Abstract
In fission yeast, the Hsk1 protein kinase is essential for the initiation of DNA replication. We have shown previously that Hsk1 forms a heterodimeric complex with the regulatory subunit, Dfp1. In this report we describe the further characterization of Dfp1. Reconstitution experiments with purified proteins indicate that Dfp1 is necessary and sufficient to activate Hsk1 phosphorylation of exogenous substrates, such as the Schizosaccharomyces pombe minichromosome maintenance protein Cdc19. The dfp1+ gene is essential for viability of S. pombe, and depletion of the Dfp1 protein significantly delays the onset of S phase. Dfp1 is a phosphoprotein in vivo and becomes hyperphosphorylated when cells are blocked in S phase by treatment with the DNA synthesis inhibitor hydroxyurea. Hyperphosphorylation in S phase depends on the checkpoint kinase Cds1. The abundance of Dfp1 varies during progression through the cell cycle. The protein is absent when cells are arrested in G1 phase. When cells are released into the cell cycle, Dfp1 appears suddenly at the G1/S transition, coincident with the initiation of DNA replication. The absence of Dfp1 before S phase is due largely, but not exclusively, to posttranscriptional regulation. We propose that cell cycle-regulated activation of Dfp1 expression at the G1/S transition results in activation of the Hsk1 protein kinase, which, in turn, leads to the initiation of DNA replication.
The Cdc7 family of protein kinases plays a central role in the regulation of DNA replication in eukaryotic cells. The prototypical member of the family, Cdc7 kinase, first was identified in Saccharomyces cerevisiae (1–3). Homologues of Cdc7 kinase since have been identified in Schizosaccharomyces pombe (Hsk1), Xenopus laevis (xeCdc7), mouse (muCdc7), and human (huCdc7), indicating that it is conserved in all eukaryotes (4–8). Genetic studies in Saccharomyces cerevisiae indicate that Cdc7 kinase is essential for the activation of origins of replication during S phase (9–11). Protein kinase activity is regulated during the cell cycle, reaching a maximum in S phase (12, 13). A number of lines of evidence suggest that Cdc7 kinase activity in S phase depends on interaction with a regulatory protein, Dbf4, which is also essential for initiation of DNA replication (12, 14, 15). Interestingly, Dbf4 appears to associate with origins of replication in vivo, suggesting that an additional role of the protein may be to localize Cdc7 kinase to its site of action (16). The critical substrates that are phosphorylated by Cdc7 protein kinase are not known with certainty. A number of experimental results have implicated the minichromosome maintenance (MCM) proteins as Cdc7 targets (5, 17, 18), but the biological relevance of MCM protein phosphorylation remains to be demonstrated.
The Hsk1 protein kinase is the fission yeast member of the Cdc7 kinase family (4). Overexpression of dominant-negative hsk1 alleles delays entry into S phase (19), as does deletion of hsk1 (4), indicating an important role for this protein kinase in regulating the initiation of DNA replication. We recently have described the purification of Hsk1 protein kinase to apparent homogeneity. The purified kinase contains two separable protein species: (i) a monomer of Hsk1 and (ii) a heterodimer of Hsk1 and Dfp1, the fission yeast homologue of Saccharomyces cerevisiae Dbf4. Monomeric Hsk1 has intrinsic autokinase activity, indicating that its kinase active site is functional in the absence of regulatory molecules. The Hsk1⋅Dfp1 heterodimer exhibits a similar level of autokinase activity, but is also capable of phosphorylating exogenous substrates. In particular, we have shown that homogeneous Hsk1⋅Dfp1 heterodimer phosphorylates the Cdc19 (Mcm2) subunit of the heteromeric MCM complex purified from fission yeast. On the basis of these observations we have proposed that the role of the regulatory subunit Dfp1 is to activate Hsk1 phosphorylation of specific substrates, such as Cdc19 (Mcm2), that are required for initiation of DNA replication at the G1/S transition. In this study we describe the further characterization of fission yeast Dfp1.
MATERIALS AND METHODS
Strains and Plasmids.
Basic fission yeast genetic and molecular biology techniques were used (20). Cells were grown in Edinburgh minimal medium plus required supplements. The genotypes of the strains used in this study are listed in Table 1. The dfp1+ gene was fused at the 3′ end to sequences encoding six histidines and three hemagglutinin (3HA) epitopes (GBY397) or six histidines and three myc epitopes (GBY395) as described previously for hsk1+ and cdc19+ (19). In the diploid strain GBY405, the entire ORF of one copy of dfp1+ was replaced with the ura4+ gene. The gene replacement was confirmed by Southern hybridization analysis. A haploid strain bearing the dfp1∷ura4+ replacement can be rescued by a plasmid-borne copy of dfp1+ (data not shown). Spore germination was performed as described (21, 22).
Table 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| GBY395 | h−leu1-32 ura4-D18 hsk1∷hsk1-6his-3HA[ura4+] dfp1∷dfp1-6his-3myc[leu1+] |
| GBY397 | h+leu1-32 ura4-D18 ade6-M210 dfp1∷dfp1-6his-3HA[ura4+] |
| GBY398 | cdc25-22 leu1-32 ura4-D18 ade6-M210 dfp1∷dfp1-6his-3HA[ura4+] |
| GBY401 | cdc10-129 ura4-D18 ade6-M210 dfp1∷dfp1-6his-3HA[ura4+] |
| GBY405 | h+/h−leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M210/ade6-M216 dfp1+/dfp1∷ura4 |
| GBY408 | cdc25-22 leu1-32 ura4-D18 hsk1∷hsk1-6his-3HA[ura4+] dfp1∷dfp1-6his-3myc[leu1+] |
| GBY410 | h+leu1-32 ura4-D18 ade6-M210 cdc10-129 pJKars41X-dfp1-6his-3myc |
| GBY411 | h−leu1-32 ura4-D18 cdc25-22 pJKars41X-dfp1-6his-3myc |
| GBY412 | h−leu1-32 ura4-D18 ade6-M210 pJKars41X-dfp1-6his-3myc |
| GBY429 | leu1-32 ura4-D18 ade6-M210 dfp1∷ura4+ pJKars81X-dfp1-6his-3myc |
| GBY434 | leu1-32 ura4-D18 ade6-M210 dfp1∷dfp1-6his-3HA[ura4+]cds1+ |
| GBY435 | leu1-32 ura4-D18 ade6-M210 dfp1∷dfp1-6his-3HA[ura4+]cds1∷ura4+ |
The plasmid pREP81X-dfp1-6his-3 myc was constructed by inserting the dfp1+ ORF fused to sequences encoding six histidines and three myc epitope tags into the multiple-cloning site of pREP81X. The plasmid pJKars41X-dfp1-6his-3 myc was constructed by inserting the attenuated, medium-strength nmt1 promoter followed by the dfp1+ ORF fused to the six histidine-triple myc tag into the multiple-cloning site of pJKars (19).
Cells were arrested in S phase by growth in 25 mM hydroxyurea for 4 hr at 30°C. G1 and G2 arrests were the result of growing cdc10–129 or cdc25–22 strains for 4 hr at 35.5°C. For mitotic arrest, cells were grown in the presence of 20 μg/ml benomyl at 30°C for 3 hr in minimal medium. Cells were synchronized by arresting a cdc25–22 culture in G2 and then rapidly cooling to 25°C, as described (22). Nitrogen starvation was carried out as described (20) for 4 hr. Flow cytometry was performed as described (19).
Expression of GST-Dfp1 Fusions in Escherichia coli.
The ORF encoding Dfp1 was cloned into pGEX4T-1 (Pharmacia). Expression was induced by the addition of isopropyl β-d-thiogalactoside, extracts were made, and the fusion proteins were purified by using glutathione-Sepharose essentially as directed by the manufacturer.
Purification of Monomeric Hsk1.
Hsk1 was purified from fission yeast as described previously (19). Hsk1 monomer was separated from Hsk1/Dfp1 heterodimer by using gel filtration, and kinase activity was assayed by using a GST-Cdc19 fusion protein as the substrate, as described (19).
Immunoblot Analysis.
Cells (2.5 × 108) were fixed in 10% trichloroacetic acid, harvested, washed once with 1 ml of 1 M Hepes, pH 7.5, and stored frozen at −80°C. Extracts were prepared by vortexing with glass beads and were fractionated on 10% SDS-polyacrylamide gels. After transfer to nitrocellulose membranes, the proteins of interest were detected with mAbs 16B12 (Babco; to detect HA-tagged proteins), 9E10 (Boehringer Mannheim; to detect myc-tagged proteins), or TAT-1 (gift of Keith Gull, University of Manchester; to detect tubulin). Immunoblots were developed with the SuperSignal chemiluminescence substrate (Pierce).
Northern Hybridization.
Total RNA was prepared by using Totally RNA reagent (Ambion). Five micrograms of RNA was fractionated on formaldehyde/agarose gels and transferred to nylon membranes (Micron Separations). Hybridizations using the dfp1+, cdc18+, or leu1+ ORF as probes were performed essentially as described (23).
Phosphatase Treatment.
S phase extract was prepared from 4 g of GBY395 cells after growth in 25 mM hydroxyurea for 4 hr at 30°C. Hsk1–6his-3HA was immunoprecipitated from 1 mg of protein, as described previously (19). Immunoprecipitates were incubated with 40 units of λ phosphatase (New England Biolabs) for 15 min at 30°C. Control reactions lacked enzyme (mock) or included the phosphatase inhibitor vanadate at 100 μM. Reactions were stopped by the addition of 0.5 volume of SDS/polyacrylamide gel loading buffer and fractionated by SDS/PAGE. After transfer to nitrocellulose membranes, Dfp1–6his-3myc was detected with 9E10 antibody.
RESULTS
Dfp1 Activates the Hsk1 Kinase.
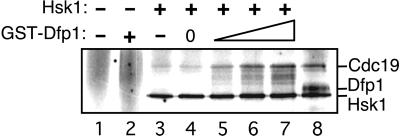
We previously have isolated monomeric Hsk1 and Hsk1⋅Dfp1 heterodimers and examined their properties (19). We found that monomeric Hsk1 has autokinase activity, whereas Hsk1⋅Dfp1 is also active in phosphorylation of exogenous substrates. On the basis of these properties, we proposed that Dfp1 activates Hsk1 phosphorylation of exogenous substrates. To test this possibility directly we reconstituted Hsk1 activation in vitro. Dfp1 was expressed in bacteria as a GST fusion protein and purified by affinity chromatography. Monomeric Hsk1 was purified from S. pombe by a combination of affinity and conventional chromatography. Increasing amounts of Dfp1p were added to Hsk1, and protein kinase activity was assayed by using Cdc19 protein (S. pombe Mcm2) as the substrate (Fig. 1). As reported previously, monomeric Hsk1 has autokinase activity, but is a poor Cdc19 kinase. Addition of Dfp1 resulted in increasing phosphorylation of Cdc19, up to the level of phosphorylation observed with purified heterodimeric Hsk1⋅Dfp1. Dfp1 increased activity of Hsk1 on Cdc19 about 7-fold, but had little effect (less than 2-fold) on Hsk1 autokinase activity. GST alone did not activate Hsk1 kinase, and GST-Dfp1 alone did not phosphorylate Cdc19, confirming that the Dfp1 moiety of the fusion protein is responsible for the observed effects on Hsk1 kinase activity. We conclude that Dfp1 is necessary and sufficient to activate Hsk1 kinase phosphorylation of exogenous substrates in vitro.
Figure 1.
Activation of Hsk1 kinase by recombinant Dfp1. Increasing amounts (0, 10, 25, or 50 ng) of GST-Dfp1 were added to purified monomeric Hsk1 (≈5 ng, lanes 4–7). Proteins were incubated under kinase assay conditions. All reactions contained 100 ng of GST-Cdc19 substrate. Control reactions contained no added protein (lane 1), GST-Dfp1 (50 ng) alone (lane 2), GST (100 ng) with Hsk1 monomer (lane 3), or heterodimeric Hsk1⋅Dfp1 (lane 8).
dfp1+ Is Essential for Viability and Regulates Progression into S Phase.
To investigate the consequences of eliminating Dfp1 expression in vivo we replaced one copy of the dfp1+ ORF with the ura4+ gene in a diploid strain. Thirteen tetrads derived by sporulation of the dfp1+/dfp1∷ura4+ diploid were dissected (Fig. 2A). The majority (11/13) of tetrads had two viable and two inviable spores, and the remaining two tetrads had only one viable spore. The viable spores were invariably uracil auxotrophs, demonstrating that dfp1+ is an essential gene. Microscopic examination revealed that the dfp1∷ura4+ spores (Fig. 2B Upper Left and Lower Right) divided once before arresting, a phenotype similar to that observed in hsk1∷ura4+ germinating spores (4).
Figure 2.
Deletion of dfp1+ delays entry into S phase. (A) Tetrad analysis of the diploid dfp1+/dfp1∷ura4+ disruption strain GBY405. (B) Representative microcolonies from one tetrad from GBY405. Microcolonies (Upper Right and Lower Left) were uracil auxotrophs and, therefore, dfp1+. Germinated spores (Upper Left and Lower Right) did not give rise to colonies. (C) DNA contents from flow cytometry of germinating spores of GBY405. Spores were germinated in medium containing uracil (+ura) to permit germination of all spores or in medium lacking uracil (−ura) to allow germination of spores bearing the dfp1 disruption only. The positions of 1C and 2C DNA contents are indicated. (D) Depletion of Dfp1. Expression of dfp1 in the strain GBY429 was repressed by the addition of thiamine to 5 μg/ml at t = 0 hr. DNA content was measured by flow cytometry every hour after repression.
The phenotype of germinating dfp1∷ura4+ spores was examined further by using a selective spore-germination protocol. Spores from the dfp1+/dfp1∷ura4+ diploid were incubated in minimal medium in the presence or absence of uracil, and the cellular DNA content was monitored by flow cytometry (Fig. 2C). Incubation of the spores in the presence of uracil allowed the germination and growth of spores containing either wild-type or dfp1∷ura4+ alleles. Under these conditions the cells initially exhibited a 1C DNA content (4 hr), but entered S phase between 6 and 7 hr as evidenced by the appearance of cells with a 2C DNA content. In contrast, when the same spores were germinated in medium lacking uracil, which allows germination of only the dfp1∷ura4+ uracil prototrophs, the cells did not enter S phase until 9 hr after germination. Approximately 50% of the dfp1∷ura4+ cells reached a 2C DNA content by 12 hr, although as indicated above, these cells are unable to progress past the two-cell stage. Thus, dfp1∷ura4+ spores are defective in initiation of DNA replication and are delayed in S phase entry for at least 2 hr relative to wild-type spores. The observed delay in S phase entry clearly implicates dfp1+ in the regulation of initiation of DNA replication. The ability of cells lacking a functional dfp1+ gene to synthesize DNA after the delay may be due to the presence of a low level of residual Dfp1 protein in the dfp1∷ura4+ spores.
To further explore the phenotype of cells lacking Dfp1 we constructed a strain (GBY429) whose only copy of dfp1+ is under the control of an attenuated thiamine-repressible nmt1 promoter. Cells were grown to midexponential phase, and thiamine was added to repress dfp1+ expression (Fig. 2D). We observed the accumulation of cells with a 1C DNA content beginning at 4 hr after thiamine addition. By 6 hr, 23% of cells exhibited a DNA content less than 2C, indicating that depletion of Dfp1 delays the initiation of DNA replication. Thus, we have confirmed by two independent methods that Dfp1 plays an important role in regulating initiation.
Dfp1 Abundance Is Cell Cycle-Regulated.
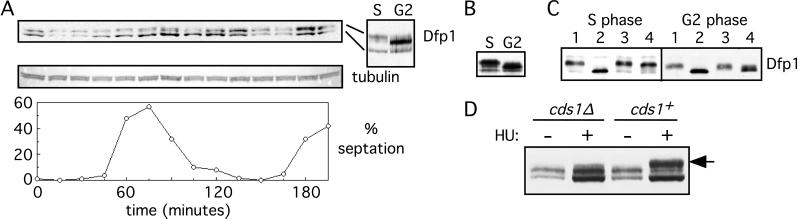
Because Dfp1 is necessary and sufficient for Hsk1 kinase activation in vitro and is required for efficient initiation of DNA replication, regulation of the expression of Dfp1 might determine the level of Hsk1 kinase activity during the cell cycle. To explore this possibility we analyzed Dfp1 expression in a synchronized population of S. pombe strain GBY398 (Fig. 3A). In this strain the chromosomal dfp1+ gene is fused to sequences encoding six histidines and three hemagglutinin epitopes. A culture of GBY398 cells was blocked in G2 phase by inactivation of Cdc25, released into the cell cycle by a temperature shift, and sampled every 15 min. Synchrony of the culture was assessed by measuring the percentage of septated cells. We found that Dfp1 is present at the cdc25 block point in G2 and then decreases after release from the G2 arrest (15 and 30 min). Immediately before the increase in septation (45 min), the level of Dfp1 increases, reaching a maximum at 90 min. Dfp1 levels remain high through the 135-min sample and then decrease before the second round of septation (150 and 165 min). Because the peak of septation corresponds to S phase in fission yeast, this suggests that Dfp1 levels decrease after release from the G2 block, likely in M and G1 phases, and then increase immediately before S phase. Dfp1 expression remains at high levels through S phase and G2 and then decreases before the next S phase. During the first cell cycle of the synchronized population we observed reproducible overall change in abundance of Dfp1 of 4-fold from G1 to G2 phase. In contrast to Dfp1, the levels of Hsk1 did not vary during cell cycle progression (data not shown).
Figure 3.
Expression and phosphorylation of Dfp1 during cell cycle progression. (A) A culture of the strain GBY398 was blocked in G2 and then released synchronously into the cell cycle. The culture was sampled every 15 min, and extracts were prepared. Dfp1 was detected by immunoblot analysis. The blot was reprobed for tubulin as a loading control. Synchrony was assessed by measuring the percentage of cells with a septum (% septation). The 60- (S) and 135-min (G2) samples are also shown side by side for comparison of the different Dfp1 isoforms. (B) Extracts were prepared from cells blocked in S phase (GBY397) or in G2 phase (GBY398) by using denaturing conditions. Proteins were immunoblotted, probing for Dfp1. (C) Native extracts were prepared from cells blocked in S or G2 phase. Hsk1 and associated Dfp1 were immunoprecipitated, and the immunoprecipitates were incubated with λ phosphatase (lanes 2), without λ phosphatase (lanes 3), or with λ phosphatase plus the inhibitor vanadate (lanes 4). The untreated controls are in lanes 1. Reaction products were subjected to immunoblot analysis, probing for Dfp1. Note that the fastest-migrating Dfp1 isoform seen in A and B is extracted inefficiently under native conditions. (D) The phosphorylation state of Dfp1 in S phase cells was analyzed in cds1+ or cds1Δ strains. Samples were taken from cultures arrested in S phase with hydroxyurea (+HU) or from asynchronous cultures (−HU). The position of the S phase Dfp1 phosphoisomer is indicated by the arrow.
Dfp1 Is a Phosphoprotein in Vivo.
We also observed the presence of three isoforms of Dfp1 in the cell cycle experiment (Fig. 3A). The abundance of the slower-mobility form reaches a maximum immediately before S phase (at 60 min, also shown in Fig. 3A lane S) and disappears when cells exit S phase into G2. In contrast, the intermediate-mobility form reaches maximum levels in G2 phase (135 min, also shown in Fig. 3A, lane G2). These differences in the mobility of Dfp1 are also apparent when extracts from cells blocked in S phase or G2 phase are compared (Fig. 3B). Again, Dfp1 has a slower mobility in S phase than in G2 phase. To identify the modification responsible for these changes in Dfp1 mobility, we immunoprecipitated Dfp1 from extracts of cells blocked in S phase or G2 phase. The immunoprecipitates were treated with λ phosphatase (λPPase), and the effect on Dfp1 mobility in SDS/PAGE was examined (Fig. 3C). Treatment of Dfp1 with λPPase changed the mobility of both the S phase and G2 phase forms of Dfp1 to a faster-migrating form. Mock treatment or phosphatase treatment in the presence of the specific inhibitor vanadate did not cause this change in mobility, indicating that Dfp1 is phosphorylated in vivo in both S phase and G2 phase, with the S phase form being hyperphosphorylated relative to the G2 form.
Hyperphosphorylation of Dfp1 in S Phase Depends on the Cds1 Kinase.
In fission yeast the Cds1 protein kinase is activated specifically during S phase and is required to maintain viability when DNA replication is blocked (24). We tested whether the phosphorylation of Dfp1 in cells blocked in S phase depends on the Cds1 protein kinase. For this purpose we used the strain GBY435 in which the chromosomal dfp1+ gene is tagged with hexahistidine-3HA and the cds1+ gene is deleted. GBY435 and an otherwise isogenic cds1+ strain were incubated in hydroxyurea for 4 hr, and extracts were prepared. After fractionation by SDS/PAGE proteins were immunoblotted to detect Dfp1 (Fig. 3D). The S phase-specific phosphoisomer, indicated by the arrow, was absent in the cds1 deletion strain. Cds1 is not required for DNA replication checkpoint-dependent arrest in hydroxyurea (24), so the absence of the hyperphosphorylated Dfp1 is not due to differences in cell cycle position between the cds1Δ and cds1+ strains. We conclude that hyperphosphorylation of Dfp1 induced by a block to DNA replication depends on the Cds1 protein kinase.
Dfp1 Expression Is Regulated by Transcriptional and Posttranscriptional Mechanisms.
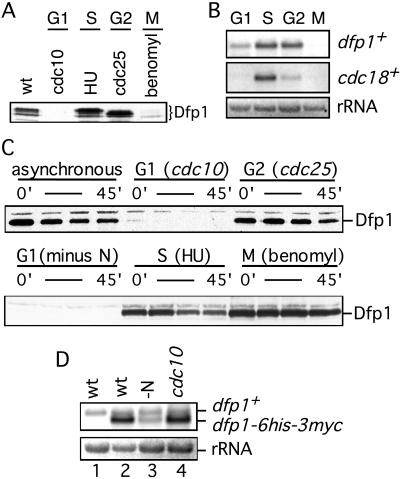
In synchronous culture the expression of Dfp1 protein is reduced during G1 phase. To confirm this observation, we blocked cell cycle progression at different points and examined Dfp1 expression by using immunoblot analysis (Fig. 4A). When cells were blocked in G1 by shifting a cdc10-129 strain to the nonpermissive temperature for 4 hr, Dfp1 protein was completely absent. Cells blocked in S phase with hydroxyurea or in G2 by shifting a cdc25-22 strain to the nonpermissive temperature contained at least 10-fold more Dfp1 than the G1 cells. Interestingly, we found that the level of Dfp1 is also reduced in mitotic cells arrested with the microtubule-depolymerizing drug benomyl.
Figure 4.
Dfp1 expression is down-regulated during G1 phase. (A) Dfp1 levels in cells blocked in G1 (GBY401), S (GBY397), G2 (GBY398), or M (GBY397) phases were compared with levels in asynchronous culture (wt, GBY397) by using immunoblot analysis. The Dfp1 species are indicated. Equal loading of all lanes was confirmed by reprobing immunoblots with anti-tubulin antibody (not shown). (B) Total RNA was prepared from cells blocked as in A and fractionated on formaldehyde-agarose gels. A Northern blot was probed for dfp1+ mRNA and cdc18+ mRNA. The ethidium bromide-stained rRNA is shown as a control for equal loading. (C) Strains (GBY410, 411, 412) expressing dfp1–6his-3myc from the repressible nmt1 promoter were blocked at the indicated cell cycle positions. Expression of dfp1–6his-3myc was repressed by the addition of thiaminee, and Dfp1 protein levels at 0, 15, 30, and 45 min after repression was analyzed by immunoblotting. Equal loading of all lanes was confirmed by reprobing immunoblots with anti-tubulin antibody (not shown). (D) RNA was prepared from a logarithmically growing culture of wild-type cells (wt, lane 1), from GBY410 in logarithmically growing culture (wt, lane 2), from GBY410 cultured in the absence of nitrogen for 4 hr (-N, lane 3), or from GBY411 cultured at 36°C for 4 hr (cdc10, lane 4). A Northern blot of this RNA was probed with the dfp1+ ORF. The positions of the dfp1+ and dfp1–6his-3myc mRNAs are indicated. The ethidium bromide-stained rRNA is shown as a control for equal loading.
To investigate the mechanism responsible for reduced expression of Dfp1 in G1 and M phases, we examined dfp1+ mRNA levels in cells blocked at specific points in the cell cycle (Fig. 4B). S phase and G2 phase cells contained similar levels of a 2.3-kb dfp1+ mRNA. G1 cells contained readily detectable dfp1+ mRNA, but the level was about 50% of that in S phase cells (average of two experiments). Thus reduced expression of dfp1+ mRNA, although contributory, cannot completely account for the more than 10-fold reduction in Dfp1 protein. Because in these experiments the block in G1 phase was achieved by incubating cdc10–129 cells at the nonpermissive temperature, it is evident that transcription of dfp1+ is not entirely dependent on cdc10+. In contrast, mRNA from cdc18+, a known Cdc10-regulated gene, was virtually absent in the same G1 cells that contained significant amounts of dfp1+ transcript. Finally, we observed that dfp1+ mRNA was absent in cells arrested in M phase (Fig. 4B). Thus, unlike cells blocked in G1 phase, the observed reduction in Dfp1 levels in cells blocked in M phase can be accounted for by a corresponding reduction in dfp1+ mRNA levels, suggesting regulation at the transcriptional level. To ensure that the relative levels of dfp1+ mRNA that we observed in G1, S, G2, and M phase were not influenced by the presence of the C-terminal epitope tag, we repeated all of our measurements with wild-type dfp1+ strains. We obtained results identical to those shown in Fig. 4B (data not shown).
To confirm that Dfp1 expression in G1 phase is regulated largely by a posttranscriptional mechanism, the dfp1 ORF with a C-terminal epitope tag was placed under the control of the heterologous nmt1+ promoter. In the absence of thiamine the nmt1+ promoter is active, and steady-state mRNA levels are largely independent of cell cycle position (see below). Cells blocked in S or G2 phase contained abundant Dfp1 (Fig. 4C). When thiamine was added to repress dfp1 transcription, the Dfp1 protein decayed with a half-life of 30–45 min. The dfp1 mRNA disappears rapidly under these conditions (data not shown), indicating that Dfp1 is a stable protein in S and G2 phases. In contrast, cells blocked in the G1 phase of the cell cycle, either by inactivation of Cdc10 or by nitrogen starvation, contained very little Dfp1 protein even before nmt1 promoter repression. The half-life of Dfp1 in G1 phase could not be determined because of the extremely low levels of the protein. These data demonstrate that the dfp1+ promoter is not required for regulated expression of Dfp1 protein at the G1/S boundary and that expression of dfp1 from a constitutive promoter is not sufficient to restore Dfp1 protein expression in G1. We conclude that Dfp1 expression is regulated posttranscriptionally in G1 phase.
We confirmed by Northern blotting that dfp1 mRNA is expressed from the nmt1+ promoter in G1 cells (Fig. 4D). The wild-type endogenous dfp1+ transcript is about 2.5 kb in size and was detectable in an asynchronous cell population or in cells blocked in G1 by inactivation of Cdc10 or nitrogen starvation, as expected. The dfp1 transcript expressed under control of the nmt1+ promoter was 2.3 kb in size because of reduction in the lengths of the 5′ and 3′ untranslated regions. Steady-state levels of this transcript in G1 cells were equal to or greater than the levels of dfp1+ mRNA expressed from the endogenous promoter. In the case of cells blocked in G1 phase by inactivation of Cdc10, the amount of dfp1 mRNA from the nmt1+ promoter was much greater than that of the endogenous wild-type mRNA despite the complete absence of epitope-tagged Dfp1 protein in the cells (Fig. 4C). These data strongly support our conclusion that Dfp1 is subject to posttranscriptional regulation as cells move from G1 into S phase. The mechanism of regulation is unknown at this point, but could affect either the rate of synthesis or degradation of Dfp1. We suspect the latter, because regulation of protein stability appears to be a common mechanism for controlling cell cycle transitions (25, 26). However, as noted above, we have not been able determine the half-life of Dfp1 in G1 phase.
In these experiments we also examined the level of Dfp1 in cells blocked in M phase. We found that the abundance of Dfp1 is normal in M phase when expression is under the control of the heterologous nmt1+ promoter (Fig. 4C). This confirms our previous conclusion that the regulation of Dfp1 expression in mitotic cells is entirely at the transcriptional level. It follows that Dfp1 expression can be regulated by two different mechanisms, a posttranscriptional mechanism operating at G1/S and a transcriptional mechanism at G2/M.
Dfp1 Is Expressed Immediately Before Initiation of DNA Replication.
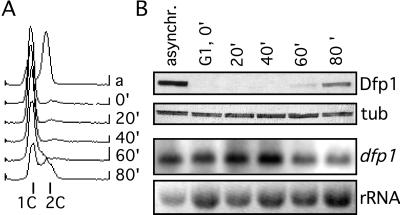
Given that the expression of Dfp1 is low in G1 phase and high in S phase, we carried out a cell-synchronization experiment to more precisely define the time of appearance of the protein. A population of cells was arrested in G1 phase by inactivation of Cdc10 and then released into the cell cycle by temperature shift. The DNA content of cells was monitored by flow cytometry, and Dfp1 expression was evaluated by immunoblot analysis (Fig. 5). In the original asynchronous population the cells exhibited a 2C DNA content and Dfp1 was expressed. After the arrest in G1 the vast majority of the cells had a 1C DNA content, and Dfp1 was undetectable. After release from cell cycle block the cells remained in G1 for 60 min, when the DNA content of the cells began to increase, indicating the onset of S phase. By 80 min, approximately 50% of the cells were clearly in S phase. Western blotting of extracts prepared from the same cell population revealed that Dfp1 first appeared at 60 min and increased significantly in amount by 80 min. Thus, the appearance of Dfp1 is exactly coincident with the initiation of DNA synthesis. As expected, there was little change in the level of dfp1+ mRNA as cells progressed from the G1 block into S phase (Fig. 5B); in fact, the modest increase in dfp1+ mRNA preceded the appearance of Dfp1 by 40 min. These data are consistent with the hypothesis that the posttranscriptional regulation of Dfp1 expression plays an important role in determining the timing of initiation.
Figure 5.
Expression of Dfp1 at the G1/S transition. Cells blocked in G1 (GBY401) were released into the cell cycle and sampled every 20 min. (A) DNA content was measured by flow cytometry. The positions of 1C and 2C DNA contents are indicated. (B) Dfp1 levels were analyzed by immunoblot analysis, probing with the anti-HA antibody 16B12 (Dfp1). The blot was reprobed with anti-tubulin antibodies as a loading control (tub). RNA levels were analyzed by Northern hybridization analysis, probing with the dfp1 ORF (dfp1). The ethidium bromide-stained rRNA is shown as a control for loading (rRNA).
DISCUSSION
The identification of Dfp1 as a subunit of Hsk1 kinase led us to speculate that Dfp1 was a regulator of Hsk1 kinase activity during cell cycle progression (19). Here we have shown that Dfp1 is sufficient to activate Hsk1 phosphorylation of Cdc19 (Mcm2) in vitro. We have also shown that cells lacking Dfp1 are severely delayed in entry into S phase, indicating an important role for Dfp1 in regulating initiation of DNA replication. Consistent with such a role, Dfp1 expression is regulated during cell cycle progression. Expression of the Dfp1 protein is reduced in G1 because of transcriptional and posttranscriptional mechanisms. Increased expression of Dfp1 at the G1/S transition leads to the rapid accumulation of Dfp1 immediately before the initiation of DNA replication. Accumulation of Dfp1 presumably results in activation of Hsk1, which is essential for initiation.
Phosphorylation of Dfp1 may also play a role in regulating Hsk1 kinase activity and, thereby, initiation. We found that Dfp1 is phosphorylated in S and G2 phases and that these Dfp1 phosphoisomers have a different migration in SDS/PAGE, suggesting that Dfp1 is phosphorylated on different residues depending on cell cycle position. Our reconstitution experiments (Fig. 1) indicate that phosphorylation of Dfp1 is not a prerequisite for activation of Hsk1 kinase. We were able to reconstitute the activated form of Hsk1 kinase by using recombinant Dfp1 produced in bacteria, which is presumably unphosphorylated. Phosphorylation of Dfp1, then, may be involved in negative regulation of kinase activity. Dfp1 phosphorylation might prevent Hsk1 kinase from triggering initiation at origins that already have fired, perhaps by causing dissociation of Dfp1 from replication origins. Alternatively, Dfp1 phosphorylation might inactivate Hsk1 and thereby prevent initiation at replication origins that fire late in S phase. Consistent with this possibility, the budding yeast Cdc7 kinase is required for late origins to fire (9, 10). Finally, Dfp1 phosphorylation may not influence Hsk1 activity directly but, rather, might alter Dfp1 stability. Phosphorylation of Dfp1 immediately before S phase could stabilize Dfp1, allowing it to accumulate and thereby activate Hsk1 kinase.
Dfp1 is hyperphosphorylated in a Cds1-dependent manner when cells are blocked in S phase. Cds1 has been implicated in the surveillance mechanisms that protect genome integrity when DNA replication is blocked or DNA is damaged during S phase (24, 27). Cds1 is unique in that it is not required to arrest the cell cycle after such insults but, rather, is important in maintaining viability during such an arrest (24). Dfp1, therefore, might function downstream of Cds1 in this pathway and play a role in the ability of cells to recover from damage that causes a block to DNA replication. This possibility is supported by a recent report of genetic and physical interactions between Dbf4 and Rad53 [the budding yeast homologues of Dfp1 and Cds1 (28)]. Cds1-dependent phosphorylation of Dfp1 might serve to block new initiation when damage to DNA is detected during S phase, inhibiting replication until damage is repaired. Additionally, stalled replication forks could be particularly unstable structures, so preventing the accumulation of additional sources of instability during a block to DNA replication might be important in recovering from DNA damage during S phase. We also have observed an S phase-specific phosphorylation of Dfp1 in cycling cells (Fig. 3A), but have not yet determined whether this modification also depends on Cds1.
Of particular significance, we found that Dfp1 protein expression is cell cycle-regulated. Levels of Dfp1 increase severalfold as cells pass from G1 into S and G2 phase in synchronous culture. This experiment likely underestimates the change in Dfp1 levels, because of the short length of G1 in fission yeast and asynchrony in the culture. When cells are blocked in G1 by two different methods, such that G1 is extended and all of the cells are at the same cell cycle position, Dfp1 is completely absent. The lack of Dfp1 expression in G1 likely serves as a mechanism to prevent premature activation of Hsk1 kinase before S phase, consistent with our finding that monomeric Hsk1 is inactive in phosphorylation of exogenous substrates (19). Additionally, if Dfp1 is required to target Hsk1 kinase to replication origins, a function that has been proposed for Dbf4 (16), the lack of Dfp1 in G1 phase might prevent the local Hsk1 concentration at replication origins from reaching critical levels. It will be of great interest to determine the effects of ectopic G1 expression of Dfp1 on cell cycle progression, although it has proven difficult to overexpress Dfp1 in G1 cells.
Taken together, our results suggest a model for Dfp1 activation of Hsk1 kinase as cells enter S phase. We propose that during G1 phase dfp1+ mRNA is expressed but Dfp1 protein does not accumulate, perhaps because it is degraded rapidly. Hsk1 in G1 phase therefore is monomeric and is inactive in the phosphorylation of exogenous substrates. At the G1/S transition expression of Dfp1 is activated and the protein accumulates. Association of Dfp1 with Hsk1 extends the substrate specificity of the kinase such that it now is able to phosphorylate its critical substrates, resulting in the initiation of DNA replication.
Acknowledgments
We thank Debbie Tien for excellent technical assistance, Dr. Susan Forsburg and Debbie Liang for advice on spore germination, and Dr. Joel Huberman and Dr. Chris Houchens for careful reading of the manuscript. This work was supported by National Institutes of Health Grant GM50806. G.W.B. is a Special Fellow of the Leukemia Society of America.
ABBREVIATION
- MCM
minichromosome maintenance
References
- 1.Culotti J, Hartwell L H. Exp Cell Res. 1971;67:389–401. doi: 10.1016/0014-4827(71)90424-1. [DOI] [PubMed] [Google Scholar]
- 2.Hollingsworth R E, Sclafani R A. Proc Natl Acad Sci USA. 1990;87:6272–6276. doi: 10.1073/pnas.87.16.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon H-J, Campbell J L. Proc Natl Acad Sci USA. 1991;88:3574–3578. doi: 10.1073/pnas.88.9.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masai H, Miyake T, Arai K. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato N, Arai K-I, Masai H. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang W, Hunter T. Proc Natl Acad Sci USA. 1997;94:14320–14325. doi: 10.1073/pnas.94.26.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hess G F, Drong R F, Weiland K L, Slightom J L, Sclafani R A, Hollingsworth R E. Gene. 1998;211:133–140. doi: 10.1016/s0378-1119(98)00094-8. [DOI] [PubMed] [Google Scholar]
- 8.Kim J M, Sato N, Yamada M, Arai K, Masai H. J Biol Chem. 1998;273:23248–23257. doi: 10.1074/jbc.273.36.23248. [DOI] [PubMed] [Google Scholar]
- 9.Bousset K, Diffley J F. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donaldson A D, Fangman W L, Brewer B J. Genes Dev. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartwell L H. J Bacteriol. 1973;115:966–974. doi: 10.1128/jb.115.3.966-974.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson A L, Pahl P M B, Harrison K, Rosamond J, Sclafani R A. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon H-J, Loo S, Campbell J L. Mol Biol Cell. 1993;4:195–208. doi: 10.1091/mbc.4.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston L H, Thomas A P. Mol Gen Genet. 1982;186:445–448. doi: 10.1007/BF00729467. [DOI] [PubMed] [Google Scholar]
- 15.Kitada K, Johnston L H, Sugino T, Sugino A. Genetics. 1992;131:21–29. doi: 10.1093/genetics/131.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowell S J, Romanowski P, Diffley J F X. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- 17.Hardy C F J, Dryga O, Seematter S, Pahl P M B, Sclafani R A. Proc Natl Acad Sci USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei M, Kawasaki Y, Young M R, Kihara M, Sugino A, Tye B K. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown G W, Kelly T J. J Biol Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- 20.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 21.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 22.Muzi-Falconi M, Kelly T J. Proc Natl Acad Sci USA. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasion S G, Brown G W, Brown L M, Ray D S. J Cell Sci. 1994;107:3515–3520. doi: 10.1242/jcs.107.12.3515. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay H D, Griffiths D J F, Edwards R, Murray J M, Christensen P U, Walworth N, Carr A M. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King R W, Deshaies R J, Peters J M, Kirschner M W. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 26.Hershko A. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 27.Murakami H, Okayama H. Nature (London) 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 28.Dohrman P R, Oshiro G, Tecklenburg M, Sclafani R A. Genetics. 1999;151:965–977. doi: 10.1093/genetics/151.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]