Abstract
The E6 and E7 oncoproteins of the high-risk human papillomavirus (HPV) types are able to immortalize human keratinocytes in vitro and likely contribute to the development of anogenital malignancies in vivo. The role of these oncoproteins in the productive viral life cycle, however, is not known. To begin to examine these possible roles, mutations in E6 were introduced in the context of the complete HPV 31 genome. Although transfected wild-type HPV 31 genomes, as well as genomes containing an E6 translation termination linker, an E6 frameshift mutation, and a point mutation in the p53 interacting domain were able to replicate in transient assays, only the wild-type genome was stably maintained as an episome. Interestingly, mutant genomes in either the E6 splice-donor site or splice-acceptor site were reduced in replication ability in transient assays; however, cotransfection of E1 and E2 expression vectors restored this function. In a similar fashion, genomes containing mutant HPV 31 E7 genes, including a translation termination mutant, two Rb-binding site mutants, a casein kinase II phosphorylation site mutant, and a transformation deficient mutant, were constructed. Although transient replication was similar to wild type in all of the E7 mutants, only the casein kinase II mutant had the ability to maintain high copies of episomal genomes. These findings suggest a role for E6 and E7 in the viral life cycle beyond their ability to extend the life span of infected cells.
High-risk genital human papillomaviruses (HPVs) are the causative agents of cervical and other anogenital cancers (1, 2). This association is due, in part, to the ability of two of the viral gene products, E6 and E7, to target cellular proteins that regulate the cell cycle (3, 4). The most notable of these are p53 and Rb. E6 facilitates the degradation of p53 through its association with an accessory protein, E6–AP, a component of the ubiquitin-degradation pathway, whereas E7 binds to and disrupts the functions of Rb (5). Numerous studies have broadened our understanding of these interactions and confirmed the role of these oncoproteins in transformation, immortalization, and the induction of genomic instability (6, 7). In contrast, little information is available on the functions of E6 and E7 in the productive viral life cycle.
The life cycle of HPV is closely associated with the differentiation program of the infected epithelial tissue (8). Consequent to infection, viral genomes replicate as episomes in basal cells coincident with cellular replication, maintaining copy number at ≈50 per cell. After cell division, the daughter cells leave the basal layer and begin to differentiate. As the cells reach the suprabasal layer, entry into S phase is induced, most likely through the action of the E7 protein (9, 10). This entry into S phase results in amplification of the viral genomes, expression of capsid proteins, and assembly of progeny virus in the outermost layer of the infected tissue (11).
The E1 and E2 proteins of papillomaviruses have been shown to activate replication and regulate transcription of early viral genes in transient assays (12–16). Recently, a genetic system has been developed and was used to show that episomal forms of HPV 31 are required for the high level of expression of late viral genes (17). In contrast, genomes with a frameshift mutation in the E1 ORF were found to be deficient in their ability to maintain episomes and, therefore, amplify DNA and express late viral transcripts. It is clear that episomal maintenance of HPV genomes is a critical component of the productive viral life cycle and persistent infection. Using this genetic system, we have examined the roles of the E6 and E7 viral proteins of high-risk HPV 31 in the productive viral life cycle. We show that E6 and E7 are both required for the maintenance of stable viral episomes; in contrast, these oncoproteins have no role in transient replication. These findings suggest a role for E6 and E7 in the viral life cycle beyond their ability to extend the life span of infected cells.
MATERIALS AND METHODS
Cell Culture.
Human foreskin keratinocytes (HFKs) were derived from neonatal human foreskin epithelium as described (10) and were maintained in serum-free keratinocyte growth medium before transfection (Keratinocyte Growth Medium; Clonetics, San Diego). SCC13 cells, a human squamous cell carcinoma line (18), and HPV 31 genome transfectants were grown in serum-containing medium (E medium; ref. 19) with J2 3T3 fibroblast feeders, which were kindly provided by the Howard Green laboratory (Harvard Medical School, Boston) and were treated with mitomycin C (Boehringer Mannheim; ref. 20).
Plasmids.
pBR322.HPV31 contains the HPV 31 genome inserted into the EcoRI site of pBR322 (17). Mutations in E6 of pBR322.HPV31 were constructed as follows: a translation termination mutant of HPV 31 E6, pBR31.E6–TTL (translation termination linker), was created by insertion of a 24-bp TTL (GTGCAGATCTTAATTAACTAACTG) into the unique HpaI site of E6. An additional mutation of E6, which abrogates expression of full-length E6, pBR31.E6–FS, was constructed by insertion of an 8-bp AatII linker (New England Biolabs) into the unique DraIII site of E6, resulting in a frameshift. Mutations at the splice-donor, pBR31.E6-SDMT, and splice-acceptor, pBR31.E6–SAM, sites of E6 were made by substitution of nucleotides 212 and 411, respectively, via recombinant PCR. pBR31.E6–YYH is a mutant that is unable to degrade p53 (21). It was created by recombinant PCR with substitutions at amino acids 45, 47, and 49.
Mutations in E7 of pBR322.HPV31 were constructed via recombinant PCR as follows: a translation termination of E7, pBR31.E7–3XTAA, was made with three stop codons engineered at amino acids 9–11. pBR31.E7–D21G, which has an Rb-binding efficiency similar to that of the “low-risk” E7 protein (22–24), and pBR31.E7–C24G, which is deficient in Rb binding (22–25), were created with substitutions at the indicated amino acids. pBR31.E7–R2P is a substitution mutation of the second amino acid of E7, which results in loss of transformation function but retention of Rb binding (22, 25). pBR31.E7–CK2 is a substitution mutation of amino acids 31 and 32, which are potential casein kinase II (CK2) phosphorylation sites (26–28).
Transfection of HFKs.
To release the viral genome, 10 μg of the given pBR31 construct (wild type or mutant) was digested with EcoRI. Restriction enzyme was heat-inactivated and genomes were unimolecularly ligated in the digestion solution with T4 DNA ligase (10 units/900 μl). DNA was precipitated with isopropyl alcohol and resuspended in TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). The entire precipitated ligation was cotransfected with 2 μg of the selectable marker pSV2neo into HFKs with LipofectAce (GIBCO/BRL) as described by the manufacturer. Cells were plated onto mitomycin C-treated fibroblast feeders in E medium 1 day after transfection. Selection began 2 days after transfection with G418 (GIBCO/BRL) as follows: 200 μg/ml G418 every 2 days for a total of 4 days, and then 100 μg/ml G418 every 2 days for 4 more days. After selection, pooled populations were expanded for analyses.
Transient Replication Assays.
Transient replication assays were carried out as described (29). Briefly, viral DNAs (3 μg of 7,912-bp HPV 31) were first cleaved from the bacterial plasmid sequences, unimolecularly ligated, and combined with carrier DNA (20 μg) and equimolar amounts of E1 and E2 expression vectors (pSG-E1 and pSG-E2; ref. 30), as indicated in the figure legends. SCC13 cells were transfected by electroporation (250 V; 960 uF; Bio-Rad GenePulser) and plated onto mitomycin C-treated fibroblast feeders. At 5 days after transfection, low-molecular-mass DNA was isolated by the Hirt method (31) and digested with DpnI to remove residual bacterially methylated input DNA and with BanII to linearize the viral genomes. After agarose gel electrophoresis and blotting to a nylon membrane (Magna; Micron Separations, Westboro, MA), replicated DNAs were detected with a 32P-labeled HPV 31 DNA probe (HpaI → EcoRI fragment) and examined by autoradiography.
Stable Replication Assays.
HPV transfectants were harvested at approximately one passage after transfection for Southern blot analyses. Total genomic DNA was prepared by resuspension of cell pellet in lysis buffer (400 mM NaCl/10 mM Tris⋅HCl, pH 7.4/10 mM EDTA) to which RNase A (50 μg/ml), proteinase K (50 μg/ml), and SDS (0.2%) was added, followed by incubation at 37°C overnight. DNA was sheared by passage through an 18-gauge needle approximately 10 times, extracted with phenol:chloroform, and then precipitated with ethanol. Total genomic DNA (10 μg) was digested with DpnI to remove any residual input DNA. Digested DNA was separated on a 0.8% agarose gel, treated, and alkaline-transferred onto DuPont GeneScreen Plus nylon membrane (NEN) as described by the manufacturer. The membrane was prehybridized in 50% (vol/vol) formamide, 4× standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), 5× Denhardt’s solution (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA), 1% SDS, 10% (vol/vol) dextran sulfate, and 0.1 mg/ml denatured herring sperm DNA for 1 h at 42°C. The HPV 31 probe was prepared by gel purification of the entire HPV 31 genome from pBR322.HPV31 digested with EcoRI and labeled with the Ready-to-go DNA labeling kit (Amersham Pharmacia). Labeled probe was purified with NucTrap columns (Stratagene), denatured, and added to fresh hybridization solution, which was incubated with membrane at 42°C overnight. Membrane was washed twice with 2× SSC/0.1% SDS for 15 min at room temperature; twice with 0.5× SSC/0.1% SDS for 15 min at room temperature; twice with 0.1× SSC/0.1% SDS for 15 min at room temperature; and once with 0.1× SSC/1% SDS for 30 min at 50°C. Hybridizing species were visualized by autoradiography.
RESULTS
HPV 31 Genomes Containing Mutations in E6.
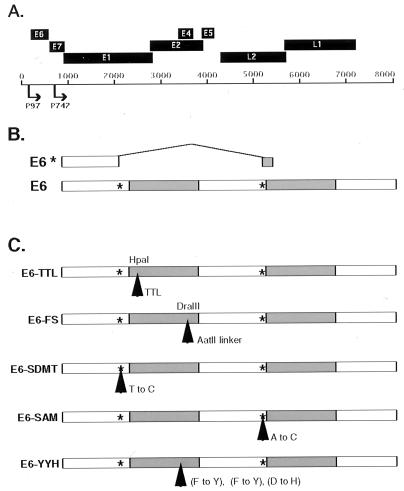
To investigate the role of the E6 oncoproteins in the productive life cycle of HPVs, a set of five mutant HPV 31 genomes was constructed in the context of pBR322.HPV31 (Fig. 1). Of these mutant genomes, two contain a translation termination mutation and a frameshift mutation (E6–TTL and E6–FS), both of which abrogate E6 translation. Previous analyses of HPV 31 early transcripts identified an intron in E6, which, when expressed, generates a putative shortened E6 product, E6* (32). To investigate the role of this intron, we also constructed pBR322.HPV31-derivative plasmids that contain mutations in either the splice-donor site, E6–SDMT, or the splice-acceptor site, E6–SAM, resulting in loss of E6* expression. These two mutants were designed to be silent in the full-length E6. Finally, a substitution mutant, which is specific to the full-length E6, was constructed based on published studies of HPV 16 E6 mutants that fail to degrade p53. Substitution of amino acids 45, 47, and 49 to tyrosine, tyrosine, and histidine, respectively, in HPV 16 E6 has been shown to inhibit degradation of p53 (21). We constructed the corresponding mutant, E6–YYH, in HPV 31 in which the same three amino acids in HPV 31 were similarly substituted.
Figure 1.
Schematic of HPV 31 E6 mutants. (A) Diagram representing the ORFs of HPV 31 and the two major promoters that drive viral expression. (B) Diagram depicting the two possible E6 ORFs expressed from the p97-initiated transcripts of HPV 31. E6 represents the full-length protein and E6* denotes a putative shortened protein expressed from a spliced transcript. The asterisks represent the splice-donor and splice-acceptor sites. (C) Diagram showing the HPV 31 E6 mutations with their mutation site(s) indicated by arrows. E6–TTL is an insertion of a TTL, and E6–FS is a frameshift mutant. Both mutants abrogate expression of full-length E6. E6–SDMT and E6–SAM indicate the two splicing mutants with substitutions at nucleotides 212 and 411, respectively, but E6–SDMT and E6–SAM are silent in E6. E6–YYH denotes the p53-binding-deficient mutant with substitutions at amino acids 45, 47, and 49. All mutants were generated in the context of the pBR322.HPV31 construct. Shaded areas represent the zinc finger regions of the protein.
Transient Replication of E6 Mutant Genomes.
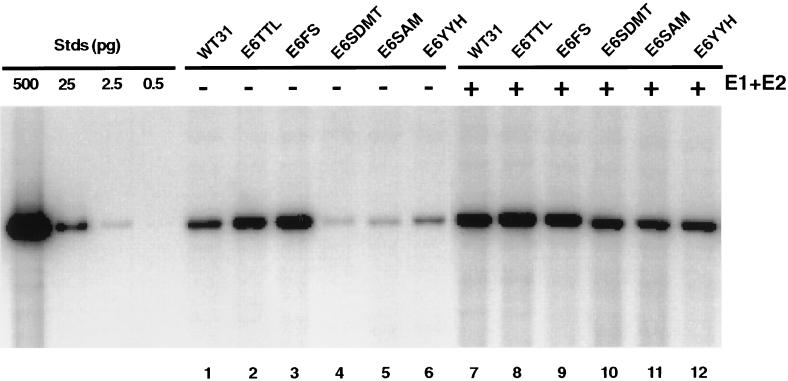
We first examined the effect of E6 mutations on the ability of intact viral genomes to replicate in transient assays. For these studies, viral DNAs were released from the bacterial vector (pBR322), unimolecularly ligated, and transfected into SCC13 keratinocytes by electroporation with the religated genomes. After 5 days, low-molecular-mass DNA was isolated and digested with DpnI to remove any residual bacterially methylated input DNA, as well as with BanII to linearize the viral genomes to facilitate quantitation. Southern analysis was then performed, and replicating DNAs were detected by autoradiography. As shown in Fig. 2, the E6 translation termination mutant (Fig. 2, lane 2) and the frameshift mutant (Fig. 2, lane 3) were found to replicate as efficiently as the wild-type genome (Fig. 2, lane 1). In contrast, the splice-site mutants (Fig. 2, lanes 4 and 5) had a significant 5-fold decrease in replication compared with wild type. The mean of three separate experiments was calculated based on quantitation by PhosphorImager analysis. The replication ability of the p53 degradation mutant (Fig. 2, lane 6) was consistently reduced compared with wild type; however, on average, the reduction was minimal in comparison to the decrease observed in the splice-site mutants. This result suggests that the p53-binding and degradation function of E6 is most likely not required for transient replication. In addition, transient replication assays were performed in HFKs for the wild-type as well as the E6–TTL mutant genomes. In these experiments, the wild-type and E6–TTL genomes replicated at comparable levels (data not shown). Taken together, these data indicate that expression of full-length E6 protein is not required for transient replication of the HPV 31 genome.
Figure 2.
Autoradiogram of Southern blot showing replicated (DpnI-resistant) viral DNAs from a transient replication assay with HPV 31 wild type (WT) and E6 mutants in SCC13 cells. Viral DNAs were excised from bacterial plasmid sequences, unimolecularly ligated, and transfected alone (lanes 1–6) or with equimolar amounts of HPV 31 E1 and E2 expression vectors (lanes 7–12). Low-molecular-mass DNA was isolated and analyzed by Southern blotting as described in Materials and Methods. DNA standards (Stds) are shown on the left and contain 500, 25, 2.5, and 0.5 pg of linearized wild-type HPV 31 DNA.
There are two possible explanations for the decreased replication ability of genomes containing mutated splice-donor or splice-acceptor sites (Fig. 2, lanes 4 and 5). It is possible that expression of the truncated E6* protein is required for replication or that, alternatively, the E6 splice sites are required for expression of transcripts that encode the replication proteins E1 and E2. To address the latter possibility, mutant genomes were cotransfected with E1 and E2 expression vectors and assayed for their ability to replicate transiently. As shown in Fig. 2 (lanes 10 and 11), when cotransfected with E1 and E2 expression vectors, the E6 splice-donor and splice-acceptor mutant genomes replicated at levels similar to those seen with the wild-type genomes (Fig. 2, lane 7). Only a modest increase in replication of wild-type genomes was observed by the addition of E1 and E2 expression vectors, consistent with the previously described autoregulation by E2 (33). Moreover, the increase observed in the E6–YYH mutant with overexpression of E1 and E2 (Fig. 2, lane 12) is similar to the increase observed for the wild-type genome (Fig. 2, lane 7). Further mutational analysis would be required to determine whether loss of the p53 degradation function affects transient replication. These data suggest that the loss of the E6 splice sites in the splice-donor and splice-acceptor mutants alters the expression of E1 and/or E2, resulting in the decreased ability of these cells to replicate transiently.
Stable Maintenance of E6 Mutant Genomes.
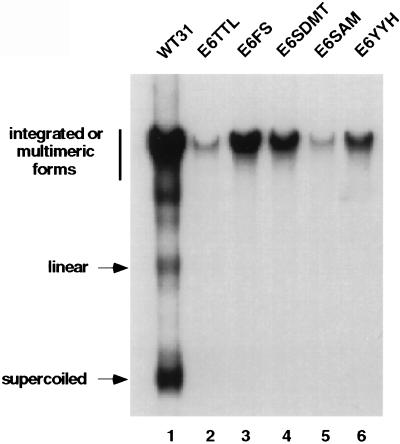
We next investigated whether E6 was necessary for stable maintenance and replication of HPV 31 genomes. HFKs were transfected via lipid with recircularized wild-type and E6 mutant HPV 31 DNAs as described in Materials and Methods. After selection for neomycin resistance, pooled colonies were harvested approximately 1 month after transfection. Southern blot analyses were performed on DpnI-digested total genomic DNA from both wild-type and E6 mutant HPV 31 cultures (Fig. 3). The cells transfected with wild-type HPV 31 were found to contain primarily episomal copies of HPV 31 DNA, consistent with previous reports (Fig. 3, lane 1); however, all of the cells transfected with genomes containing mutations in E6 had only integrated copies (Fig. 3, lanes 2–6). These experiments were repeated three times with similar results. As expected, the splice-site mutants that were deficient in transient viral replication were also unable to replicate stably, resulting in integration. Interestingly, the translation termination and frameshift mutants and the p53 degradation mutant, which were able to replicate efficiently in transient assays, were unable to replicate stably, as evidenced by the lack of supercoiled DNA just 1 month after transfection. In addition, senescence occurred typically between 1 and 2 months for the E6–TTL, E6–FS, and E6–YYH mutant cells in which the cells were passaged two or three times. In contrast, the E6–SDMT and E6–SAM cells had slightly extended life spans.
Figure 3.
Autoradiogram showing Southern analysis of stably transfected HFKs with HPV 31 wild type (WT) and E6 mutants. Total genomic DNAs were digested with DpnI to remove residual input DNA. The migration of the various DNA forms is indicated on the left. Similar results were seen in three independent transfections.
Construction of E7 Mutants.
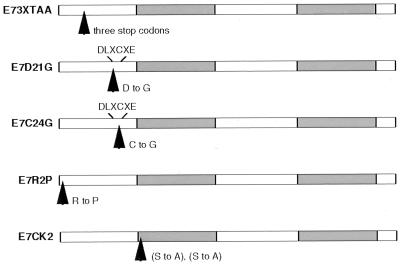
It was next important to determine whether the second viral oncoprotein, E7, had a function in the HPV viral life cycle beyond its ability to extend the life span of infected cells. The binding of E7 to Rb is the best characterized function of E7; therefore, we constructed mutants that would determine whether this activity had a role in the maintenance of stable episomes. As with the E6 mutants, mutants of E7 were generated in the context of the HPV 31 genome construct pBR322.HPV31 (Fig. 4). E7–3XTAA contains an insertion of three tandem translation termination codons, whereas E7–D21G contains a substitution in the Rb-binding region (DLXCXE) and binds Rb with a reduced affinity similar to the affinity observed in low-risk E7 (22–24). E7–C24G also contains a substitution in the Rb-binding region, but this mutation results in a total loss of Rb binding (22–25). E7–R2P is a mutant that has lost the ability to transform but retains Rb binding (22, 25), and E7–CK2 contains an altered CK2 phosphorylation site (26–28).
Figure 4.
Schematic of HPV 31 E7 mutants. Mutation sites are indicated by arrows. E7–3XTAA represents a translation termination mutant with a substitution of amino acids 9–11 with three stop codons. E7–D21G and E7–C24G indicate mutations in the DLXCXE Rb-binding motif at the indicated amino acids. E7–R2P represents a transformation-defective mutant at amino acid 2, and E7–CK2 denotes a potential CK2 phosphorylation site mutant with substitutions of amino acids 32 and 33. All mutants were generated in the context of the pBR322.HPV31 construct. Shaded areas indicate the zinc finger regions of the protein.
Transient Replication of E7 Mutant Genomes.
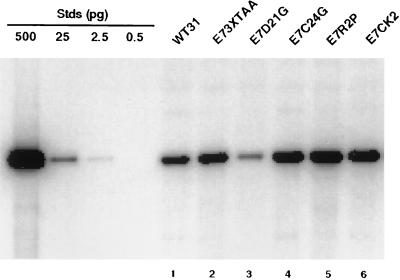
As with the E6 mutants, we first examined the effect of the E7 mutants on transient viral replication. Viral DNAs were prepared as described above and transfected into SCC13 cells. After 5 days, low-molecular-mass DNA was isolated from cells and digested with DpnI and BanII. Southern analysis was then performed, and replicating DNAs were detected by autoradiography. As shown in Fig. 5, all of the E7 mutants (Fig. 5, lanes 2–6) have DNA levels comparable to wild type (Fig. 5, lane 1), indicating that E7 does not significantly affect transient viral replication. Although replication in the E7–D21G mutant is slightly decreased in this experiment (Fig. 5, lane 3), analysis of the data from three separate experiments with E7–D21G indicates no statistical difference in transient replication ability compared with wild type. Again, transient replication assays were performed with wild-type and E7–3XTAA mutant genomes in HFKs and similar results were found (data not shown).
Figure 5.
Autoradiogram of Southern analysis of replicated (DpnI-resistant) viral DNAs from a transient replication assay with HPV 31 wild type (WT) and E7 mutants in SCC13 cells. Transfections of viral DNAs and Southern analysis were carried out as described in the legend to Fig. 2. DNA standards (Stds) are shown on the left and contain 500, 25, 2.5, and 0.5 pg of linearized wild-type HPV 31 DNA.
Stable Maintenance of E7 Mutant Genomes.
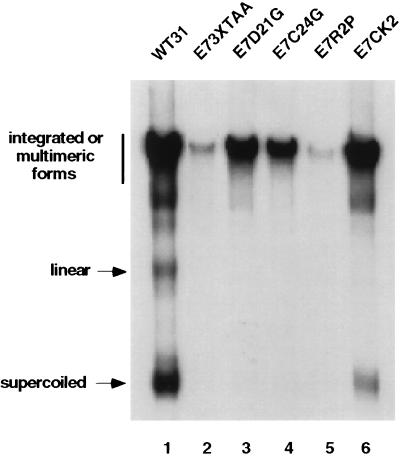
Although transient replication was unaffected in the E7 mutants, we next examined whether mutations in E7 alter stable replication. Viral DNAs were prepared as described above and transfected into primary HFKs. After selection, cells were cultured for approximately 4 weeks followed by isolation of total genomic DNA. DNAs were digested with DpnI and subjected to Southern analysis (Fig. 6). Only the wild-type genome (Fig. 6, lane 1), and the E7–CK2 mutant genome (Fig. 6, lane 6) had high levels of supercoiled DNA, indicative of episomal maintenance. All other mutants (Fig. 6, lanes 2–5) had predominately integrated DNA. These experiments were repeated three times with similar results. Surprisingly, we detected low levels of episomal DNA (less than 5% of wild type) in the E7–D21G and the E7–C24G mutants, each in separate experiments (data not shown). However, we could not correlate this low level of plasmid maintenance with extended life span as is seen with the wild type and E7–CK2 mutant. These data suggest an essential role for the E7 protein in stable viral replication and genome maintenance.
Figure 6.
Autoradiogram of Southern analysis of stably transfected HFKs with HPV 31 wild type (WT) and E7 mutants. Total genomic DNAs were prepared and digested as described in Materials and Methods. The migration of the various DNA forms is indicated on the left. These experiments were repeated with three independent transfections, and each yielded similar results. The wild-type sample (lane 1) is the same sample shown in Fig. 3, lane 1.
DISCUSSION
In the present study, we have shown that expression of both the E6 and E7 oncoproteins from the high-risk HPV type 31 is necessary for the stable maintenance of episomes after introduction into HFKs. In contrast, loss of E6 or E7 function has no effect in transient replication studies. These observations suggest distinct requirements for these two modes of replication and are consistent with previous findings. Although the E1 and E2 gene products are both required in trans for transient and stable replication of papillomavirus plasmids (16), we and others have shown that transient and stable replication differ in their cis requirement for E2-binding sites (33, 34). Our current studies extend these differential requirements to the E6 and E7 oncoproteins and suggest that their action is necessary for viral episomes to be stably maintained.
We observed that wild-type HPV 31 episomes were detected in transfected cells after just 1 month after transfection, whereas E6 and E7 mutant genomes had only integrated copies in this same period of time. Several models could explain these observations. In the first model, episomal forms of the E6 and E7 mutant genomes may be unable to properly segregate on cell division and, as a result, are rapidly lost from the cell. Only genomes that have integrated into host sequences would be retained after several cell divisions. Such a model has been suggested recently for the action of Epstein–Barr virus-encoded nuclear antigen 1 in the stable maintenance of Epstein–Barr virus plasmids (35). A second model is based on the observation that cells transfected with genomes containing mutations in E6 and E7 eventually undergo senescence. It is possible that senescent cells are unable to maintain episomes, resulting in the detection of only integrated forms. The failure to detect episomal copies of HPV DNA after transfection into immortal tumor cell lines suggests that this model alone may not explain E6 and E7 function (W.G.H., unpublished work). A third model proposes that the action of E6 and E7 on their respective cellular targets is directly required for stable maintenance.
Insight into the mechanism of E6 and E7 action is provided through mutational analysis. In addition to mutations that completely abrogate E6 function, we analyzed a mutant of E6, E6–YYH, which is unable to degrade p53 (21). Genomes expressing this mutation were incapable of stable episomal maintenance, implicating the p53-degradation function of E6 in stable replication. There are many identified binding partners of E6, including E6–AP (36), E6–BP (37), paxillin (38), hDLG (39), IRF-3 (40), and E6TP1 (41). In addition, E6 activates the expression of the catalytic subunit of telomerase hTERT (42). Because it is difficult to identify mutants of E6 that are completely inactive in one function but not another, it is likewise difficult by mutational analysis alone to determine which of the activities of E6 is important in stable replication. To investigate whether abrogation of p53 function is sufficient to allow for stable replication, expression vectors for a dominant-negative form of p53 were cotransfected together with the E6–TTL mutant genomes into normal HFKs, but no stable episomes were detected (data not shown). Similarly, cotransfection with expression vectors for hTERT failed to yield stable episomes (data not shown). It is possible that these transfection assays do not mimic the state in established cell lines with activated telomerase or mutant p53; therefore, it is premature to conclude that these two activities do not play a role in stable replication.
In addition to analysis of point mutants, we examined the effects of splice-site mutations in E6 on replication. Two major types of E6 transcripts are expressed from the P97 early promoter of HPV 31 (32). The first is an unspliced message, and the second contains an intron in E6 that encodes a putative E6* protein. This E6* protein is a shortened version of E6; however, its synthesis in cells has yet to be established convincingly. In our studies, mutations of either the E6 splice-donor or splice-acceptor sites abrogated transient replication ability, which could be restored by cotransfection of expression vectors for E1 and E2. This observation provides genetic evidence that either of these two replication proteins—or perhaps both—are expressed from promoters upstream of E6 that use the intron in E6. This finding is consistent with recent studies that identified HPV 31 E1 transcripts initiated at P97 in undifferentiated cells (43). Further support is provided by studies from Remm et al. (44), who established that translation of the HPV 18 E1 protein is significantly enhanced by the presence of an upstream intron in E6 and that synthesis of E1 protein is reduced on mutation of the splice site. It is technically difficult to determine E1 and E2 protein levels, because these proteins have not been detected by immunoprecipitation or Western analyses in cell lines that stably maintain episomes. Furthermore, analysis of E1 and E2 transcripts is not feasible with the splice-site mutant genomes, as transfection results in integration of genomes, leading to altered transcripts that are not comparable to wild-type transcripts. We cannot formally exclude the possibility that the reduction in transient replication is caused by E6* action, but the ability to overcome this defect by the addition of E1 and E2 expression vectors makes it unlikely.
In a similar manner, we examined genomes containing several point mutations in E7, including those at the N terminus, the Rb-binding domain, and the putative CK2 phosphorylation site. Genomes containing the E7–R2P mutation were consistently found to be integrated into host chromosomes, whereas E7–CK2 mutant genomes were always stably maintained as episomes. A variable phenotype, however, was observed with the two mutations in the Rb-binding domain: E7–D21G and E7–C24G. We detected viral episomes in these Rb-binding mutant cell lines at a low frequency, each in two separate experiments, despite the fact that the majority of sequences were found to be integrated. We could not correlate these rare occurrences with extended life span, as is found in cells that stably maintain episomes. For our studies, we were able to examine only pooled cultures, as the cells undergo senescence before the expansion required for cloning. However, it is possible that the small number of episomes observed are present in only a subset of cells and that these cells may have acquired mutations in the Rb pathway, such as in the p16 gene (45). The E7–R2P mutation retains the ability to bind Rb but is likely to have additional deficiencies, suggesting that Rb-binding activity alone is not sufficient for stable replication (22, 25). Moreover, the ability to maintain episomes with E7–CK2 mutant genomes suggests that this modification of E7 is not required for this phase of the viral life cycle.
Studies from other laboratories have also examined the requirements for stable replication of papillomaviruses. The stable maintenance of bovine papillomavirus type 1 origin containing plasmids has been observed previously in Chinese hamster ovary cell lines, which express high levels of E1 and E2 from heterologous promoters (34). In addition, a recent study in which a spontaneously immortalized human keratinocyte line, BC-1-Ep/SL, was transfected with HPV 16 genomes containing a TTL insertion in E7 has shown stable maintenance of episomes. These cells, however, fail to amplify viral DNA on differentiation in raft cultures, indicating that E7 functions at a later stage in the viral life cycle (P. Lambert, personal communication). Because Chinese hamster ovary and BC-1-Ep/SL cells are immortal, the cellular changes induced by HPV 31 E6 and E7 may be induced redundantly by mutations in cellular genes. Our studies show that E6 and E7 are required for episomal maintenance in undifferentiated normal human keratinocytes; therefore, we were unable to identify additional roles in differentiation-induced viral function with the mutations we examined.
We conclude that both HPV type 31 oncoproteins E6 and E7 are required for stable, but not transient, replication in human keratinocytes. Our data provide evidence for a role of the HPV oncoproteins beyond their well documented role in transformation. Importantly, our findings suggest that E6 and E7 significantly contribute to the productive life cycle of HPVs.
Acknowledgments
We thank Kathy Rundell for critically reviewing the manuscript, the members of the Laimins’ laboratory for helpful discussions, and Victoria Hope and Andrew James for technical advice. This work was supported by the Carcinogenesis Training Grant 5T32 CA09560-12 to J.T.T., a Gramm Fellowship Award from Northwestern University to J.T.T., postdoctoral Grant F32CA73087 to W.G.H., and a Midwest Sexually Transmitted Disease Center Grant to L.A.L. from the National Institute of Allergy and Infectious Diseases.
ABBREVIATIONS
- HPV
human papillomavirus
- HFK
human foreskin keratinocyte
- CK2
casein kinase II
- TTL
translation termination linker
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Howley P M. In: Fundamental Virology. 3rd Ed. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 947–978. [Google Scholar]
- 2.Pfister H. Obstet Gynecol Clin North Am. 1996;23:579–595. [PubMed] [Google Scholar]
- 3.Huibregtse J M, Beaudenon S L. Semin Cancer Biol. 1996;7:317–326. doi: 10.1006/scbi.1996.0041. [DOI] [PubMed] [Google Scholar]
- 4.Jones D L, Munger K. Semin Cancer Biol. 1996;7:327–337. doi: 10.1006/scbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- 5.Kubbutat M H G, Vousden K H. Semin Virol. 1996;7:295–304. [Google Scholar]
- 6.Galloway D A, McDougall J K. Semin Canc Biol. 1996;7:309–315. doi: 10.1006/scbi.1996.0040. [DOI] [PubMed] [Google Scholar]
- 7.Thomas J T, Laimins L A, Ruesch M N. Papillomavirus Rep. 1998;9:59–64. [Google Scholar]
- 8.Laimins L A. In: Human Tumor Viruses. McCance D J, editor. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 201–223. [Google Scholar]
- 9.Cheng S, Schmidt-Grimminger D, Murant T, Broker T, Chow L. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 10.Halbert C, Demers G, Galloway D. J Virol. 1992;66:2125–2134. doi: 10.1128/jvi.66.4.2125-2134.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laimins L A. Semin Virol. 1996;7:305–313. [Google Scholar]
- 12.Chiang C M, Dong G, Broker T R, Chow L T. J Virol. 1992;66:5224–5231. doi: 10.1128/jvi.66.9.5224-5231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Vecchio A M, Romanczuk H, Howley P M, Baker C C. J Virol. 1992;66:5949–5958. doi: 10.1128/jvi.66.10.5949-5958.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demeret C, Le Moal M, Yaniv M, Thierry F. Nucleic Acids Res. 1995;23:4777–4784. doi: 10.1093/nar/23.23.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frattini M G, Laimins L A. Proc Natl Acad Sci USA. 1994;91:12398–12402. doi: 10.1073/pnas.91.26.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ustav M, Stenlund A. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frattini M G, Lim H B, Laimins L A. Proc Natl Acad Sci USA. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rheinwald J G, Beckett M A. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 19.Meyers C, Frattini M, Hudson J, Laimins L. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 20.Meyers C, Laimins L A. Curr Top Microbiol Immunol. 1994;186:199–215. doi: 10.1007/978-3-642-78487-3_11. [DOI] [PubMed] [Google Scholar]
- 21.Crook T, Tidy J A, Vousden K H. Cell. 1991;67:547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- 22.Banks L, Edmonds C, Vousden K. Oncogene. 1990;5:1383–1389. [PubMed] [Google Scholar]
- 23.Edmonds C, Vousden K. J Virol. 1989;63:2650–2656. doi: 10.1128/jvi.63.6.2650-2656.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heck D, Yee C, Howley P, Munger K. Proc Natl Acad Sci USA. 1992;89:4442–4446. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe S, Kanda T, Sato H, Furuno A, Yoshike K. J Virol. 1990;64:207–214. doi: 10.1128/jvi.64.1.207-214.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbosa M, Edmonds C, Fisher C, Schiller J, Lowy D, Vousden K. EMBO J. 1990;9:153–160. doi: 10.1002/j.1460-2075.1990.tb08091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Firzlaff J, Galloway D, Eisenman R, Luscher B. New Biol. 1989;1:44–53. [PubMed] [Google Scholar]
- 28.Firzlaff J M, Luscher B, Eisenman R N. Proc Natl Acad Sci USA. 1991;88:5187–5191. doi: 10.1073/pnas.88.12.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubert W G, Kanaya T, Laimins L A. J Virol. 1999;73:1835–1845. doi: 10.1128/jvi.73.3.1835-1845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frattini M G, Laimins L A. Virology. 1994;204:799–804. doi: 10.1006/viro.1994.1596. [DOI] [PubMed] [Google Scholar]
- 31.Hirt B. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 32.Hummel M, Hudson J B, Laimins L A. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stubenrauch F, Lim H B, Laimins L A. J Virol. 1998;72:1071–1077. doi: 10.1128/jvi.72.2.1071-1077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piirsoo M, Ustav E, Mandel T, Stenlund A, Ustav M. EMBO J. 1996;15:1–11. [PMC free article] [PubMed] [Google Scholar]
- 35.Aiyar A, Tyree C, Sugden B. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huibregtse J M, Scheffner M, Howley P M. EMBO J. 1991;10:4126–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J J, Reid C E, Band V, Androphy E J. Science. 1995;269:529–530. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 38.Tong X, Howley P M. Proc Natl Acad Sci USA. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Proc Natl Acad Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronco L V, Karpova A Y, Vidal M, Howley P M. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Q, Srinivasan S, Boyer S N, Wazer D E, Band V. Mol Cell Biol. 1999;19:733–744. doi: 10.1128/mcb.19.1.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klingelhutz A J, Foster S A, McDougall J K. Nature (London) 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 43.Klumpp D J, Laimins L A. Virology. 1999;257:239–246. doi: 10.1006/viro.1999.9636. [DOI] [PubMed] [Google Scholar]
- 44.Remm M, Remm A, Ustav M. J Virol. 1999;73:3062–3070. doi: 10.1128/jvi.73.4.3062-3070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Nature (London) 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]