C D, Wolfe R S. J Biol Chem. 1974;249:4879–4885. [PubMed] [Google Scholar]
- 19.Ensign S A. Appl Environ Microbiol. 1996;62:61–66. doi: 10.1128/aem.62.1.61-66.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balch W E, Wolfe R S. J Bacteriol. 1979;137:256–263. doi: 10.1128/jb.137.1.256-263.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chistoserdova L, Vorholt J A, Thauer R K, Lidstrom M E. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 22.Vorholt J A, Chistoserdova L, Lidstrom M E, Thauer R K. J Bacteriol. 1998;180:5351–5356. doi: 10.1128/jb.180.20.5351-5356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Ginkel C G, de Bont J A M. Arch Microbiol. 1986;145:403–407. [Google Scholar]
- 24.Matthews R G, Goulding C W. Curr Opin Chem Biol. 1997;1:332–339. doi: 10.1016/s1367-5931(97)80070-1. [DOI] [PubMed] [Google Scholar]
- 25.LeClerc G M, Grahame D A. J Biol Chem. 1996;271:18725–18731. doi: 10.1074/jbc.271.31.18725. [DOI] [PubMed] [Google Scholar]
- 26.Sauer K, Thauer R K. Eur J Biochem. 1997;249:280–285. doi: 10.1111/j.1432-1033.1997.t01-1-00280.x. [DOI] [PubMed] [Google Scholar]
- 27.Gunsalus R P, Wolfe R S. J Biol Chem. 1980;255:1891–1895. [PubMed] [Google Scholar]
- 28.Noll K M, Rinehart K L, Jr, Tanner R S, Wolfe R S. Proc Natl Acad Sci USA. 1986;83:4238–4242. doi: 10.1073/pnas.83.12.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellermann J, Hedderich R, Böcher R, Thauer R K. Eur J Biochem. 1988;172:669–677. doi: 10.1111/j.1432-1033.1988.tb13941.x. [DOI] [PubMed] [Google Scholar]
- 30.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer R K. Science. 1997;278:1457–1462. doi: 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]