Abstract
Alterations in normal protein biogenesis and the resulting accumulation of improperly folded proteins in the endoplasmic reticulum (ER) trigger a stress response that up-regulates the expression of ER chaperones, while coordinately repressing overall protein synthesis and causing cell-cycle arrest. Activation of this unfolded protein response (UPR) in mouse NIH 3T3 fibroblasts with the glycosylation inhibitor tunicamycin led to a decline in cyclin D- and E-dependent kinase activities and to G1 phase arrest. Cyclin D1 protein synthesis was rapidly inhibited by tunicamycin treatment. However, the drug did not significantly affect the mitogen-dependent activities of the extracellular signal-activated protein kinases ERK1 and ERK2 or the level of cyclin D1 mRNA until much later in the response. Therefore, the UPR triggers a signaling pathway that blocks cyclin D1 translation despite continuous mitogenic stimulation. Enforced overexpression of cyclin D1 in tunicamycin-treated cells maintained cyclin D- and E-dependent kinase activities and kept cells in cycle in the face of a fully activated UPR. Translational regulation of cyclin D1 in response to ER stress is a mechanism for checkpoint control that prevents cell-cycle progression until homeostasis is restored.
Keywords: endoplasmic reticulum stress, cyclin-dependent kinases
Folding and maturation of all secretory pathway proteins occur in the endoplasmic reticulum (ER), a calcium-rich, oxidizing environment in which nascent chains are modified by the addition of asparagine-linked oligosaccharide chains and the formation of intrachain and interchain disulfide bonds (1). Pharmacological alterations in the ER redox potential, glycosylation machinery, or calcium levels can disrupt normal ER protein biogenesis, resulting in accumulation of misfolded proteins within the ER and triggering a complex chain of events termed the unfolded protein response (UPR; refs. 2 and 3). Activation of the mammalian UPR is characterized in part by increased transcription of at least seven genes encoding ER molecular chaperones, such as BiP/GRP78 (2), as well as induction of C/EBP homologous protein (CHOP), a transcription factor also known as growth arrest and DNA damage gene product-153 or GADD153 (4, 5). The increased synthesis of ER chaperones, which serve to correct protein misfolding, occurs concomitantly with a marked decrease in the rate of overall protein synthesis (6) and with arrest in the G1 phase of the cell division cycle (7, 8). Inhibition of protein synthesis lowers the overall rate of protein traffic into the ER, thus limiting damage to this organelle. The fact that this process is counterbalanced by an increased synthesis of specific ER chaperones highlights the specificity of this aspect of the UPR.
Mammalian cells contain at least three ER transmembrane signaling proteins that are thought to be the proximal effectors of the UPR. Ern1 and 2 (ER to nucleus) consist of an ER luminal domain that is believed to “sense” ER stress, a single membrane spanning segment, and a cytosolic tail containing both an essential serine/threonine kinase module and an RNase domain (5, 9). Experiments involving overexpression of both wild-type Ern and dominant-negative Ern mutants suggest that induction of ER chaperone genes and CHOP involves Ern activation (5, 9). The third ER transmembrane signaling protein, PERK, has an ER luminal domain and a cytosolic serine/threonine kinase domain that shares homology with the cytosolic RNA-dependent protein kinase (PKR; ref. 10). The UPR-mediated down-regulation of protein synthesis is accompanied by increased phosphorylation of eIF-2α, which impedes the formation of functional 40S translation–initiation complexes and inhibits translation (6). PERK is activated by ER stress in vivo and phosphorylates eIF-2α in vitro, suggesting that the UPR may use PERK to coordinate the more global repression of protein synthesis with Ern-induced transcriptional up-regulation of selected genes (10).
The mechanism underlying UPR-induced cell-cycle arrest has been largely unexplored. In principle, ER-stress conditions could indirectly impede cell-cycle progression by interfering with the proper maturation of growth factor receptors or other modulators of mitogenic signaling (11). Alternatively, ER stress may directly induce a checkpoint response that prevents cells from completing their cell division cycle under conditions that compromise the proper folding and assembly of proteins. In general, cell-cycle progression requires the activity of regulatory cyclins and their catalytic partners, the cyclin-dependent kinases (CDKs). Progression through G1 phase initially depends on holoenzymes composed of one or more of the D type cyclins (D1, D2, and/or D3) in association with either CDK4 or CDK6; this step is followed by activation of the cyclin E- and A-dependent kinase CDK2 as cells approach the G1/S transition (12). Cell-cycle arrest in response to mitogen deprivation or antiproliferative cytokines can be achieved through degradation of unstable cyclin subunits, by specific posttranslational modifications of the CDK subunits, or via association of active cyclin-bound CDKs with polypeptide CDK inhibitors (CKIs; refs. 13 and 14). Although the Cip/Kip CKIs (including p21Cip1, p27Kip1, and p57Kip2) act as potent inhibitors of cyclin E–CDK2 and cyclin A–CDK2, they positively regulate cyclin D–CDK assembly and remain bound to catalytically active cyclin D–CDK complexes (15, 16). Mitogen withdrawal inhibits cyclin D1 transcription and accelerates the turnover of the protein, leading to the rapid disassembly of cyclin D-dependent kinase complexes and to the release of Cip/Kip proteins from this latent pool. In turn, the mobilized CKIs can inhibit cyclin E–CDK2 and cyclin A–CDK2, thus preventing S phase entry and resulting in G1 phase arrest usually within a single cell cycle (14). Conversely, enforced ectopic expression of D type cyclins under experimental conditions in which their assembly with CDK4 is promoted resequesters Cip/Kip proteins, reactivates CDK2, and enables S phase entry (17).
Here, we show that the UPR initiates a rapid block in translation of cyclin D1 mRNA, resulting in the loss of cyclin D-dependent kinase activity. This loss, in turn, leads to inhibition of cyclin E- and A-dependent CDK2 and to cell-cycle arrest in G1 phase. Enforced expression of cyclin D1 can override UPR-induced cell-cycle arrest, underscoring its importance as a physiologic target of UPR signaling.
METHODS
Cells and Culture Conditions.
NIH 3T3 cells were maintained in DMEM supplemented with 10% (vol/vol) FCS, antibiotics, and glutamine (GIBCO). NIH 3T3 cells engineered to overexpress Flag epitope-tagged cyclin D1 or a stable mutant, cyclin D1(T286A) (18) were maintained in complete medium containing 7.5 μg/ml puromycin. Either 3 × 105 cells (100-mm diameter culture dishes) or 1.5 × 106 cells (150-mm diameter dishes) were seeded in complete medium and allowed to reach ≈75% confluence. Cells were shifted to fresh medium supplemented with 10% (vol/vol) FCS containing tunicamycin (Sigma) or thapsigargin (Sigma) at concentrations given in the figure legends or were washed twice with PBS and shifted to medium containing 0.1% FCS. Cells harvested by trypsinization thereafter were stained with propidium iodide and analyzed by flow cytometry to determine their DNA content (19).
Immunoprecipitation, Immunoblotting, and Protein Kinase Assays.
Cells were lysed in EBC buffer (50 mM Tris⋅HCl, pH 7.5/120 mM NaCl/0.5% Nonidet P-40/1 mM PMSF/20 units/ml of aprotinin/0.4 mM NaF). Proteins were resolved on denaturing polyacrylamide gels, transferred to nitrocellulose (Micron Separations, Westborough, MA), and reacted with primary antibodies. Immunoblotting of cyclin D1, CDK4, p27Kip1, p21Cip1, CHOP, and BiP was performed as described (17, 20); rabbit antiserum was raised against recombinant hamster CHOP protein. CDK2-p27Kip1 and CDK2-p21Cip1 complexes were precipitated from clarified lysates normalized for total protein concentration. Lysates were incubated with antiserum to the C terminus of mouse CDK2 (21) together with 30 μl of protein A Sepharose (Amersham Pharmacia). After three washes in EBC buffer, proteins in immune complexes were resolved on denaturing polyacrylamide gels, transferred to nitrocellulose membranes, and probed with antibodies to p27Kip1 or p21Cip1. Sites of antibody binding were visualized by using protein A-conjugated horseradish peroxidase (EY Laboratories) with chemiluminescence detection (enhanced chemiluminescence kit; Amersham Pharmacia). For extracellular signal-regulated protein kinase (ERK) analysis, cells were washed with PBS and lysed directly on the dish in gel sample buffer. Lysates representing equal cell equivalents were resolved on 10% denaturing polyacrylamide gels, and ERKs were analyzed by immunoblotting with a rabbit antibody specific for activated ERK1 and ERK2 (p44 and p42 MAPK) doubly phosphorylated at Thr-202 and Tyr-204 (9101S, New England Biolabs) or with rabbit antibody recognizing total ERK protein (SC-93, Santa Cruz Biotechnology). Sites of antibody binding were detected with an anti-rabbit Ig-conjugated horseradish peroxidase (New England Biolabs), followed by chemiluminescence as described above.
Assays for cyclin D1-dependent kinase were performed as described (22). For detection of CDK2 activity, cells were lysed in Tween-20 IP buffer (50 mM Hepes, pH 7.5/10 mM MgCl/2.5 mM EGTA/1 mM EDTA/1 mM DTT and the protease and phosphatase inhibitors indicated above). Clarified lysates (500 μg per sample) were incubated for 1 h at 4°C with antiserum directed to the CDK2 C terminus plus 30 μl of protein A Sepharose. Immune complexes were washed four times with IP buffer and two times with kinase buffer (50 mM Hepes, pH 7.5/10 mM MgCl/1 mM DTT/20 μM unlabeled ATP and protease and phosphatase inhibitors). Reactions initiated by the addition 10 μCi of [γ-32P]ATP (6,000 Ci/mmol; NEN) were incubated at 30°C for 25 min (with linear incorporation kinetics). Labeled proteins were denatured in sample buffer and separated on denaturing polyacrylamide gels before autoradiography.
Isolation and Analysis of RNA.
Total RNA was extracted from cultured cells by using the RNeasy kit (Qiagen, Valencia, CA) and analyzed by Northern blotting by using random primer labeled hamster BiP and mouse CHOP cDNA probes and hybridization procedures previously described (20). cDNA probes used under the same conditions included a 1.3-kb EcoRI fragment from mouse cyclin D1 (23) and human β-actin (CLONTECH).
Biosynthetic Labeling.
Subconfluent cells in 60-mm diameter culture dishes were incubated in methionine- and cysteine-free DMEM (Life Technologies, Gaithersburg, MD) for the final 30 min of tunicamycin treatment and then shifted to medium containing 150 μCi/ml Tran-35S-label (ICN) for the indicated time periods. Proteins were immunoprecipitated from cell lysates as described above. Radiolabeled proteins were electrophoretically separated on denaturing 10% polyacrylamide gels, visualized by autoradiography by using Amplify reagent (Amersham Pharmacia), and quantitated by image analysis.
RESULTS
Tunicamycin Triggers Cyclin D1 Loss and G1 Phase Arrest.
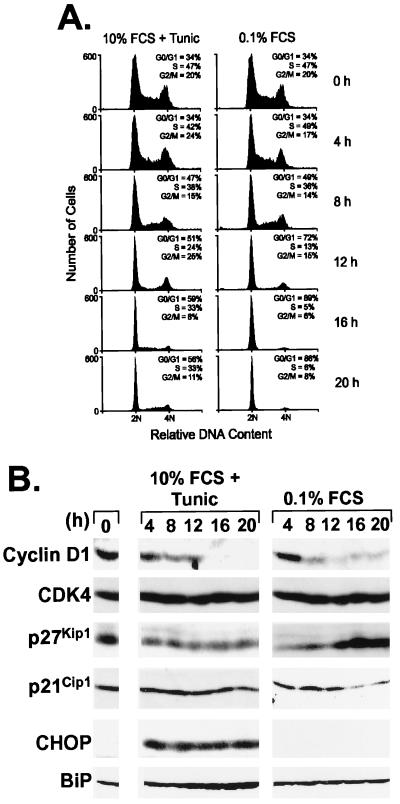
The ability of tunicamycin, a glycosylation inhibitor, to activate the UPR in a variety of cell types is well established (2). To assess the nature and kinetics of cell-cycle arrest induced by tunicamycin with that induced by growth factor withdrawal, mouse NIH 3T3 fibroblasts were either treated with 0.5 μg/ml tunicamycin in complete growth medium containing 10% (vol/vol) FCS or deprived of mitogens by placing them in medium containing 0.1% FCS but lacking the drug. Greater than 85% of the cells arrested in G0/G1 within 16 h of serum starvation, as indicated by the accumulation of cells with unreplicated (2N) DNA (Fig. 1A, right column). Treatment of cells with medium containing serum and tunicamycin also resulted in a marked accumulation of cells in the G0/G1-phase (Fig. 1A Left) in agreement with previous reports (8), although cell-cycle arrest was less complete than that observed with serum starvation. After 20 h of exposure to the drug (equivalent to ≈1 cell cycle), the vast majority of cells remained viable as judged by the absence of apoptotic cells containing less than 2N DNA content. However, longer drug treatment decreased cell viability, thus confounding analysis of cell-cycle dynamics beyond a single cycle.
Figure 1.
Loss of cyclin D1 correlates with tunicamycin-induced G1 arrest. (A) NIH 3T3 cells treated with 0.5 μg/ml tunicamycin in complete serum-containing medium (Left) or transferred to medium containing 0.1% FCS but no drug (Right) were assayed for DNA content by flow cytometry at the indicated times. Cells with a 2N DNA content (abscissa) are in G0/G1, whereas those with a 4N DNA content have completed S phase and are in G2 or M phase. The S phase fraction is represented by cells whose DNA content is between 2N and 4N. (B) Cells treated as above were lysed, and after separation of equal amounts of lysate proteins on denaturing polyacrylamide gels and transfer to nitrocellulose membranes, the indicated proteins were detected by direct immunoblotting by using cognate antibodies. All proteins were visualized by enhanced chemiluminescence, and exposures for Left and Right are matched.
The major cyclin D-dependent kinase in NIH 3T3 cells is D1–CDK4, which is activated in mid G1 phase before the appearance of cyclin E–CDK2 (22). These cells also express p21Cip1 and p27Kip1 but little p57Kip2 (16). After serum starvation or exposure to tunicamycin, we assayed the levels of cyclin D1, CDK4, and the two Cip/Kip proteins. The levels of cyclin D1 declined rapidly in both cell populations, whereas neither treatment resulted in significant changes in the abundance of CDK4 (Fig. 1B). Consistent with previous observations that p27Kip1 accumulates as mitogen-starved cells exit the cycle (14), the level of p27Kip1 increased within 16 h after serum withdrawal. In contrast, p21Cip1 synthesis is induced by mitogens (14), and its level decreased rapidly in serum-deprived cells (Fig. 1B Right). Markedly different responses occurred in tunicamycin-treated cells in which the amount of p27Kip1 declined, while p21Cip1 levels remained relatively constant (Fig. 1B Left). Thus, tunicamycin and serum starvation had opposing effects on p21Cip1 and p27Kip1 expression. As expected, tunicamycin treatment, but not serum starvation, induced the accumulation of both BiP and CHOP (Fig. 1B). These results suggested that G1 arrest induced by tunicamycin correlated more closely with a loss of cyclin D1 than with accumulation of CKIs of the Cip/Kip family. We also found that thapsigargin, an inhibitor of the ER Ca2+-ATPase and a potent activator of the UPR, induced a rapid loss of cyclin D1 in NIH 3T3 cells (data not shown). Loss of cyclin D1 was noted previously in human cancer A2780 and HT-29 cells treated with 2-deoxyglucose, glucosamine, and the calcium ionophore A23187, all of which induce ER stress and growth arrest (24). Thus, decreased expression of cyclin D1 occurs in response to ER stress induced in different cell types by various chemical stimuli.
A second class of CDK inhibitors, the INK4 proteins, can also be induced selectively in response to particular antiproliferative signals (25). Because formation of INK4 binary complexes with CDK4 leads to destabilization of cyclin D1 (26, 27), we considered the possibility that tunicamycin might trigger cyclin D1 loss through such a mechanism. NIH 3T3 cells have sustained a loss of the chromosomally linked loci encoding p16INK4a and p15INK4b, so we confined our analyses to the other two INK4 family members, p18INK4c and p19INK4d. Neither of these proteins accumulated during tunicamycin treatment (data not shown). Therefore, the observed loss of cyclin D1 was not secondary to INK4 protein induction.
Enforced Expression of Cyclin D1 Prevents Tunicamycin-Induced Growth Arrest.
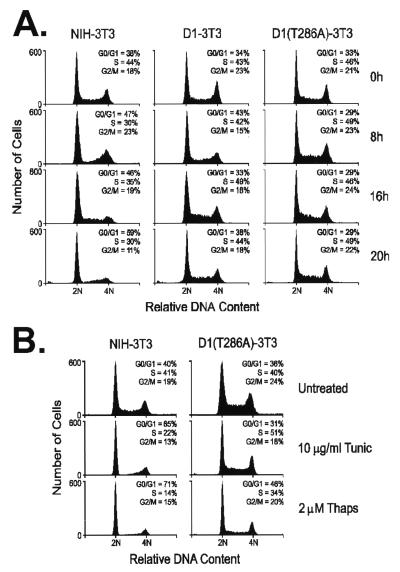
We next compared the effects of tunicamycin treatment on NIH 3T3 cells to the response of two NIH 3T3 derivatives previously engineered to overexpress constitutively either wild-type cyclin D1 or a mutated form containing an alanine for threonine substitution at codon 286 [D1(T286A)] (18). Phosphorylation of Thr-286 by glycogen synthase kinase-3β (GSK-3β) triggers the ubiquitination and proteasomal degradation of cyclin D1, so that the nonphosphorylatable D1 mutant is highly stable (t1/2 ≈ 3–4 h vs. t1/2 ≈ 25 min for wild-type D1; refs. 18 and 28). Overexpression of neither form of cyclin D1 is sufficient to render cells resistant to arrest on serum withdrawal (17, 18), because the cyclin D–CDK assembly normally requires mitogenic signaling (22). However, unlike parental NIH 3T3 cells (Fig. 2A Left), cells overexpressing wild-type (Fig. 2A Center) or mutant cyclin D1 (Fig. 2A Right) did not arrest in response to tunicamycin. Therefore, the reduction in the level of endogenous cyclin D1 observed after tunicamycin treatment of NIH 3T3 cells (Fig. 1B) must be mechanistically important in establishing G1 phase arrest. The remaining experiments shown below were performed with both D1-3T3 and D1(T286A)-3T3 cells; however, because virtually indistinguishable results were obtained, the pertinent data are illustrated for D1(T286A)-3T3 cells only. Overexpression of cyclin D1(T286A) prevented cell-cycle arrest at higher concentrations of tunicamycin, as well as by thapsigargin (Fig. 2B), indicating a general role for cyclin D1 loss in UPR-induced arrest.
Figure 2.
Enforced overexpression of cyclin D1 prevents tunicamycin-induced growth arrest. (A) After treatment of parental NIH 3T3 (Left), D1-3T3 (Middle), or D1(T286A)-3T3 (Right) cells with 0.5 μg/ml tunicamycin in complete medium, cells were harvested at the indicated times and assayed for DNA content by flow cytometry. (B) NIH 3T3 (Left) and D1(T286A)-3T3 (Right) cells were treated with the indicated doses of tunicamycin and thapsigargin for 20 h and then assayed for DNA content by flow cytometry.
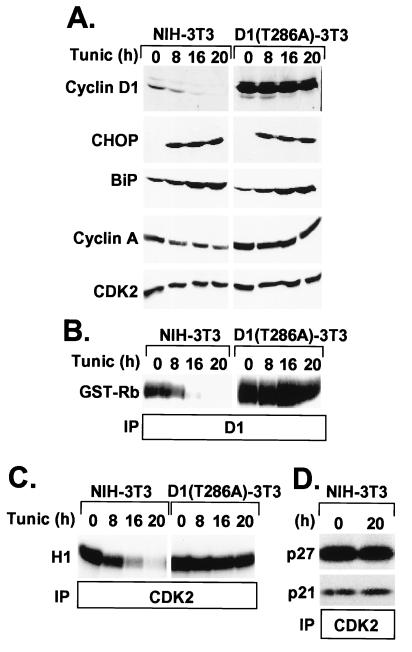
In contrast to NIH 3T3 cells in which cyclin D1 protein was undetectable after 16 h of drug exposure, the steady-state level of D1(T286A) was unchanged through 20 h of treatment (Fig. 3A). This result reflects the large pool of ectopically overexpressed D1(T286A) and its reduced rate of turnover as compared with endogenous cyclin D1. The observed induction of BiP and CHOP indicated that tunicamycin activated the UPR in both cell lines (Fig. 3A). Depletion of cyclin D1 in tunicamycin-treated NIH 3T3 cells was accompanied by the loss of retinoblastoma protein kinase activity in anti-D1 immune complexes (Fig. 3B) and the accumulation of hypophosphorylated retinoblastoma protein in the cells over the same time course (data not shown). However, the kinase activity of enzyme complexes recovered from D1(T286A)-3T3 cells was not diminished during drug treatment (Fig. 3B).
Figure 3.
Constitutive expression of cyclin D1(T286A) prevents loss of cyclin A and CDK2 catalytic activity. (A) Parental NIH 3T3 (Left) or D1(T286A)-3T3 (Right) cells treated with 0.5 μg/ml tunicamycin in complete serum-containing medium were assayed for expression of cyclin D1, BiP, CHOP, cyclin A, and CDK2 by immunoblotting. Immune complexes recovered with antibodies to cyclin D1 (B) or to CDK2 (C) were assayed for protein kinase activity by using retinoblastoma protein or histone H1 as substrates, respectively. (D) Lysates from untreated NIH 3T3 cells (0) or those treated with tunicamycin for 20 h (20) were precipitated with antibodies to CDK2. Denatured immune complexes were then separated on gels and blotted with antibodies to p27Kip1 or p21Cip1 (indicated at left) to score for complexes containing the CKIs. Although cyclin A levels fell in drug-treated cells (A), the recovery of equivalent amounts of Cip/Kip proteins after tunicamycin treatment suggests that the ratio of CKIs to cyclin bound CDK2 was increased, consistent with the observed inhibition of CDK2 kinase activity (C).
When CDK2 immune complexes (containing both cyclins E and A) were recovered from both cell types and tested for their ability to phosphorylate histone H1, CDK2 kinase activity was inhibited efficiently in NIH 3T3 cells but remained unaffected in the D1(T286A)-3T3 cells (Fig. 3C). This result underscores the dependency of cyclin E- and A-dependent kinases on formation of cyclin D–CDK4 complexes. In contrast to the profound loss of cyclin D1 in parental NIH 3T3 cells, the amounts of cyclin E (data not shown) and cyclin A were reduced by ≈50% after tunicamycin treatment, whereas CDK2 expression was unaffected (Fig. 3A, Cyclin A and CDK2). In the D1(T286A)-3T3 cells, however, the levels of all three proteins remained constant during the course of tunicamycin treatment (Fig. 3A; data for cyclin E not shown). The observed reduction in cyclin E and A levels in drug-treated NIH 3T3 cells would have been expected to yield some decrease in CDK2 activity, but other mechanisms must contribute to its complete elimination.
We reasoned that the remaining cyclin E–CDK2 and A–CDK2 complexes in tunicamycin-treated cells were sequestered in an inactive state by virtue of their association with either p21Cip1 or p27Kip1. Only a 3-fold increase in the ratio of bound Cip/Kip proteins to cyclin E–CDK2 is sufficient to compromise its catalytic activity severely and prevent S phase entry (29). The amounts of p21Cip1 or p27Kip1 coprecipitating with CDK2 were unchanged by tunicamycin treatment (Fig. 3D), despite the fact that cyclin A and p27Kip1 levels had both declined (Figs. 3A and 1B). Together, these data are compatible with the notion that cyclin–CDK2 complexes remaining after 20 h of tunicamycin treatment are maintained in an inactive state because of their increased association with p21Cip1 and p27Kip1.
Inhibition of Mitogenic Signaling Is Not Responsible for UPR-Mediated Cyclin D1 Loss.
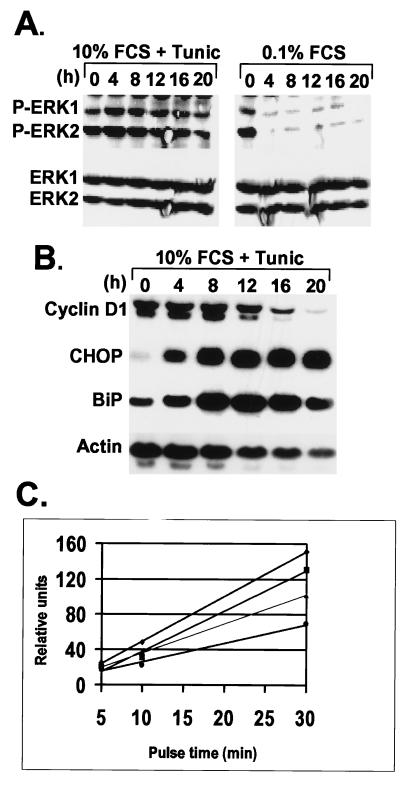
In NIH 3T3 cells, the activities of the extracellular signal-regulated kinases 1 and 2 (ERK1 and ERK2) are required for cyclin D1 expression. Mitogen-dependent activation of ERKs involves the sequential activation of Ras, Raf-1, and the mitogen-activated protein kinase kinases, which ultimately phosphorylate ERKs on conserved Thr and Tyr residues to generate the catalytically active enzymes. Lysates of NIH 3T3 cells cultured in either 10% (vol/vol) FCS plus tunicamycin or in medium containing 0.1% FCS were analyzed by immunoblotting with antibodies that either specifically recognize phosphorylated, active ERK isoforms (Fig. 4A Upper) or ones that detect both phosphorylated and nonphosphorylated ERK species (Fig. 4A Lower). Serum starvation resulted in the rapid loss of phosphorylated ERKs, but ERK1 and ERK2 remained phosphorylated throughout the 20-h course of tunicamycin treatment with only a slight decrease after 16 h. Hence, tunicamycin-induced cell-cycle arrest does not result from inhibition of mitogen-dependent ERK activation.
Figure 4.
Tunicamycin does not inhibit mitogenic signaling but blocks cyclin D1 translation. (A) After treatment of NIH 3T3 cells with tunicamycin or with medium containing 0.1% FCS, the levels of phosphorylated ERKs (Upper) were compared with the total ERK pool (Lower) by immunoblotting with antibodies that detect phosphorylated or all ERK isoforms, respectively. (B) After tunicamycin treatment of NIH 3T3 cells for the same time intervals, mRNAs encoding cyclin D1, CHOP, BiP, or actin were detected by Northern blotting performed with the cognate radiolabeled cDNA probes. (C) NIH 3T3 cells were left untreated (diamonds) or treated with 0.5 μg/ml tunicamycin for 2 h (squares), 4 h (triangles), or 12 h (circles) and then pulse-labeled for the indicated periods of time with Tran-35S-label (ICN). Cyclin D1 was immunoprecipitated from lysates, separated on a denaturing gel, and the rate of cyclin D1 synthesis was quantified by scanning the autoradiographs.
We therefore expected that tunicamycin treatment would not prevent cyclin D1 transcription, which depends to a great extent on persistent ERK activity. Northern blotting indicated that cyclin D1 mRNA levels remained unchanged through 8 h of tunicamycin treatment, during which the expected accumulation of CHOP and BiP mRNAs were observed (Fig. 4B). Only after more prolonged treatment did cyclin D1 mRNA decline (Fig. 4B). Because the depletion of cyclin D1 protein was apparent within 4 h of tunicamycin treatment (Figs. 1B and 3A), the acute stress-induced loss of the protein cannot be attributed to decreased D1 transcription.
Cyclin D1 proteasomal degradation is also a mitogen-regulated process (18). As discussed above, phosphorylation of Thr-286 by GSK-3β targets cyclin D1 for degradation via the 26S proteasome; however, because GSK-3β activity is down-regulated in mitogen-stimulated cells, cyclin D1 has its normal turnover rate of ≈25 minutes. Tunicamycin treatment neither increased GSK-3β activity nor accelerated cyclin D1 turnover (negative data not shown).
Tunicamycin Inhibits Translation of Cyclin D1.
It seemed likely that cyclin D1 loss in cells undergoing ER stress might be a direct result of the UPR-induced translational repression. In a series of metabolic labeling experiments performed with NIH 3T3 cells treated for various times with tunicamycin, a progressive reduction in the rate of cyclin D1 synthesis was observed that could be detected as early as 2 h after tunicamycin addition (Fig. 4C). Therefore, repression of cyclin D1 translation closely correlates with the rapid depletion of cyclin D1 protein in cells challenged by ER stress. Because tunicamycin provokes stress by inhibiting glycosylation within the ER lumen, whereas cyclin D1 is synthesized on non-membrane-bound polyribosomes, inhibition of cyclin D1 translation must involve signaling from the ER to the cytoplasmic protein synthesis machinery.
DISCUSSION
Pharmacological activation of the mammalian UPR leads to a reduced rate of cyclin D1 translation and to a rapid loss of cyclin D-dependent kinase activity. Concomitant inhibition of cyclin E- and A-dependent kinase activity depends secondarily on the release of Cip/Kip proteins from disrupted cyclin D–CDK complexes and their mobilization into complexes containing CDK2. Inhibition of both classes of G1 CDKs results in cell-cycle arrest. As shown here, the enforced expression of cyclin D1 in tunicamycin-treated cells was itself sufficient to prevent the loss of both CDK4- and CDK2-associated kinase activity and could thereby maintain the stressed cells in cycle. Under these conditions, other hallmarks of the UPR, such as BiP and CHOP induction, continued unabated, indicating that the ER-stress-induced transcriptional response was fully active in these cycling cells.
Accumulation of D type cyclins during G1 phase depends on persistent mitogenic stimulation. Conversely, growth factor withdrawal prevents cyclin D1 gene expression, and the relative instability of the protein ensures that cyclin D1 levels fall precipitously, thereby enabling mitogen-deprived cells to exit the cycle quickly. Although agents that interfere with protein biogenesis in the ER might conceivably hinder maturation of growth factor receptors, thereby compromising mitogenic signaling and braking the cell cycle (11, 24), several lines of evidence suggest that this interference is not the mechanism responsible for rapid growth arrest during the UPR. First, mitogen-dependent activation of ERKs was not inhibited by tunicamycin. Second, when ectopically expressed under conditions of ER stress, cyclin D1 continued to assemble into active cyclin–CDK complexes, a process that also depends on ERK signaling (17, 22). Third, although serum depletion normally results in p27Kip1 induction and loss of mitogen-responsive p21Cip1 (14), tunicamycin-induced stress led instead to p27Kip1 loss, whereas p21Cip1 levels remained constant. Finally, although mitogen withdrawal rapidly diminishes cyclin D1 transcription (23) and accelerates cyclin D1 turnover (18), the UPR affects neither of these processes acutely but instead leads to a rapid decrease in the rate of cyclin D1 translation. Therefore, growth factor deprivation and ER stress trigger cyclin D1 loss via distinct signaling pathways.
Clotrimazole, an antiproliferative agent that depletes cellular calcium stores and inhibits calcium influx, decreases protein translation by inducing phosphorylation and activation of PKR (30). The resulting inhibitory phosphorylation of eIF-2α blocks formation of the 40S translation initiation complex and decreases the synthesis of cyclins D1, E, and A. Overexpression of cyclin D1 partially overrides clotrimazole-induced cell-cycle arrest (30) in a manner similar to that observed in our experiments with tunicamycin-treated cells. Although PKR is activated when intracellular calcium stores are depleted (6, 30), cells nullizygous for PKR still respond to ER stress by reducing translation (10), implying compensation through an additional signaling pathway. Inhibition of cyclin D1 translation during the UPR might instead result from PERK-mediated phosphorylation of eIF-2α (10). Nonetheless, it remains unclear whether PERK, being localized to the ER membrane, has the capacity to influence cytoplasmic protein synthesis. The initiation factor eIF-4E, which serves as the cap binding protein and is a known positive regulator of cyclin D1 translation (31), represents another possible UPR target. Mitogens stimulate the efficiency of cyclin D1 translation by promoting the phosphorylation and activation of eIF-4E (32), and eIF-4E over-expression in NIH 3T3 cells increases cyclin D1 protein levels without altering its mRNA (31). Thus, the ER-stress pathway could inactivate eIF-4E through the action of an as-yet undefined UPR-regulated phosphatase or an eIF-4E kinase inhibitor.
Regardless of the exact signaling mechanism, the ability of cells overexpressing cyclin D1 to bypass ER-stress-mediated growth arrest highlights the critical nature of cyclin D1 as a proximal target of the UPR. The fact that activation of the UPR is not sufficient to induce the complete loss of other proteins necessary for maintenance of cell-cycle progression (e.g., CDK2, CDK4, and cyclin A)—whereas other proteins (e.g., BiP and CHOP) are concurrently induced—underscores the specificity of this response. At the very least, the effect of UPR-induced translational repression on the level of individual proteins must vary according to their pool size and half-life, as well as the efficiency of translation of their respective mRNAs. Cyclin D1 synthesis is particularly sensitive to translational repression, making it an excellent target for rapidly influencing the cell-cycle machinery. Indeed, the depletion of the short-lived cyclin D1 protein allows the UPR to “short-circuit” cell-cycle progression well before ER-stress conditions interfere with mitogenic signaling pathways and reduce cyclin D1 mRNA. Therefore, UPR-mediated repression of protein translation seems to serve at least two functions. The global response reduces ER traffic, thus limiting damage. Growth arrest depends on blocking the synthesis of cyclin D1, a specific rate-limiting regulator of G1 phase progression. The latter process likely represents a UPR-activated G1 checkpoint that provides time for reestablishment of normal cellular homeostasis.
Acknowledgments
The authors thank Dr. Richard Ashmun for his expertise and assistance with flow cytometric analyses and Dr. Jason Weber for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM54068 and Cancer Center CORE Grant CA21765 (to J.W.B. and L.M.H.), the Howard Hughes Medical Institute (to C.J.S. and J.A.D.), and the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital.
ABBREVIATIONS
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- CDK
cyclin-dependent kinase
- PKR
RNA-dependent protein kinase
- CKIs
CDK inhibitors
- ERK
extracellular signal-regulated protein kinase
- GSK
glycogen synthase kinase
References
- 1.Hammond C, Helenius A. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 2.Lee A S. Curr Opin Cell Biol. 1992;4:267–273. doi: 10.1016/0955-0674(92)90042-b. [DOI] [PubMed] [Google Scholar]
- 3.Sidrauski C, Chapman R, Walter P. Trends Cell Biol. 1998;8:245–249. doi: 10.1016/s0962-8924(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 4.Wang X-Z, Lawson B, Brewer J W, Zinszner H, Sanjay A, Mi L-J, Boorstein R, Kreibich G, Hendershot L M, Ron D. Mol Cell Biol. 1996;16:4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X-Z, Harding H P, Zhang Y, Jolicoeur E M, Kuroda M, Ron D. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brostrom C O, Brostrom M A. Prog Nucleic Acid Res Mol Biol. 1998;58:79–125. doi: 10.1016/s0079-6603(08)60034-3. [DOI] [PubMed] [Google Scholar]
- 7.Melero J A, Fincham V. J Cell Physiol. 1978;95:295–306. doi: 10.1002/jcp.1040950307. [DOI] [PubMed] [Google Scholar]
- 8.Carlberg M, Larsson O. Anticancer Res. 1993;13:167–171. [PubMed] [Google Scholar]
- 9.Tirasophon W, Welihinda A A, Kaufman R J. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harding H P, Zhang Y, Ron D. Nature (London) 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 11.Cai B, Tomida A, Mikami K, Nagata K, Tsuruo T. J Cell Physiol. 1998;177:282–288. doi: 10.1002/(SICI)1097-4652(199811)177:2<282::AID-JCP10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 12.Sherr C J. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 13.Morgan D O. Nature (London) 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 14.Sherr C J, Roberts J M. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 15.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 16.Cheng M, Olivier P, Diehl J A, Fero M, Roussel M F, Roberts J M, Sherr C J. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng M, Sexl V, Sherr C J, Roussel M F. Proc Natl Acad Sci USA. 1998;95:1091–1096. doi: 10.1073/pnas.95.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diehl J A, Cheng M, Roussel M F, Sherr C J. Genes Dev. 1998;12:192–229. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quelle D E, Ashmun R A, Shurtleff S E, Kato J-Y, Bar-Sagi D, Roussel M, Sherr C J. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 20.Brewer J W, Cleveland J L, Hendershot L M. EMBO J. 1997;16:7207–7216. doi: 10.1093/emboj/16.23.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka M, Kato J, Fisher R P, Morgan D O, Sherr C J. Mol Cell Biol. 1994;14:7265–7275. doi: 10.1128/mcb.14.11.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J-Y. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushime H, Roussel M F, Ashmun R A, Sherr C J. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 24.Tomida A, Suzuki H, Kim H D, Tsuruo T. Oncogene. 1996;13:2699–2705. [PubMed] [Google Scholar]
- 25.Ruas M, Peters G. BBA Rev Cancer. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 26.Serrano M, Hannon G J, Beach D. Nature (London) 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 27.Bates S, Parry D, Bonetta L, Vousden K, Dickson C, Peters G. Oncogene. 1994;9:1633–1640. [PubMed] [Google Scholar]
- 28.Diehl J A, Zindy F, Sherr C J. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 29.Reynisdóttir I, Polyak K, Iavarone A, Massagué J. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 30.Aktas H, Fluckiger R, Acosta J A, Savage J M, Palakurthi S S, Halperin J A. Proc Natl Acad Sci USA. 1998;95:8280–8285. doi: 10.1073/pnas.95.14.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenwald I B, Lazaris-Karatas A, Sonenberg N, Schmidt E V. Mol Cell Biol. 1993;13:7358–7363. doi: 10.1128/mcb.13.12.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonenberg N, Gingras A-C. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]