Abstract
The T domain of diphtheria toxin is known to participate in the pH-dependent translocation of the catalytic C domain of the toxin across the endosomal membrane, but how it does so, and whether cellular proteins are also required for this process, remain unknown. Here, we report results showing that the T domain alone is capable of translocating the entire C domain across model, planar phospholipid bilayers in the absence of other proteins. The T domain therefore contains the entire molecular machinery for mediating transfer of the catalytic domain of diphtheria toxin across membranes.
Many toxic proteins of bacterial and plant origin are known to act by enzymically modifying substrates within the cytosol of mammalian cells, but the mechanism by which any of these toxins crosses a membrane to gain access to its substrates is not yet understood. These intracellularly acting toxins are generally bipartite proteins, containing the enzymic and receptor-binding functions on separate polypeptides, designated A and B, respectively (1). In some toxins, the B moiety also serves a second function, in mediating translocation of the A moiety across membranes.
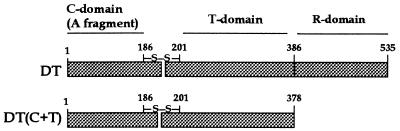
Diphtheria toxin (DT), the earliest example of an AB toxin, is a single, 535-residue polypeptide containing three folding domains: the amino-terminal C, or catalytic, domain (residues 1–185); the intermediate T, or transmembrane, domain (residues 202–378); and the carboxyl-terminal R, or receptor-binding, domain (residues 386–535) (Fig. 1). The catalytic domain is connected to the T domain by an arginine-rich loop and a readily reducible disulfide bridge (linking C186 to C201).
Figure 1.
(Upper) Linear diagram of the whole diphtheria toxin (DT) molecule (protease nicked). (Lower) Linear diagram of the DT(C+T) construct.
After binding to its cell-surface receptor via the R domain, DT is proteolytically cleaved within the arginine-rich loop, yielding two disulfide-linked fragments: fragment A (corresponding to domain C) and fragment B (corresponding to domains T and R). The receptor-bound toxin is endocytosed and trafficked to an acidic vesicular compartment, where it undergoes a conformational change that allows the T domain to insert into the membrane and the catalytic domain to be translocated to the cytosol. The C186–C201 disulfide is reduced at some stage in this process, allowing the catalytic domain to be released into the cytosol. There, it catalyzes the ADP-ribosylation of elongation factor 2, causing inhibition of protein synthesis and cell death. (For a general review of diphtheria toxin, see ref. 2.)
The crystallographic structure (3, 4) of native DT shows the T domain to consist of a bundle of 10 α-helices, which resembles the channel-forming domains of certain colicins (5–8) and α-helical domains found in certain mammalian proteins involved in regulation of apoptosis (9). Studies in planar lipid bilayers have shown that under low pH conditions (pH ≤ 6) in the cis compartment (the compartment to which protein is added), the isolated T domain is able to form cation-selective channels in the membrane (10); similar channels are formed by whole DT and by a truncated mutant, DT(C+T), lacking the R domain (10, 11). Channels are also formed in plasma membranes at low pH by the whole toxin or the B fragment (12, 13). Two long hydrophobic helices (TH8 and TH9) buried within the native T domain insert into the bilayer when the domain converts to a transmembrane form (14–17). The conformation of the transmembrane T domain and its role in translocation are poorly understood, however. Moreover, the question of whether the entire translocation machinery is built into the T domain, or alternatively, one or more cellular proteins are required for translocation, has remained unanswered.
In recent studies with the isolated T domain, we obtained evidence that the amino terminus of the domain is translocated across planar lipid bilayers as channels are formed (18). The phenomenon that enabled us to demonstrate this was the rapid closure of channels at cis-negative voltages when a hexahistidine (H6) tag was fused to the T domain’s amino terminus. The rapid channel closure was inhibited by addition of Ni2+ (which binds to polyhistidines) to the trans compartment, or by trans streptavidin when a residue near the 6 histidines was biotinylated. These results show that the H6 tag had been translocated from the cis to the trans side of the membrane. Rapid channel closure at negative voltages is therefore caused by the H6 tag interacting with the channel from the trans side, probably plugging the lumen.
Inspired by these results with the isolated T domain, we have now done analogous experiments with whole DT and with DT(C+T), which lacks the carboxyl-terminal R domain (Fig. 1).
MATERIALS AND METHODS
Mutagenesis.
The whole DT gene with an E148S active site mutation in the C domain was amplified by PCR using the plasmid pWHS105 (19) as a template, and cloned into the pET22b vector (Novagen) by using BamHI and XhoI restriction endonuclease sites, resulting in pET22b-DT. This expresses DT as a fusion protein with a carboxyl-terminal H6 tag (LGHHHHHH) and an amino-terminal tag (MDIGIDSDP). The experiment shown in Fig. 2A was conducted with this DT.
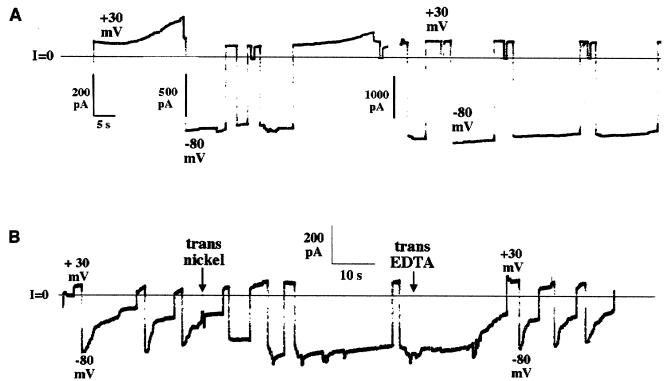
Figure 2.
The effect of the amino-terminal H6 tag on the voltage-gating properties of DT channels. Before the start of each record, DT lacking (A) or containing (B) an amino-terminal H6 tag was added to the cis solution to a concentration of 60 ng/ml (A) or 390 ng/ml (B), and the current across the membrane began to rise linearly with the voltage held at +30 mV. (A) DT-induced conductance is seen rising at +30 mV; when the voltage was switched to −80 mV, the conductance remained on. The solutions on both sides of the membrane contained 1 M KCl, 2 mM CaCl2, and 1 mM EDTA; the cis solution contained 30 mM Mes (pH 5.3), and the trans solution contained 50 mM Hepes (pH 7.2). In this experiment, DT contained a carboxyl-terminal H6 tag; results were similar when using DT without any H6 tag. (B) DT (with amino-terminal H6 tag)-induced conductance turned on at +30 mV as in A, but in contrast turned off rapidly at −80 mV. At the first arrow, NiSO4 was added to the trans solution to a concentration of 20 μM. After the nickel addition, channel conductance failed to turn off at −80 mV. At the second arrow, EDTA was added to the trans solution to a concentration of 2 mM. With the voltage held at −80 mV, the conductance decreased dramatically over several seconds. Subsequent switching of voltage between +30 and −80 mV produced the rapid turn-off of conductance seen before the nickel addition. The solutions were the same as in A, but lacked EDTA. The records were filtered at 10 Hz by the chart recorder.
The plasmid pWHS105 was used to express whole DT with the same E148S active-site mutation and an amino-terminal H6 tag: MGSSHHHHHHSSGLVPR∗GSHM (thrombin cleavage site indicated by ∗). The data shown in Fig. 2B were obtained with this DT preparation. An Arg → Glu substitution mutation in the thrombin cleavage site was introduced into pWHS105 by site-directed mutagenesis using PCR, resulting in pET15b-DT(R to Q), and thereby making the amino-terminal histidine tag resistant to trypsin or thrombin digestion. This allowed specific nicking of DT within the disulfide-bonded loop between the C and T domains without losing the histidine tag.
A DNA segment coding for residues 1–378 of DT (the C and T domains, with R domain deleted) was prepared by PCR with pWHS105 as a template. This was cloned into the pET15b vector (Novagen) by using NdeI and EcoRI restriction endonuclease sites, resulting in pET15b-DT(C+T); the expressed DT(C+T) is a fusion protein with an amino-terminal H6 tag sequence identical to that from pWHS105. pET15b-DT(C+T) was further mutagenized to introduce an Arg → Glu substitution mutation in the thrombin cleavage site as described above, resulting in pET15b-DT(C+T)(R to Q). The data shown in Fig. 4 were obtained with the DT expressed from this plasmid.
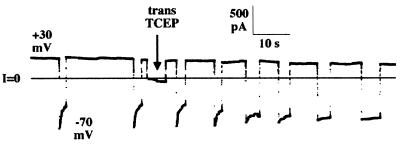
Figure 4.
Trans addition of the membrane-impermeant disulfide reducing agent TCEP eliminates rapid turn-off by protease-nicked, amino-terminal H6-tagged DT(C+T) channels. Before the start of the record, nicked H6-tagged DT(C+T) was added to the cis solution to a concentration of 110 ng/ml, and the voltage was held at +30 mV. The solutions on both sides of the membrane contained 1 M KCl, 2 mM CaCl2, and 1 mM EDTA; the cis solution contained 5 mM Mes (pH 5.3), and the trans solution contained 5 mM Hepes (pH 7.2). After channel activity appeared, the pHs of the cis and trans solutions were brought to 6.0 with Mes (final concentrations 35 mM cis, 60 mM trans). Channel conductance turned off rapidly at −70 mV. Four minutes before the start of the record, TCEP (pH 6.2) was added to the cis solution to a concentration of 100 mM, with no effect on channel conductance at both +30 and −70 mV. At the arrow, TCEP (pH 6.2) was added to the trans solution to a concentration of 100 mM. Subsequently, channel conductance failed to turn off at −70 mV. (TCEP addition to the trans solution, without prior addition to the cis solution, gave the same results as in this experiment.) The record was filtered at 10 Hz by the chart recorder.
A DNA fragment obtained by restriction digestion of pET15b-DT(C+T)(R to Q) with NcoI and EcoRI restriction endonucleases, which codes for the thrombin-resistant amino-terminal H6 tag and the amino acids 1–378 of DT, was cloned into the pET22b vector, resulting in pET22b-DT(C+T)(R to Q). This was further mutagenized (20) to introduce C186S, C201S, and N58C (or S146C) substitution mutations. The resulting plasmids, pET22b-DT(C+T)(R to Q)/58C/186S/201S or pET22b-DT(C+T)(R to Q)/146C/186S/201S, allowed the expression of single-cysteine mutants of DT(C+T) with a protease-resistant amino-terminal H6 tag. The data shown in Fig. 3 were obtained with these single-cysteine DT(C+T) constructs after biotinylation with a thiol reactive reagent, N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)proprionamide, essentially following described methods (21). All of the oligonucleotides used for PCR were from Integrated DNA Technologies (Coralville, IA). The nucleotide sequences of all the DT constructs were confirmed by DNA sequencing.
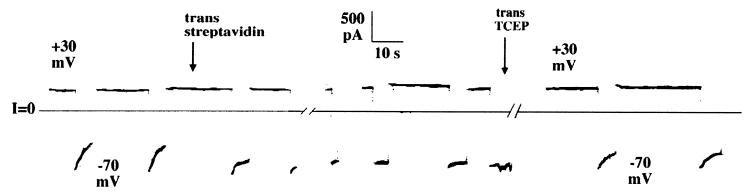
Figure 3.
Streptavidin added to the trans side prevents rapid turn-off by amino-terminal H6-tagged DT(C+T) channels to which biotin is linked at residue 146 via a disulfide bond; subsequent trans addition of the membrane-impermeant disulfide reducing agent TCEP reverses the trans streptavidin effect by cleaving the biotin. Native cysteines at position 186 and 201 were mutated to serines; residue 146 was mutated to cysteine and then biotinylated. Before the start of the record, H6-tagged DT(C+T)(R to Q)/146C/186S/201S) was added to the cis solution to a concentration of 400 ng/ml, and the voltage was held at +30 mV. The solutions on both sides of the membrane contained 1 M KCl, 2 mM CaCl2, 1 mM EDTA, and 30 mM Mes; the pH of the cis solution was 5.3 and that of the trans solution was 6.0. Channel conductance turned off rapidly at −70 mV. At the first arrow, streptavidin was added to the trans solution to a concentration of 30 μg/ml. Subsequently, channel conductance failed to turn off at −70 mV. The first break lasted 35 seconds. At the start of the second break, as indicated by the second arrow, the membrane-impermeant disulfide reducing agent TCEP (pH 6.3) was added to the trans solution to a concentration of 60 mM. The second break lasted 6 min. After the TCEP addition, channel conductance turned off rapidly at −70 mV. The record was filtered at 10 Hz by the chart recorder. Records similar to this one were obtained with the biotinylated DT(C+T)(R to Q)/58C/186S/201S) mutant.
Protein Expression.
DT, or DT(C+T) mutants, were expressed periplasmically (pET22b-derived plasmids) or cytoplasmically (pET15b-derived plasmids) in E. coli BL21(DE3) (Novagen). Cell extracts were prepared from the periplasm (22) or cytoplasm, and proteins were purified by using Ni2+ affinity chromatography according to the Novagen manual followed by gel filtration chromatography. Proteins were concentrated with centrifugal concentrators (Amicon). Single-cysteine mutants of DT(C+T) were stored in the presence of 5–10 mM DTT.
Nicking.
Whole DT was nicked by treatment with thrombin (Novagen, 1 unit/1 mg DT in 0.5–1 ml volume) for 8 hours at room temperature; the reaction was stopped with Benzamidine Sepharose 6B (Amersham Pharmacia). DT(C+T) was nicked by incubation with 1% (wt/wt) of porcine trypsin (Sigma) for 30 min in the presence of 1 mM NAD+ (Boeringer Mannheim); the reaction was stopped with soybean trypsin inhibitor (Sigma). Nicking was verified by SDS/PAGE under reducing or nonreducing conditions. Nicked proteins were further purified by using Ni2+ affinity chromatography. All proteins were stored frozen at −20 or −80°C.
Bilayer Experiments.
Planar lipid (asolectin) bilayer membranes were made and recorded from as previously described (18). The sign of the voltages is that of the cis solution.
RESULTS
In both whole DT and DT(C+T), the H6 tag was attached to the amino terminus of the catalytic domain. The presence of this H6 tag caused an effect on the voltage gating of DT or DT(C+T) channels that was qualitatively similar to that observed with H6-tagged T domain alone. Thus, channels formed by amino-terminally H6-tagged DT or DT(C+T) close rapidly at negative voltages (Fig. 2B), whereas channels formed by these proteins lacking the H6 tag do not (Fig. 2A). [The response to positive and negative voltages of a membrane treated with DT or DT(C+T) (with no attached amino-terminal histidine tag) is complicated by the dual effect of voltage on channel behavior (11, 23). These complicated aspects of the voltage-dependent gating, which have not been completely worked out, need not concern us. The only relevant point for our purposes is that channels formed by DT or DT(C+T), with no amino-terminal histidine tag, never show a rapid closure at the negative voltages used here.]
Localization of the Amino Terminus of the C Domain to the Trans Side.
Addition of Ni2+ to the trans compartment prevented the rapid closure of channels formed by amino-terminally H6-tagged DT (Fig. 2B). Ni2+ in the cis compartment had no effect. Thus the amino terminal H6 tag has been translocated from the cis to the trans side of the membrane.
Localization of Residues Within the C Domain to the Trans Side.
Biotin was attached to a cysteine introduced at either residue 58 or 146 in the catalytic domain. The addition to the trans solution of streptavidin, which binds tightly to biotin, interfered with the rapid H6 tag-induced closure (Fig. 3), thereby demonstrating that these residues are also on the trans side. This effect of streptavidin was not seen with unbiotinylated DT(C+T), nor was it seen if free biotin was added to the trans compartment before streptavidin.
Localization of the Carboxyl Terminus of the C Domain to the Trans Side.
Addition of tris-(2-carboxyethyl)phosphine (TCEP) (24), a membrane-impermeant disulfide reducing agent, to the trans compartment abolished rapid channel closure by trypsin-nicked H6-DT or H6-DT(C+T), whereas cis TCEP had no effect (Fig. 4). There was no effect of TCEP if these proteins had not been nicked. These results imply that severing both the peptide and disulfide linkages between the C and T domains allows the C domain to diffuse away from the membrane, thereby eliminating the effect of the H6 tag on the T domain channel.
DISCUSSION
We have shown that associated with channel formation in planar phospholipid bilayers by the T domain of diphtheria toxin, its catalytic domain is translocated across the membrane. That the N-terminal H6 tag of the catalytic domain is on the trans side of the membrane after DT channel formation was demonstrated by showing that addition of Ni2+ to the trans compartment prevented rapid channel closure. Thus the H6 tag, and presumably the amino terminal end of the catalytic domain, is translocated from the cis to the trans side of the membrane on channel formation by DT.
Moreover, not just the amino terminal end of the catalytic domain is translocated to the trans side of the membrane, but apparently the entire domain. This is evidenced by experiments in which biotin was attached to a cysteine introduced at either residue 58 or 146 in the catalytic domain. The addition to the trans solution of streptavidin, which binds tightly to biotin, interfered with the rapid H6 tag-induced closure (Fig. 3), thereby demonstrating that these residues are also on the trans side.
If the catalytic domain is translocated to the trans side of the membrane and, as reported earlier (18), the amino terminus of the isolated T domain is similarly translocated, it is likely that the region connecting them also resides on the trans side. If this is the case, the C186–C201 disulfide linking the carboxyl terminus of the C domain to the amino terminus of the T domain (see Fig. 1) should be accessible to a membrane-impermeant reducing agent added to the trans compartment. Indeed, we found that addition of TCEP (24), a membrane-impermeant disulfide reducing agent, to the trans compartment abolished rapid channel closure by trypsin-nicked H6-DT or H6-DT(C+T), whereas cis TCEP had no effect (Fig. 4). Also, as expected, there was no effect of TCEP if these proteins had not been nicked. These results are consistent with the notion that severing both the peptide and disulfide linkages between the C and T domains allows the C domain to diffuse away from the membrane, thereby eliminating the effect of the H6 tag on the T domain channel. The ability of the C186–C201 disulfide bond to be translocated in this system lends credence to the hypothesis that reduction of the disulfide in vivo occurs on contact with cytosolic reducing agents, such as glutathione (25).
There is a quantitative difference between our results with whole DT and DT(C+T) on the one hand, and our previous results with T domain alone. Whereas T domain channels with an amino-terminal histidine tag close rapidly and almost completely at negative voltages as small as −5 mV, DT and DT(C+T) channels with an amino terminus histidine tag do not show significant closure at voltages less negative than −60 mV; even at −90 mV, the rate and completeness of closure is not as great as that seen at −30 mV for the corresponding T domain channels. Because the channels formed by whole DT, DT(C+T), and T domain are identical in their permeability properties (and therefore in their basic structure) to those formed by the 57-aa carboxyl terminus of the T domain (26), the quantitative difference in closure rates caused by the amino-terminal histidine tag is probably a consequence of differences in length and folding of the swinging arm to which the histidine tag is attached.
The fact that H6-tagged forms of DT and DT(C+T) gave similar results indicates that the R domain is not required for the translocation. Records essentially identical to that depicted in Fig. 2B were obtained when both the arginine-rich loop and the C186–C201 disulfide connecting the C and T domains were left intact, when the loop had been nicked by limited proteolysis, or when the disulfide had been eliminated by reduction or mutation. As expected, when the loop was nicked and the disulfide was reduced, rapid closure was not observed, because the catalytic domain with its amino terminal H6 tag was no longer attached to the T domain.
The results presented here argue that when the T domain inserts into the membrane, the entire catalytic domain (which is covalently linked to it) is translocated to the trans side of the membrane. It might be argued that the driving force for the translocation process we have observed comes from the interaction of the amino-terminal H6 tag with the applied electric field. However, this possibility is negated by the finding that translocation still occurs with H6-tagged constructs, and channels still form with untagged constructs, at 0, or even slightly negative (−10 mV) voltages, when there is a transmembrane pH gradient (pH 5.3 cis; pH 7.2 trans). Our results are consistent with those of London and coworkers (27), which indicate that the catalytic domain of DT lacking a H6 tag can be translocated across liposomal membranes when the toxin inserts under low pH conditions.
The mechanism of translocation by the T domain remains to be elucidated. Interestingly, translocation still occurs with an intact disulfide bridge between residues 186 and 201. It is not clear at this point what role the channel per se plays in the process. Whatever the mechanism, the data presented indicate that all of the translocation machinery is contained within the T domain, and thus that translocation does not absolutely require any other part of the toxin or any cellular proteins, such as a receptor. (It remains possible, however, that chaperonins facilitate refolding of the catalytic domain in the cytosol.) Probably, a similar mechanism applies to protein translocation across lipid bilayers seen with colicin Ia (21), and we anticipate that similar mechanisms of translocation may occur in certain other AB toxins, such as botulinum toxin, which contains an α-helical translocation domain (28), and possibly in eukaryotic proteins, such as Bcl-xL, which contain α-helical domains resembling the T domain of DT (9).
Acknowledgments
We thank Carl Davis, Can Cui, and Rachel Legmann for their help in constructing DT plasmids, and Karen Jakes, Paul Kienker, and Stephen Slatin for their helpful comments on the manuscript.
ABBREVIATIONS
- DT
diphtheria toxin
- T domain
transmembrane or translocation domain
- C domain
catalytic domain
- H6 tag
hexahistidine tag
- TCEP
tris-(2-carboxyethyl)phosphine
References
- 1.Gill D M. In: Bacterial Toxins and Cell Membranes. Jeljaszewicz J, Wadstrom T, editors. London: Academic; 1978. pp. 291–322. [Google Scholar]
- 2.Madshus I, Stenmark H. Curr Top Microbiol Immunol. 1992;175:1–26. doi: 10.1007/978-3-642-76966-5_1. [DOI] [PubMed] [Google Scholar]
- 3.Choe S, Bennett M J, Fujii G, Curmi P M G, Kantardjieff K A, Collier R J, Eisenberg D. Nature (London) 1992;357:216–222. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- 4.Bennett M J, Eisenberg D. Protein Sci. 1994;3:1464–1475. doi: 10.1002/pro.5560030912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker M W, Pattus F, Tucker A D, Tsernoglou D. Nature (London) 1989;337:93–96. doi: 10.1038/337093a0. [DOI] [PubMed] [Google Scholar]
- 6.Wiener M, Freymann D, Ghosh P, Stroud R M. Nature (London) 1997;385:461–464. doi: 10.1038/385461a0. [DOI] [PubMed] [Google Scholar]
- 7.Elkins P, Bunker A, Cramer W, Stauffacher C. Structure (London) 1997;5:443–458. doi: 10.1016/s0969-2126(97)00200-1. [DOI] [PubMed] [Google Scholar]
- 8.Vetter I R, Parker M W, Tucker A, Lakey J, Pattus F, Tsernoglou D. Structure (London) 1998;6:863–874. doi: 10.1016/s0969-2126(98)00088-4. [DOI] [PubMed] [Google Scholar]
- 9.Muchmore S W, Sattler M, Liang H, Meadows R P, Harlan J E, Yoon H S, Nettesheim D, Chang B S, Thompson C B, Wong S-L, Ng S-C, Fesik S W. Nature (London) 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 10.Kagan B L, Finkelstein A, Colombini M. Proc Natl Acad Sci USA. 1981;78:4950–4954. doi: 10.1073/pnas.78.8.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donovan J J, Simon M I, Draper R K, Montal M. Proc Natl Acad Sci USA. 1981;78:172–176. doi: 10.1073/pnas.78.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eriksen S, Olsnes S, Sandvig K, Sand O. EMBO J. 1994;13:4433–4439. doi: 10.1002/j.1460-2075.1994.tb06765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanzrein M, Falnes P O, Sand O, Olsnes S. J Membr Biol. 1997;156:141–148. doi: 10.1007/s002329900196. [DOI] [PubMed] [Google Scholar]
- 14.Oh K J, Zhan H, Cui C, Hideg K, Collier R J, Hubbell W L. Science. 1996;273:810–812. doi: 10.1126/science.273.5276.810. [DOI] [PubMed] [Google Scholar]
- 15.Huynh P, Cui C, Zhan H, Oh K J, Collier R J, Finkelstein A. J Gen Physiol. 1997;110:229–242. doi: 10.1085/jgp.110.3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kachel K, Ren J, Collier R J, London E. J Biol Chem. 1998;273:22950–22056. doi: 10.1074/jbc.273.36.22950. [DOI] [PubMed] [Google Scholar]
- 17.Zhan H, Elliott J L, Shen W H, Huynh P, Finkelstein A, Collier R J. J Membr Biol. 1999;167:173–181. doi: 10.1007/s002329900481. [DOI] [PubMed] [Google Scholar]
- 18.Senzel L, Huynh P, Jakes K, Collier R J, Finkelstein A. J Gen Physiol. 1998;112:317–324. doi: 10.1085/jgp.112.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen W H, Choe S, Eisenberg D, Collier R J. J Biol Chem. 1994;269:29077–29084. [PubMed] [Google Scholar]
- 20.Howorka S, Bayley H. BioTechniques. 1998;25(5):764–766. doi: 10.2144/98255bm03. [DOI] [PubMed] [Google Scholar]
- 21.Qiu X-Q, Jakes K S, Kienker P K, Finkelstein A, Slatin S L. J Gen Physiol. 1996;107:313–328. doi: 10.1085/jgp.107.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ausubel F, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1988. [Google Scholar]
- 23.Hoch D H. Ph.D. thesis. Yeshiva University: Albert Einstein College of Medicine; 1985. [Google Scholar]
- 24.Burns J A, Butler J C, Moran J, Whitesides G M. J Org Chem. 1991;56:2648–2650. [Google Scholar]
- 25.Falnes P O, Madshus I H, Sandvig K, Olsnes S. J Biol Chem. 1992;267:12284–12290. [PubMed] [Google Scholar]
- 26.Silverman J A, Mindell J A, Zhan H, Finkelstein A, Collier R J. J Membr Biol. 1994;137:17–28. doi: 10.1007/BF00234995. [DOI] [PubMed] [Google Scholar]
- 27.Jiang J X, Chung L A, London E. J Biol Chem. 1991;266:24003–24010. [PubMed] [Google Scholar]
- 28.Lacy D B, Tepp W, Cohen A C, DasGupta B R, Stevens R C. Nat Struct Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]