Abstract
Mitochondria have emerged as central regulators of apoptosis. Here, we show that TID1, a human homolog of the Drosophila tumor suppressor lethal (2) tumorous imaginal discs, l(2)tid, encodes two mitochondrial matrix proteins, designated hTid-1L and hTid-1S. These splice variants are both highly conserved members of the DnaJ family of proteins, which regulate the activity of and confer substrate specificity to Hsp70 proteins. Both hTid-1L and hTid-1S coimmunoprecipitate with mitochondrial Hsp70. Expression of hTid-1L or hTid-1S have no apparent capacity to induce apoptosis but have opposing effects on apoptosis induced by exogenous stimuli. Expression of hTid-1L increases apoptosis induced by both the DNA-damaging agent mitomycin c and tumor necrosis factor α. This activity is J domain-dependent, because a J domain mutant of hTid-1L can dominantly suppress apoptosis. In sharp contrast, expression of hTid-1S suppresses apoptosis, whereas expression of a J domain mutant of hTid-1S increases apoptosis. Hence, we propose that TID1 gene products act to positively and negatively modulate apoptotic signal transduction or effector structures within the mitochondrial matrix.
The Drosophila l(2)tid gene has been classified as a tumor suppressor and encodes Tid56, a 56-kDa protein that is processed to a 50-kDa mitochondrial localized protein (1–3). Null mutants of Tid56 exhibit a lethal phenotype in which cells of the imaginal discs fail to differentiate and grow into lethal tumors. TID1, a human homolog of l(2)tid, encodes hTid-1, a 52-kDa protein with strong homology to Tid56 (4).
hTid-1 and Tid56 are members of the DnaJ family of proteins. DnaJ proteins act as cochaperones and specificity factors for DnaK proteins and their eukaryotic homologs, the Hsp70 family (5–8). This protein family is characterized by a J domain, a highly conserved tetrahelical domain that binds to Hsp70 chaperones and activates their ATPase activity. The canonical J domain protein, DnaJ, was cloned from Escherichia coli as a mutant that cannot support the replication of bacteriophage λ (9, 10). DnaJ/Hsp70 systems are involved in protein folding (11), protein degradation, assembly and disassembly of multiprotein complexes (7), and translocation of proteins across membranes (12).
The hyperproliferative phenotype of l(2)tid mutant embryos suggests that the Tid56 protein is involved in regulation of cell growth or death. Given the mitochondrial localization of Tid56 and the important role of mitochondria in regulating apoptosis (13, 14), the tumorous imaginal discs phenotype may reflect a failure of imaginal disc cells to properly integrate stimuli of cell death and survival. Several mitochondrial activities have been implicated in transducing, amplifying, and repressing apoptotic signals, including the release of cytochrome c and Apoptosis-Inducing Factor from the mitochondrial intermembrane space, the production of reactive oxygen species, and the loss of inner membrane potential. In addition, mitochondrial localization is important for the function of many of the Bcl-2 family of apoptosis regulators.
Here, we report that TID1 encodes two mitochondrial matrix localized splice variants of 43 and 40 kDa, which we have named hTid-1L and hTid-1S, respectively. Both hTid-1L and hTid-1S retain their respective J domains and coimmunoprecipitate with mitochondrial Hsp70 (mtHsp70). Expression of these proteins does not induce apoptosis, but surprisingly, expression of each of the two splice variants has opposing effects on a cell’s ability to respond to an exogenous apoptotic stimulus. hTid-1L expression increases apoptosis triggered by both tumor necrosis factor α (TNFα) and the DNA-damaging agent mitomycin c (MMC). A J domain mutant of hTid-1L is able to suppress apoptosis to levels well below control cells. In sharp contrast, hTid-1S is able to suppress apoptosis, and a J domain mutant of hTid-1S increases apoptosis. Expression of hTid-1L and hTid-1S affect cytochrome c release from the mitochondria and caspase 3 activation, both of which are downstream of the mitochondria in TNFα signaling. However, hTid-1L and hTid-1S do not affect the rate of caspase 8 activation, which is upstream of the mitochondria. Hence, hTid-1L and hTid-1S are two mitochondrial matrix-localized proteins that can regulate apoptotic signal transduction and may comprise a mechanism by which the mitochondria amplify or dampen apoptotic signals.
METHODS
Cell Lines and Reagents.
U20S cells were cultured in DMEM containing 10% fetal calf serum supplemented with 50 units/ml penicillin and 50 μg/ml streptomycin. Inducible hTid-1L, hTid-1S, H121QL, and H121QS were expressed in U20S cells from a plasmid containing a muristerone-inducible promoter (Invitrogen). Inducible cells lines were cultured under constant selection with 50 μg/ml Zeocin (Invitrogen) and 300 μg/ml G418. Gene expression was induced with 1 μM muristerone (Invitrogen) for 24 hours. SAOS-2 cells were cultured in DMEM containing 15% fetal calf serum and 50 units/ml penicillin and 50 μg/ml streptomycin.
Antibodies.
The mAbs against hTid-1 (RS-13 and RS-11) were produced by J. DeCaprio (Dana-Farber Cancer Institute, Boston, MA) by using standard methods and a glutathione S-transferase-hTid-1 fusion protein as the antigen. The cytochrome oxidase subunit 1 (COx1)- and cytochrome oxidase subunit 2 (COx2)-specific mAbs were purchased from Molecular Probes (A-6405 and A-6404, respectively). The anti-cytochrome c mAbs (65981A) and pro-caspase 8 mAbs (66231A) were purchased from PharMingen. The anti-pro-caspase 3 mAbs (C31720) were purchased from Transduction Laboratories (Lexington, KY). The anti-mtHsp70 mAbs (MA3–028) were purchased from Affinity Bioreagents (Golden, CO). The anti-Hsc70 mAbs were purchased from StressGen (Victoria, Canada) (SPA815).
PCR Cloning of hTid-1S.
Primers of sequence 5′-cgagacagatgtggagggga-3′ and 5′-gaataatttaaacacact-3′ were used to amplify TID1-related sequences from a human fetal brain cDNA library (CLONTECH).
Subcellular and Submitochondrial Fractionation.
For subcellular fractionation, SAOS-2 cells were trypsinized, washed in PBS, suspended in sucrose buffer (10 mM Tris⋅HCl pH 7.5/1 mM EDTA/0.25 M sucrose/1 μg/ml each aprotinin and leupeptin/0.01% PMSF), and homogenized by 20 strokes of a Teflon tissue homogenizer (Glas-Col, Terre Haute, IN). Nuclei were pelleted at 500 × g. Mitochondria were pelleted at 10,000 × g. hTid-1, COx1, and cytochrome c were visualized by Western blot.
For proteinase protection assays, U2OS cells were trypsinized and homogenized in sucrose buffer, and mitochondria were isolated as described above. Mitochondrial pellet was resuspended in hypotonic buffer (5 mM Tris⋅HCl/5 mM KCl/1.5 mM MgCl2/0.1 mM EDTA/1 mM DTT, pH 7.4) for 20 minutes on ice. The sample was split into three fractions. The first fraction was left untreated. The second fraction was treated with proteinase K (50 μg/ml) for 20 minutes on ice. PMSF (0.03%) and EGTA (1 mM) were added and the fraction was subjected to sonication for 90 seconds on ice in a Sonic Dismembranator 550 (Fisher Scientific). The third fraction was subjected to sonication and treated with proteinase K as described above. The three samples were then analyzed by using SDS/PAGE and immunoblot for the presence of hTid-1, COx2, and mtHsp70.
For digitonin extraction, SAOS-2 cells were trypsinized, washed in PBS, and resuspended in sucrose buffer, and mitochondria were isolated as described above. Mitochondria were resuspended in sucrose buffer containing the indicated concentration of digitonin (Sigma) for 1 minute on ice. Fractions were then diluted 1:5 and centrifuged for 15 minutes at 10,000 × g. Pellets and supernatants were then analyzed by Western blot for the presence of hTid-1, COx1, and cytochrome c.
Immunoprecipitation.
One 10-cm plate of SAOS-2 cells was trypsinized, washed in PBS, and lysed in 1% NP40/150 mM NaCl/50 mM Tris⋅HCl, pH 8.0/1 μg/ml each aprotinin and leupeptin/0.01% PMSF on ice. The sample was split into four fractions and incubated for 1 hour with either anti-hTid-1, anti-mtHsp70, or anti-Hsc70 mAbs. Immune complexes were collected on protein G agarose beads (GIBCO) and washed three times in 0.1% NP40 lysis buffer. Samples were then separated by SDS/PAGE, transferred to poly(vinylidene difluoride) membrane, and probed with either anti-mtHsp70, anti-Hsc70, or anti-hTid-1 mAbs. Proteins were visualized by ECL (Amersham Pharmacia) by using x-ray film.
Apoptosis Assays.
Inducible U2OS cells were induced with 1 μM muristerone for 24 hours, or went uninduced and were treated with either the indicated concentration of mitomycin c (Sigma) for 24 hours or the indicated concentration of TNFα plus 30 μg/ml cycloheximide. Cells were fixed by exposure to methanol vapor for 10 minutes followed by immersion in methanol for at least 10 minutes. Cells were stained with 1 μg/ml Hoechst 33258 and 0.1% lowfat milk (Carnation) for 7 minutes and rinsed in water. Apoptotic nuclei were counted by using fluorescence microscopy. For transient-transfection experiments, U2OS cells were transfected by the calcium phosphate technique by using BES-buffered saline (15). Six micrograms of the appropriate hTid-1 construct was transfected with 6 μg of green fluorescent protein DNA. Twenty-four hours after transfection, Hoechst 33342 was added to the media at 1 μg/ml final concentration for 7 minutes. Media was then slowly removed. Cells expressing green fluorescent protein were counted and scored for apoptotic nuclei.
Caspase Activation and Cytochrome c Release Assays.
U2OS cells were induced for 24 hours with 1 μM muristerone and treated for the indicated time with 10 ng/ml TNFα plus 30 μg/ml cycloheximide. Cells were trypsinized, washed in PBS, and resuspended in lysis buffer (1% NP40/50 mM Tris⋅HCl/150 mM NaCl, pH 8.0/1 μg/ml each aprotinin and leupeptin/0.01% PMSF), and protein concentration was analyzed by using the Bradford method (Bio-Rad). Sixty micrograms of total cell lysate was then analyzed by Western blot analysis using mAb specific for pro-caspase 3 and pro-caspase 8.
Caspase activity was measured by using fluorescent caspase 8 and caspase 3 activity assays (CLONTECH). Inducible U20S cells were induced with muristerone for 24 hours and treated with 10 ng/ml TNFα plus 30 μg/ml cycloheximide for 4.5 hours. Cells were trypsinized, combined with apoptotic cells in the tissue-culture media, counted, and assayed for caspase activity by the manufacturer’s protocol on a fluorescent plate reader (excitiation 380 nm, emission 530 nm). For caspase 8 activity, 1 × 106 cells of each line were used, and for caspase 3 activity, 3.5 × 105 cells of each line were used.
For cytochrome c release assays, U2OS cells were induced with muristerone for 24 hours and treated with 10 ng/ml TNFα plus 30 μg/ml cycloheximide for 4.5 hours. Cells were then trypsinized, resuspended in sucrose buffer, homogenized in a Teflon tissue homogenizer, and centrifuged at 10,000 × g for 10 minutes as described above. Postmitochondrial supernatant (100 μg of protein) was then analyzed by Western blot analysis for the presence of cytoplasmic cytochrome c. Cytochrome c levels were quantitated by using NIH image software.
RESULTS AND DISCUSSION
TID1 Encodes Two Mitochondria-Localized Splice Variants, hTid-1L and hTid-1S.
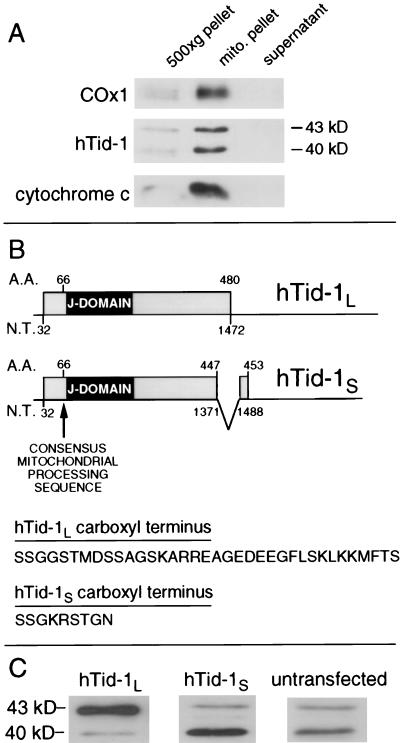
To detect endogenous TID1 related proteins, human osteosarcoma SAOS-2 cells were fractionated and analyzed by immunoblot by using hTid-1-specific mAbs. The major proteins detected by two independent mAbs, which we have named hTid-1L and hTid-1S, have apparent molecular masses of 43 kDa and 40 kDa, respectively. Both hTid-1L and hTid-1S fractionate with the mitochondrial proteins cytochrome c and COx1 (Fig. 1A). This result is consistent with immunofluorescence and immunoelectron microscopy experiments, which show that like Tid56, TID1-encoded proteins colocalize with mitochondria (T.D.-M., J.S., M. Grace, L. Y. Lee, and K.M., unpublished data).
Figure 1.
TID1 encodes two mitochondrial localized proteins, hTid-1L and hTid-1S. (A) SAOS-2 cells were homogenized, and nuclei were pelleted at 500 × g. Mitochondria were pelleted at 10,000 × g. Supernatant, 500 × g pellet, and 10,000 × g pellet were analyzed by immunoblot for the presence of COx1, hTid-1, and cytochrome c. (B) hTid-1L and hTid-1S are splice variants of TID1. hTid-1L mRNA encodes a protein with a predicted molecular mass of 52 kDa, which is cleaved at its amino terminus to form hTid-1L. Mature hTid-1L migrates with an apparent molecular mass of 43 kDa on SDS/PAGE. hTid-1S is encoded by an mRNA in which an exon encoding the carboxyl-terminal 33 aa of hTid-1L is removed and replaced with an exon from the 3′-untranslated region of hTid-1L mRNA, which encodes 6 aa and a stop codon. hTid-1S mRNA encodes a protein with a predicted molecular weight of 49.5 kDa, which is cleaved at its amino terminus to form hTid-1S. Mature hTid-1S migrates with an apparent molecular mass of 40 kDa on SDS/PAGE. Both hTid-1L and hTid-1S have a consensus mitochondrial cleavage site at amino acid position 66. (C) Expression of a cDNA of hTid-1L (Left) or hTid-1S (Center) gives rise to proteins of 43 and 40 kDa, respectively, that comigrate on SDS/PAGE with the endogenous hTid-1 polypeptides from untransfected U2OS cells (Right).
Evidence from expressed sequence tag database searches suggested that hTid-1L and hTid-1S may represent the protein products derived from two alternatively spliced mRNAs (4). PCR analysis of a human embryonic brain-derived cDNA library revealed two TID1 cDNAs. The long form matched the TID1 cDNA originally cloned, whereas the shorter form represents the alternatively spliced form of TID1 predicted from analysis of the expressed sequence tag database. In the alternatively spliced cDNA, an exon encoding the carboxyl-terminal 33 aa and the stop codon from the original clone is replaced with an exon located within the 3′-untranslated region of the original clone that encodes 6 aa and a stop codon (Fig. 1B). Expression of the originally published TID1 cDNA clone, including the 3′-untranslated region, leads to the production of both hTid-1L and hTid-1S (data not shown). Expression of a TID1 cDNA in which the 3′-untranslated region has been removed leads to the production of only the 43-kDa band, which comigrates with endogenous hTid-1L on SDS/PAGE. Expression of the alternatively spliced form in cells leads to production of a 40-kDa band that comigrates with endogenous hTid-1S (Fig. 1C). Hence, we conclude that hTid-1L and hTid-1S are encoded by alternatively spliced mRNAs of the TID1 gene. Most mitochondrial matrix proteins encoded by nuclear DNA are cleaved at their amino terminus on entering the mitochondria. Both hTid-1L and hTid-1S have a predicted mitochondrial processing sequence (LRP-GV) (16) that would result in cleavage at amino acid 66 on entry into the mitochondria. The predicted mature hTid-1L and hTid-1S proteins consist of 415- and 388-aa residues and have predicted molecular masses of 45.6 and 42.7 kDa, respectively. Hence, the mature hTid-1L and hTid-1S represent cleavage products of cytoplasmic pre-proteins.
hTid-1L and hTid-1S Are Localized to the Mitochondria Matrix and Interact with mtHsp70.
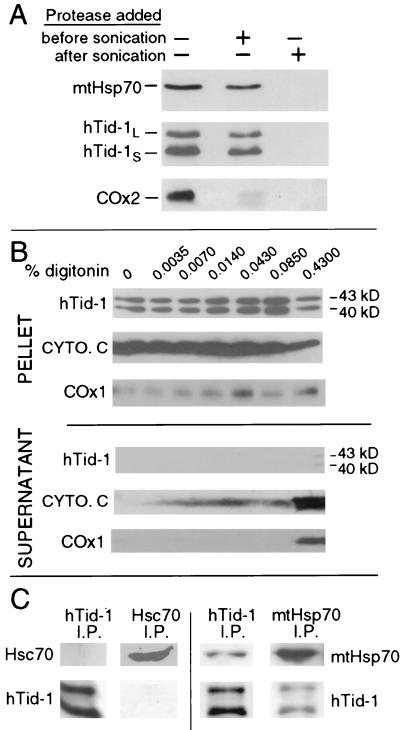
To determine the submitochondrial localization of hTid-1L and hTid-1S, mitochondria were subjected to a proteinase protection assay. Mitochondria were swelled in hypotonic buffer, which causes the outer membrane to rupture, and then treated with proteinase K before or after sonication, which ruptures the inner membrane. The samples were then analyzed by immunoblotting for the presence of hTid-1, matrix-localized mtHsp70 and the mitochondrial inner membrane protein COx2 (Fig. 2A). Addition of proteinase K before sonication left hTid-1L, hTid-1S, and mtHsp70 intact, but led to the proteolytic digestion of COx2, indicating that the intermembrane space was exposed to protease. Addition of proteinase K after sonication, however, led to complete proteolytic digestion of hTid-1L, hTid-1S, and mtHsp70. These results indicate that, like mtHsp70, hTid-1L and hTid-1S are mitochondrial matrix-localized proteins, because they are only vulnerable to proteinase after physical disruption of the inner mitochondrial membrane.
Figure 2.
hTid-1L and hTid-1S are localized to the mitochondrial matrix and form complexes with mtHsp70. (A) U2OS cells were homogenized, and mitochondria were isolated. Mitochondria were swelled in hypotonic buffer to burst the outer mitochondrial membrane. Samples went untreated or were treated with proteinase K before or after sonication, which disrupts the mitochondrial inner membrane. Samples were analyzed by immunoblot for the presence of matrix-localized mtHsp70, hTid-1, and the mitochondrial inner membrane protein COx2. COx2 is digested before sonication, indicating that the inner mitochondrial membrane is exposed to protease. hTid-1L, hTid-1S, and mtHsp70 are only digested when proteinase K is added after the mitochondrial inner membrane is disrupted by sonication. (B) Digitonin extraction of mitochondrial proteins. Mitochondria were isolated from SAOS-2 cells and treated with the indicated amount of digitonin. The intermembrane space protein cytochrome c (cyto. c), and the integral inner membrane protein COx1 are extracted from the mitochondria, whereas hTid-1L and hTid-1S are held in the pellet by a digitonin-resistant membrane. (C) Endogenous hTid-1L and hTid-1S coimmunoprecipitate with mtHsp70. Immunoprecipitation experiments were performed from U20S cells by using mAbs specific for either hTid-1, mtHsp70, or Hsc70. Immune complexes were analyzed by SDS/PAGE and Western blot with hTid-1-, mtHsp70-, or Hsc70-specific mAbs.
To confirm that hTid-1L and hTid-1S reside within the mitochondrial matrix, we extracted intermembrane proteins from isolated mitochondria with digitonin. Digitonin can selectively solubilize mitochondrial outer membranes while leaving inner membranes intact (17). Mitochondria were incubated with increasing amounts of digitonin, centrifuged, and analyzed for the presence of hTid-1, cytochrome c, and COx1 in both the pellet and supernatant (Fig. 2B). Cytochrome c levels in the supernatant increased with higher digitonin concentrations, as expected for an intermembrane space protein. The integral inner membrane protein COx1 was extracted only with the highest concentration of digitonin. In contrast, hTid-1L and hTid-1S were not efficiently extracted, even at the highest concentration, indicating that both hTid-1L and hTid-1S are protected by the digitonin-resistant inner mitochondrial membrane and thus reside in the mitochondrial matrix.
J domain proteins have been shown to interact with Hsp70-family proteins and activate their ATPase activity. Because hTid-1 proteins have extremely well conserved J domains, we suspected that they may be interacting with the mitochondrial Hsp70 homolog, mtHsp70 (GRP75), which is also localized to the mitochondrial matrix. We performed coimmunoprecipitation–immunoblot experiments by using mAbs specific for hTid-1, mtHsp70, or the constitutively expressed nonmitochondrial Hsp70 homolog Hsc70 as a control. hTid-1-specific mAbs immunoprecipitate endogenous hTid-1L and hTid-1S in complex with mtHsp70 from human U2OS cells. (Fig. 2C). In contrast, Hsc70 did not coimmunoprecipitate with hTid-1 proteins. The reverse experiment shows that mtHsp70 specific mAbs immunoprecipitate mtHsp70 in complex with hTid-1L and hTid-1S. Hsc70 antibodies did not coimmunoprecipitate hTid-1 proteins. These results demonstrate that endogenous hTid-1L and hTid-1S interact specifically with a mitochondrial matrix-localized Hsp70 homolog and suggest that they may function as specificity factors in an Hsp70-like chaperone system in the mitochondrial matrix.
hTid-1L and hTid-1S Have Opposing Effects on Apoptosis.
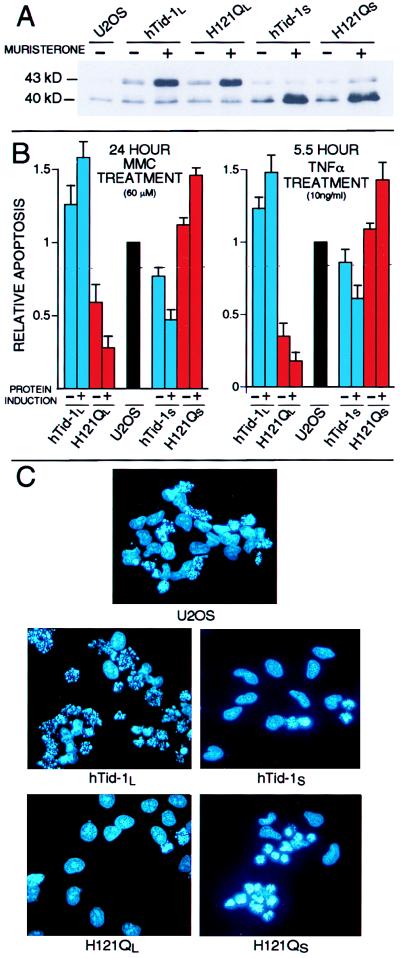
Mitochondria are central regulators and amplifiers of apoptotic signal transduction (13, 14, 18). On induction of apoptosis, mitochondria typically undergo a series of changes that are hallmarks of and functionally important for many forms of programmed cell death. Among these changes are the release of the caspase-activating protein cytochrome c (19–21) and apoptosis inducing factor (22, 23) from the mitochondrial intermembrane space, the production of a burst of reactive oxygen species, and a dramatic permeability transition of the mitochondrial inner membrane (13). In addition, Bcl-2 and related apoptotic regulatory proteins localize to mitochondrial membranes and functionally regulate the mitochondrial permeability transition pore as well as cytochrome c release (20, 24, 25). Given that hTid-1L and hTid-1S are localized to the mitochondrial matrix and are homologs of a Drosophila tumor suppressor, we tested whether expression of these proteins could affect apoptosis. We created a series of U20S cells lines that express either wild-type hTid-1L or hTid-1S, or J domain mutants of these proteins (H121QL or H121QS, respectively) from a muristerone-inducible promoter (Fig. 3A). This mutation of a highly conserved histidine residue is known to abrogate J domain-mediated activation of Hsp70 proteins in other systems (26, 27). Because these mutations should not affect the ability of the protein to interact with substrate, they are predicted to act as dominant-negative forms of hTid-1L and hTid-1S. The inducible system allowed for some basal expression of our hTid-1 constructs in the absence of muristerone, but induction of protein expression in these cells with muristerone produced protein levels ≈5- to 10-fold above basal expression levels. On protein induction, no cytoplasmic hTid-1 proteins were detected, demonstrating that all induced proteins are targeted to the mitochondria.
Figure 3.
hTid-1L and hTid-1S regulate apoptosis induced by mitomycin c and TNFα. (A) U2OS cells that express hTid-1L, hTid-1S, or J domain mutants (H121QL and H121QS, respectively) from a muristerone-inducible promoter were treated with muristerone for 24 hours (+) or went untreated (−) and were analyzed by immunoblot for the presence of hTid-1 proteins. (B) U2OS cells which express hTid-1L, hTid-1S or J domain mutants from a muristerone inducible promoter were either treated with muristerone (+) or mock-treated (−) for 24 hours and treated with 60 μM MMC for 24 hours (Left), or 10 ng/ml TNFα plus 30 μg/ml cycloheximide for 5.5 hours (Right), fixed, and stained with Hoechst. Apoptotic nuclei were counted and the numbers were compared with control cells. Rates of apoptosis in U2OS cells ranged from 20 to 30% for cells treated with MMC, and from 40 to 50% for cells treated with TNFα. The average of at least three independent experiments is shown. Error bars are ± 1 SD. (C) Fluorescence micrographs of Hoechst-stained U20S cells that inducibly express the indicated protein after 24-hour treatment with 60 μM MMC. Apoptotic cells display condensed and fragmented chromatin.
Induction of expression of protein per se did not elicit any detectable apoptosis in any of the four cell lines. However, when these cell lines were treated with either the DNA-damaging agent MMC or TNFα, the cell line expressing hTid-1L showed markedly increased levels of apoptosis relative to control cells, whereas cells expressing the J domain mutant of hTid-1L (H121QL) showed decreased levels of apoptosis compared with control cells (Fig. 3 B and C). In contrast, cells expressing hTid-1S showed decreased levels of apoptosis relative to control cells, whereas cells expressing the corresponding J domain mutant (H121QS) showed increased levels of apoptosis. The various hTid-1 constructs had the greatest effects on enhancing or repressing an apoptotic response when protein was induced with muristerone. However, a more modest effect was also seen in the absence of protein induction. We attribute these effects to basal expression from the inducible promoters (Fig. 3A). Similar results were obtained with multiple independent U2OS cell lines expressing each of the four forms of hTid-1. In addition, a similar pattern of apoptosis modulation was observed in transient-transfection experiments with U2OS cells (data not shown).
These results show that the two splice variants of TID1 have opposing effects on apoptosis. hTid-1L has proapoptotic activities, whereas hTid-1S has antiapoptotic activities. Significantly, these activities are J domain-dependent, because a mutation that is known to abrogate J domain-mediated activation of Hsp70 proteins in other systems (26, 27) is able to reverse the effects of the wild-type proteins, most likely by interfering with the activities of mitochondrial substrates that play important roles in propagating apoptotic signals. More specifically, we propose that because each of the mutant proteins has a different effect on apoptotic responses, each of the wild-type splice variants must have distinct cellular substrates and activities.
hTid-1L and hTid-1S Affect Cytochrome c Release and Caspase 3 Activation but Do Not Affect Caspase 8 Activation.
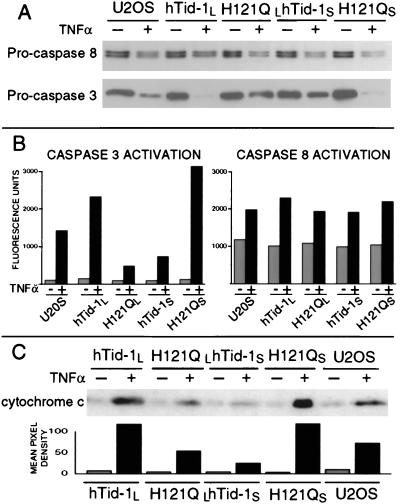
The finding that hTid-1L and hTid-1S can modulate apoptotic signal transduction at the cellular level in response to diverse stimuli led us to examine biochemical markers to localize the effects to the mitochondria and its known downstream targets. In TNFα signaling, pro-caspase 8 is cleaved and activated at the TNFα receptor complex (28). Active caspase 8 cleaves Bid, which then localizes to the mitochondria and elicits a proapoptotic response, including the release of cytochrome c (29, 30). Caspase 8 is therefore upstream of the mitochondria. Cleavage and activation of pro-caspase 3 is regulated by the release of cytochrome c from the mitochondria (13, 20, 21, 31) and is thus downstream of the mitochondria. Therefore, we examined the extent of pro-caspase 8 and pro-caspase 3 cleavage and activation in the four hTid-1-inducible cell lines on treatment with TNFα by immunoblot and fluorogenic activity assays (Fig. 4 A and B). We found that cleavage and activation of pro-caspase 8 occurs at similar levels in the four cell lines. However, pro-caspase 3 was cleaved and activated more efficiently in the cell line expressing hTid-1L than in control cells and less efficiently in the H121QL lines than in control cells. In contrast, hTid-1S-expressing cells showed decreased cleavage and activation of pro-caspase 3 relative to control cells, whereas the lines expressing H121QS showed increased activation of pro-caspase 3. Expression of hTid-1 proteins does not interfere with the normal turnover of pro-caspase 3 or 8, because their half-lives are similar in cells treated with cycloheximide alone (data not shown). These results indicate that expression of hTid-1L and hTid-1S affect apoptosis downstream of caspase 8 and upstream of caspase 3, which is consistent with a role as mitochondrial modulators of apoptosis.
Figure 4.
hTid-1L and hTid-1S affect the rates of caspase 3 activation and cytochrome c release but not the rate of caspase 8 activation. Inducible U2OS cells expressing hTid-1L, hTid-1S, or J domain mutants (H121QL and H121QS, respectively) were treated with 10 ng/ml TNFα and cycloheximide for 4.5 hours (+) or went untreated (−). (A) Whole-cell lysates were analyzed by immunoblot for pro-caspase 8 and pro-caspase 3. (B) Lysates were analyzed for ability to cleave fluorogenic caspase 8 (IETD-AFC) or caspase 3 substrates (DEVD-AFC). (C) Cells were suspended in sucrose buffer and homogenized. Samples were centrifuged at 10,000 × g, and cytoplasmic extracts were analyzed by immunoblot for the presence of cytochrome c. Mean pixel densities of cytochrome c Western blot analysis are shown Lower.
We next examined the rate of cytochrome c release from mitochondria on treatment with TNFα in the four inducible cell lines (Fig. 4C). Immunoblot analysis of cytoplasmic extracts of cells treated with TNFα indicate that more cytochrome c is released from the mitochondria of cells expressing hTid-1L or H121QS during apoptosis than control cells. In contrast, mitochondria from cells expressing hTid-1S or H121QL release less cytochrome c than control cells. These results are consistent with the effects seen at the level of cell death and caspase 3 activation and further localize the activity of TID1-encoded proteins to the mitochondria.
To date, specific mitochondrial factors implicated in apoptotic function have been localized to either the outer or inner mitochondrial membrane, the intermembrane space, or part of a membrane-bound complex.
hTid-1L and hTid-1S represent a class of mitochondrial matrix-localized proteins able to modulate this process. The opposing effects of the splice variants suggest a possible regulatory mechanism in which the relative abundance of hTid-1L and hTid-1S, or their cellular substrates, enable the mitochondria to either amplify or dampen apoptotic signals. Because expression of dominant-negative forms of hTid-1L and hTid-1S specifically dampen and enhance apoptotic responses, respectively, we suggest that each of the wild-type proteins has specific substrates and activities. Hence, hTid-1S is not simply a dominant negative form of hTid-1L, but rather a protein with discrete activities and substrates. In addition, the different activities of the mutant splice variants rule out titration of a common binding partner, such as mtHsp70, as a mechanism of action.
The mechanism underlying the l(2)tid hyperproliferative phenotype is unclear. However, the emergence of mitochondria as regulators of apoptosis suggests that the l(2)tid imaginal disc tumors may result from a defect in mitochondrial control of apoptosis. TID1 is a highly conserved human homolog of l(2)tid and encodes two splice variants that exhibit opposing effects of apoptosis.
Hence, we propose that hTid-1L and hTid-1S modulate apoptotic effector structures in the inner mitochondrial membrane, such as components of the permeability transition pore, which is regulated by the proapoptotic Bcl-2 family member Bax (25), or the F0F1 ATPase, which is implicated in Bax-mediated cell death (32). Alternatively, hTid-1L and hTid-1S may be part of an intramitochondrial signaling pathway that integrates disparate apoptotic initiating stimuli.
Acknowledgments
We thank Dr. J. DeCaprio (Dana-Farber Cancer Institute, Boston, MA) for the anti-hTid-1 antibodies and Dr. B. Schilling for the preparation of the hTid-1 immunogen. We thank Dr. Stanley Korsmeyer, Nicola Tolliday, and Dr. Lily Yeh Lee for their critical comments on the manuscript, John Daniel and other members of the Münger lab for their support and suggestions, and Miranda Grace for excellent technical assistance. This work was supported by Grants VM97 and RPG-94-011-04-VM from the American Cancer Society. J.S. is a Ryan Fellow. T.D.-M. is supported by the Charles A. King Trust (Fleet Investment Services, Boston, MA). K.M. is the recipient of a Junior Faculty Research Award (JFRA-597) from the American Cancer Society and is a Ludwig Scholar.
ABBREVIATIONS
- MMC
mitomycin c
- TNFα
tumor necrosis factor α
- mtHsp70
mitochondrial Hsp70
- COx1
cytochrome oxidase subunit 1
- COx2
cytochrome oxidase subunit 2
References
- 1.Kurzik-Dumke U, Gundacker D, Renthrop M, Gateff E. Dev Genet. 1995;16:64–76. doi: 10.1002/dvg.1020160110. [DOI] [PubMed] [Google Scholar]
- 2.Kurzik-Dumke U, Debes A, Gundacker D. In: Guidebook to Molecular Chaperones and Protein-Folding Catalysts. Gething M J, editor. Oxford: Oxford Univ. Press; 1997. pp. 117–121. [Google Scholar]
- 3.Kurzik-Dumke U, Debes A, Kaymer M, Dienes P. Cell Stress Chaperones. 1998;3:12–27. doi: 10.1379/1466-1268(1998)003<0012:mlateo>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schilling B, De-Medina T, Syken J, Vidal M, Munger K. Virology. 1998;247:74–85. doi: 10.1006/viro.1998.9220. [DOI] [PubMed] [Google Scholar]
- 5.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 6.Caplan A J, Cyr D M, Douglas M G. Mol Biol Cell. 1993;4:555–563. doi: 10.1091/mbc.4.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cyr D M, Langer T, Douglas M G. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 8.Misselwitz B, Staeck O, Rapoport T A. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 9.Bardwell J C, Tilly K, Craig E, King J, Zylicz M, Georgopoulos C. J Biol Chem. 1986;261:1782–1785. [PubMed] [Google Scholar]
- 10.Ohki M, Tamura F, Nishimura S, Uchida H. J Biol Chem. 1986;261:1778–1781. [PubMed] [Google Scholar]
- 11.Georgopoulos C, Welch W J. Annu Rev Cell Biol. 1993;9:601–734. doi: 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- 12.Pfanner N, Craig E A, Meijer M. Trends Biochem Sci. 1994;19:368–372. doi: 10.1016/0968-0004(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 13.Kroemer G, Zamzami N, Susin S A. Immunol Today. 1997;18:44–51. doi: 10.1016/s0167-5699(97)80014-x. [DOI] [PubMed] [Google Scholar]
- 14.Green D R, Reed J C. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.Gavel Y, von Heijne G. Protein Eng. 1990;4:33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- 17.Hartl F U, Schmidt B, Wachter E, Weiss H, Neupert W. Cell. 1986;47:939–951. doi: 10.1016/0092-8674(86)90809-3. [DOI] [PubMed] [Google Scholar]
- 18.Marchetti P, Castedo M, Susin S A, Zamzami N, Hirsch T, Macho A, Haeffner A, Hirsch F, Geuskens M, Kroemer G. J Exp Med. 1996;184:1155–1160. doi: 10.1084/jem.184.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed J C. Cell. 1997;91:559–562. doi: 10.1016/s0092-8674(00)80442-0. [DOI] [PubMed] [Google Scholar]
- 20.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 22.Susin S A, Zamzami N, Marzo I, Snow B E, Brothers G M, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, et al. Nature (London) 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 23.Susin S A, Zamzami N, Castedo M, Hirsch T, Marchetti P, Macho A, Daugas E, Geuskens M, Kroemer G. J Exp Med. 1996;184:1331–1341. doi: 10.1084/jem.184.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T I, Jones D P, Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 25.Marzo I, Brenner C, Zamzami N, Jurgensmeier J M, Susin S A, Vieira H L, Prevost M C, Xie Z, Matsuyama S, Reed J C, Kroemer G. Science. 1998;281:2027–2031. doi: 10.1126/science.281.5385.2027. [DOI] [PubMed] [Google Scholar]
- 26.Tsai J, Douglas M G. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 27.Wall D, Zylicz M, Georgopoulos C. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- 28.Nagata S. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Zhu H, Xu C J, Yuan J. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 30.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 31.Kluck R M, Martin S J, Hoffman B M, Zhou J S, Green D R, Newmeyer D D. EMBO J. 1997;16:4639–4649. doi: 10.1093/emboj/16.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama Q, Xu Q, Velours J, Reed J C. Mol Cell. 1998;1:327–336. doi: 10.1016/s1097-2765(00)80033-7. [DOI] [PubMed] [Google Scholar]