Abstract
Apoptosis signal-regulating kinase 1 (ASK1) is a pivotal component of a signaling pathway induced by many death stimuli, including tumor necrosis factor α, Fas, and the anticancer drugs cisplatin and paclitaxel. Here we report that ASK1 proapoptotic activity is antagonized by association with 14-3-3 proteins. We found that ASK1 specifically bound 14-3-3 proteins via a site involving Ser-967 of ASK1. Interestingly, overexpression of 14-3-3 in HeLa cells blocked ASK1-induced apoptosis whereas disruption of the ASK1/14-3-3 interaction dramatically accelerated ASK1-induced cell death. Targeting of ASK1 by a 14-3-3-mediated survival pathway may provide a novel mechanism for the suppression of apoptosis.
Apoptosis is a physiological process of cell death that plays a critical role in normal development as well as in the pathophysiology of a variety of diseases (1, 2). Extensive research in recent years has identified key molecules participating in a conserved pathway that mediates the highly ordered process of apoptotic cell death (for review, see refs. 3–5). It has also become clear that cell death pathways are intimately connected with cell survival pathways to ensure that cell death occurs only when needed. This association is achieved in part through specific targeting of the key components of proapoptotic signaling cascades by antiapoptotic mechanisms. For example, the release of cytochrome c from mitochondria and its activation of effector caspases are blocked by the actions of Bcl-2 and Bcl-XL (6–9). Activated Akt/protein kinase B promotes cell survival in part by phosphorylating critical proapoptotic proteins such as Bad (10, 11) and caspase-9 (12).
Apoptosis signal-regulating kinase 1 (ASK1), a Ser/Thr kinase, is a pivotal component of a signal-transduction pathway induced by a variety of stress stimuli (13–17). The kinase activity of ASK1 is stimulated by tumor necrosis factor (TNF) α via members of the TNF receptor-associated factor (TRAF) family (14, 18) and by Fas ligation via the Daxx protein (17). Catalytically inactive mutants of ASK1 block cell death induced by TNFα and Fas, suggesting the importance of ASK1 in transmitting signals to the death machinery (14, 17). ASK1 appears to be a general mediator of cell death because it is responsive to a variety of additional death signals, including oxidative stress (15, 19) and treatment with the chemotherapeutic drugs cisplatin and paclitaxel (16, 20). Consistent with this hypothesis, overexpression of ASK1 is sufficient to induce apoptosis in many cell types (14, 17). Regulation of ASK1 by both proapoptotic and antiapoptotic signals may provide a critical point of control for cell death and cell survival. Here we report that the proapoptotic activity of ASK1 is antagonized by its binding to 14-3-3 proteins.
MATERIALS AND METHODS
Mutagenesis.
Mutations K709R and S967A were introduced into ASK1 by using the QuikChange site-directed mutagenesis kit (Stratagene) with pcDNA3-HA-ASK1 (14) as a template. The primers used were 5′-gtcagaattgctattagggaaatcccagagagagac-3′ for K709R and 5′-ctcaggagtatagccttgccggtacctgtgc-3′ for S967A.
Cell Cultures, Transfection, and Immunoprecipitation.
HeLa, HEK293, and COS-7 cells were grown in DMEM with 10% fetal bovine serum and transfected with plasmid DNA by using Lipofectamine reagent (Life Technologies, Grand Island, NY). For immunoprecipitation, 4 × 105 cells were lysed in lysis buffer (1% Nonidet P-40/10 mM Hepes, pH 7.4/150 mM NaCl/5 mM NaF/2 mM Na3VO4/5 mM Na4P2O7/10 μg/ml aprotonin/10 μg/ml leupeptin/1 mM PMSF). Hemagglutinin (HA)-tagged fusion proteins were immunoprecipitated with an anti-HA mAb (12CA5; ref. 21) by using protein G Sepharose 4 Fast Flow (Amersham Pharmacia). After washing three times with lysis buffer, the HA immunoprecipitates (IPs) were resuspended in Hepes buffer (50 mM, pH 7.5) to assay for ExoS activation and for kinase activity. Endogenous 14-3-3 and ASK1 proteins were immunoprecipitated by using antibodies raised against 14-3-3 and ASK1, respectively (sc-629 and sc-9731; Santa Cruz Biotechnology). For the cell death assays described below, cells were transfected by using Fugene 6 (Roche Molecular Biochemicals).
Exoenzyme S (ExoS) Activation Assay.
HA-IPs were added to a reaction mixture containing (in a final volume of 25 μl) 0.2 M sodium acetate (pH 6.0), 20 nM purified ExoS, 30 μM soybean trypsin inhibitor, 30 μM [adenylate 32P-phosphate]NAD+ (Amersham Pharmacia), and 1.5 μM BSA. The reactions were carried out essentially as described (22). The specific incorporation of [32P]ADP ribose into the substrate soybean trypsin inhibitor catalyzed by activated ExoS was quantified in a Beckman scintillation counter (model LS6500). Purified ExoS (23) is a gift from J. Barbieri (Medical College of Wisconsin).
Solid-Phase Binding Experiments.
Hexahistidine (His)-tagged 14-3-3ζ and its mutant derivatives were immobilized on Ni2+-charged iminodiacetic acid-Sepharose 6B beads as described (24). Radiolabeled ASK1 proteins were generated from pcDNA3–HA–ASK1 in the presence of [35S]methionine (Amersham Pharmacia) by using the TNT in vitro transcription/translation system (Promega). For the in vitro binding assays, immobilized His–14-3-3 (5 μg) was mixed with 35S-labeled ASK1 in binding buffer (1% Nonidet P-40/137 mM NaCl/1 mM MgCl2/40 mM Tris⋅HCl, pH 8.0). The 14-3-3 complexes were extensively washed with binding buffer before resolution with SDS/12.5% PAGE. Radiolabeled proteins in the 14-3-3 complexes were visualized by using a PhosphorImager (Molecular Dynamics). For the peptide competition assay, immobilized 14-3-3ζ (0.2 μg) was incubated with peptides in a total reaction volume of 200 μl for 30 min before the addition of 35S-labeled ASK1. Peptides were synthesized at the Emory Microchemistry Facility. To test the requirement of phosphorylation for the ASK1–14-3-3 interaction, 35S-labeled ASK1 (5 μl) was treated with potato acid phosphatase (0.4 unit; Sigma) at 30°C in Pipes buffer (50 mM Pipes, pH 6.0/1 mM DTT) with or without phosphatase inhibitors (10 mM NaF/10 mM Na3VO4/10 mM sodium 2-glycerophosphate).
ASK1 Kinase Assays.
The kinase activity of immunoprecipitated HA–ASK1 was determined essentially as described (14, 19) in a reaction mixture containing 20 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, and 100 μM [γ-32P]ATP (Amersham Pharmacia) with or without 1 μg of myelin basic protein. After incubation, reaction mixtures were resolved with SDS/PAGE. Phosphorylation of ASK1 (autokinase activity) or myelin basic protein (trans-kinase activity) was quantified by using a PhosphorImager.
Cell Death Assays.
For the attachment-based viability assay, HeLa cells in 35-mm dishes were transiently transfected with a lacZ vector as a marker (pcDNA3.1/His B/lacZ; Invitrogen) together with plasmids coding for indicated proteins (Fig. 3; 0.25 μg of lacZ vector with 0.75 μg of ASK1 vectors or pcDNA3). Twenty-four hours after transfection, the medium was replaced with serum-free DMEM. After additional incubation to reach the indicated times, attached cells were harvested for the β-galactosidase (β-gal) assay (24). The β-gal activity was expressed as relative units with the control cell activity defined as 1. For nuclear morphology examination, HeLa cells were grown on glass coverslips and cotransfected with an enhanced green fluorescent protein (eGFP) marker plasmid (pTJM9; 0.25 μg) and vectors expressing test proteins (0.75 μg of pcDNA3–HA–ASK1 plus 0.25 μg of pcDNA–Flag–14-3-3 or a lacZ vector). Twelve hours after transfection, the medium was changed to serum-free DMEM. Twenty-four hours later, cells on the coverslips were mounted on glass slides by using Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). The fraction of eGFP-positive cells (transfected) that had condensed and fragmented nuclei was determined by using a fluorescence microscope. Expression of vector-encoded proteins was confirmed by immunoblotting by using anti-HA (12CA5) or anti-Flag (M2) antibodies.
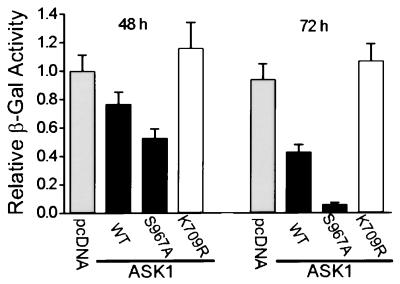
Figure 3.
The mutation S967A accelerates ASK1-induced cell death. HeLa cells were cotransfected with expression vectors for HA-tagged ASK1wt, ASK1S967A, or ASK1K709R and a vector for β-gal. At the indicated times, attached cells were harvested and assayed for β-gal activity. This activity reflects the viability of transfected cells only, because dead cells will detach from the culture dish. The control sample was transfected with an amount of pcDNA3 equal to the amount of ASK1 DNA. Results shown are mean ± standard error (n = 3) and are representative of three independent experiments.
DNA Fragmentation Analysis.
For monitoring the integrity of DNA, HeLa cells (1 × 106) were collected for DNA isolation. Total DNA was extracted with the Puregene DNA isolation kit (Gentra Systems). A portion of the DNA was analyzed by using 1.5% agarose gel electrophoresis.
RESULTS AND DISCUSSION
ASK1 Specifically Interacts with 14-3-3 Proteins.
On examination of the primary structure of ASK1, we found that the sequence C-terminal to the kinase domain of ASK1 contains a potential recognition motif for 14-3-3, RSIS967LP (25–27). 14-3-3 is a family of dimeric proteins that recognize and bind to phosphoserine motifs, such as those found in Raf-1 (25) and Bad (28). 14-3-3 binding plays an important role in regulating the function of Raf-1 (29, 30) and Bad (10, 11, 28). Interestingly, the potential 14-3-3-binding motif in ASK1 is conserved among its homologues from human, mouse, and Drosophila even though the overall sequence similarity in the C-terminal fragment is limited (31), suggesting the potential importance of this motif for ASK1 function. To test whether ASK1 is associated with 14-3-3 proteins in cells, we performed coimmunoprecipitation experiments. Human ASK1 proteins were transiently expressed in HeLa cells as HA-tagged fusions. HA–ASK1 IPs were isolated and then used to test whether expressed ASK1 interacts with endogenous 14-3-3 proteins. We had previously demonstrated that 14-3-3 proteins are essential cofactors for the catalytic activity of ExoS from Pseudomonas aeruginosa (22). This specific biochemical activity provides a sensitive and simple functional assay for the detection of 14-3-3 proteins (32, 33). As shown (Fig. 1A), incubation of HA–ASK1wt (wild type) IP with ExoS stimulated the catalytic activity of ExoS, indicating the presence of 14-3-3 in ASK1 immunocomplexes. The HA IP from cells expressing a control protein showed no detectable 14-3-3 activity. The catalytic activity of ASK1 is not required for 14-3-3 binding because the mutation K709R, which abolishes its kinase activity (14), showed no effect on ASK1–14-3-3 association. In reciprocal experiments, the presence of exogenously expressed HA–ASK1wt and HA–ASK1K709R was detected in 14-3-3 immunocomplexes from HeLa lysates (Fig. 1B). Similar results were obtained from HEK293 and COS-7 cells, confirming the ASK1–14-3-3 complex formation in vivo (data not shown). Furthermore, in untransfected Jurkat T cells, endogenous 14-3-3 was found in the ASK1 complex isolated with an anti-ASK1 antibody, whereas no 14-3-3 was detectable in the control ADP-ribosylation factor IP (ref. 34; data not shown).
Figure 1.
Human ASK1 specifically binds 14-3-3 in vivo and in vitro. HeLa cells were transiently transfected with plasmids coding for HA-tagged ASK1wt, ASK1S967A, ASK1K709R, or the control CAB1 (Clan of ARF-Binder 1; A. L. Boman and R. A. Kahn, unpublished data). CAB1 is an ADP ribosylation factor-associated protein that does not bind 14-3-3. Forty-eight hours after transfection, cell lysates were prepared. (A) ASK1 immunocomplexes contain 14-3-3 proteins. ASK1 complexes were immunoprecipitated by using an anti-HA antibody and assayed for the presence of 14-3-3 by its ability to activate the ADP ribosyltransferase activity of ExoS. The 14-3-3-dependent ExoS activity was expressed as pmol of ADP ribose incorporated into the substrate per min per pmol of ExoS. Data shown are representative of three experiments. Error bars represent standard error (n = 3). (B) 14-3-3 immunocomplexes contain ASK1. Endogenous 14-3-3 proteins were isolated from the same HeLa cell lysates as in A by using anti-14-3-3 serum. The 14-3-3 IPs were blotted for HA-ASK1 by using an anti-HA antibody (Upper). Lower shows similar expression levels of the HA-tagged proteins in total cell lysates. (C) Interaction of ASK1 with 14-3-3ζ is disrupted by binding-site mutations of 14-3-3ζ. Immobilized His-tagged 14-3-3ζ or control β-gal proteins (5 μg each) were incubated with 35S-labeled ASK1. After washing, bound proteins were resolved by using SDS/PAGE, and ASK1 was revealed by autoradiography (Upper). Similar amounts of immoblized proteins were used as revealed by Coomassie blue staining (Lower). (D) Peptide ligands of 14-3-3 inhibit ASK1–14-3-3 interactions. Peptides were preincubated with immobilized His–14-3-3ζ (0.2 μg) before adding 35S-labeled ASK1. After washing, 14-3-3ζ-bound ASK1 was quantified by PhosphorImager. The percentage of ASK1 bound to 14-3-3 relative to peptide-free samples is plotted against increasing concentrations of the test peptides. Error bars represent standard error (n = 3).
Previous studies have identified a site in 14-3-3 that mediates its interaction with diverse proteins (24, 35–38). Lys-49 and Val-176 of 14-3-3ζ are critical components of this site (24, 37). We tested whether ASK1 is specifically associated with 14-3-3 through this site by using an in vitro assay. Of the seven 14-3-3 isoforms found in mammalian cells, we chose 14-3-3ζ for this assay because it is widely expressed in various tissues (39). Consistent with the coimmunoprecipitation data, ASK1 was detected in complex with immobilized 14-3-3ζwt in vitro (Fig. 1C). The point mutations K49E and V176D, both of which disrupt the interaction of 14-3-3ζ with Raf-1 and several other ligands (24, 37, 40), abolished its interaction with ASK1. A more conservative mutation, K49R, which has no effect on 14-3-3 binding to previously studied ligands, showed no effect on 14-3-3ζ–ASK1 interaction. These results suggest that ASK1, like other 14-3-3 ligands, binds 14-3-3 through a common site. This conclusion is further supported by experiments with 14-3-3-binding peptides to disrupt 14-3-3–ASK1 interaction. Our cocrystallization studies have localized two peptide ligands in the binding groove of 14-3-3, a phosphorylated peptide derived from Raf-1, pS-Raf-259, and an unphosphorylated synthetic peptide, R18 (38). Indeed, pS-Raf-259 and R18 both effectively inhibited the interaction of ASK1 with 14-3-3ζ, with IC50 values of 2.0 μM and 0.2 μM, respectively (Fig. 1D). As a control, a nonphosphorylated Raf-259 peptide, which essentially cannot bind 14-3-3 (25), exhibited little inhibition (IC50 > 200 μM). These data demonstrate that ASK1 specifically interacts with 14-3-3, suggesting a possible involvement of 14-3-3 in the regulation of ASK1 function.
The Interaction of ASK1 with 14-3-3 Involves Ser-967.
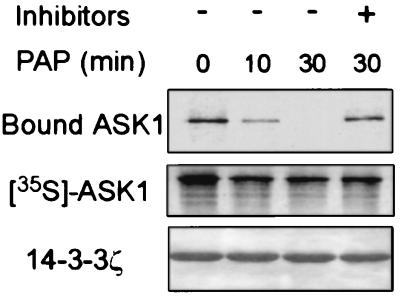
Many interactions with 14-3-3 are mediated by target phosphorylation (25, 26, 41, 42). To determine whether the ASK1–14-3-3 interaction is phosphorylation-dependent, we incubated 35S-labeled ASK1 with potato acid phosphatase for various lengths of time before adding immobilized 14-3-3ζ protein. As shown in Fig. 2, phosphatase treatment decreased the amount of ASK1 bound to 14-3-3ζ in a time-dependent manner, suggesting that dephosphorylation of ASK1 diminished its ability to bind 14-3-3. In support of this conclusion, treatment of intact cells with a permeable phosphatase inhibitor, okadaic acid, increased the amount of ASK1 complexed with 14-3-3 (data not shown). These data are consistent with the notion that the association of ASK1 with 14-3-3 requires phosphorylation. We then tested the possible involvement of the RSIS967LP motif of ASK1 in 14-3-3 binding by making mutant ASK1S967A, which lacks the predicted phosphorylation site that regulates 14-3-3 interaction. The effect of the mutation S967A on ASK1–14-3-3 association was tested by using coimmunoprecipitation experiments as shown in Fig. 1 A and B. HA–ASK1S967A IP contained <2% of the 14-3-3 activity of HA–ASK1wt IP, suggesting that S967A disrupted the ASK1–14-3-3 interaction. In a reciprocal experiment, 14-3-3 IP contained HA–ASK1wt but not HA–ASK1S967A (Fig. 1B). Consistent with these results, S967A abolished the interaction of ASK1 with 14-3-3 in an in vitro binding assay (data not shown). Together, these data demonstrate that Ser-967 is a critical component of the 14-3-3 interaction site of ASK1 and suggest that phosphorylation of this residue may be the major mode of regulation of 14-3-3 binding to ASK1. Because some 14-3-3 activity was detected in ASK1S967A IP (Fig. 1A), it is possible that secondary sites of ASK1 also contribute to 14-3-3 binding.
Figure 2.
Interaction of ASK1 with 14-3-3 is phosphorylation-dependent. 35S-labeled ASK1 was treated with potato acid phosphatase (PAP) for the indicated times with or without phosphatase inhibitors, and then incubated with 14-3-3ζ-coated beads. After washing, 14-3-3ζ-bound ASK1 was subjected to SDS/PAGE and autoradiography (Top). Middle shows phosphatase-treated ASK1. The amount of 14-3-3ζ in the beads was revealed by Coomassie blue staining (Bottom).
Disruption of 14-3-3–ASK1 Association Accelerates ASK1-Induced Apoptosis.
ASK1 has been shown to transmit TNFα- and Fas-induced death signals and promote apoptotic cell death (14, 15, 17). It is possible that 14-3-3 binding modulates ASK1-mediated apoptotic signaling. To test this model, we examined the effect of disrupting 14-3-3 interaction on ASK1-induced cell death. HeLa cells were cotransfected with a lacZ expression vector and plasmids expressing either HA–ASK1wt or its mutant derivatives. The extent of death among transfected cells was determined by decreased β-gal activity of the attached cells (Fig. 3). In agreement with previous reports (14, 15), overexpression of ASK1wt induced cell death, as evidenced by a time-dependent decrease in β-gal activity as compared with the control vector. ASK1-induced cell death was specific because expression of a catalytically inactive ASK1 mutant, K709R, did not lead to increased cell death. Interestingly, expression of ASK1S967A, a 14-3-3-binding defective mutant, drastically decreased the β-gal activity to a level one-seventh of that of ASK1wt-expressing cells at 72 hours. The effect of S967A on ASK1-induced cell death was not the result of differential expression of wt and mutant proteins because similar levels of ASK1 proteins were detected by immunoblotting (data not shown). The S967A effect is not limited to HeLa cells, because expression of ASK1S967A also enhanced cell death in other cell lines tested, such as HEK293 and COS-7 (data not shown). In addition, both the trypan blue exclusion assay and the 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye reduction assay (43) used with total cell populations gave results consistent with those obtained from the β-gal assay (data not shown). These data strongly suggest that association of 14-3-3 inhibits the death-promoting activity of ASK1. We then used both morphological and biochemical markers of apoptosis to characterize the nature of ASK1-induced cell death. HeLa cells were transiently transfected with expression plasmids for ASK1 or its mutant derivatives and a marker plasmid expressing eGFP. After 36 hours, cells were stained with DAPI and examined by using fluorescence microscopy. Cells cotransfected with eGFP and a control vector showed normal nuclear morphology with homogeneous DAPI staining (Fig. 4A). In contrast, expression of ASK1wt or ASK1S967A induced the appearance of irregularly stained nuclei showing chromatin condensation and nuclear fragmentation characteristic of apoptotic cells. Cells producing catalytically inactive ASK1K709R showed normal nuclear staining. The fraction of transfected cells that had apoptotic nuclear morphology was quantitated in a blind fashion (Fig. 4B Upper), which indicated that ASK1S967A expression led to an increased proportion of apoptotic cells as compared with ASK1wt. A hallmark of apoptosis in many cell types is the cleavage of genomic DNA into oligonucleosomal fragments. As shown in Fig. 4B Lower, expression of ASK1wt or ASK1S967A produced laddering of fragmented DNA like that seen in apoptotic cells. On the other hand, expression of the catalytically inactive ASK1K709R caused no obvious DNA laddering. Taken together, these experiments suggest that disruption of ASK1–14-3-3 association leads to enhanced proapoptotic activity of ASK1. The binding of 14-3-3 may contribute to suppression of the proapoptotic activity of ASK1 under normal survival conditions.
Figure 4.
14-3-3 binding inhibits ASK1-induced apoptosis. HeLa cells were cotransfected with an eGFP expression vector and test plasmids as indicated. Thirty-six hours posttransfection, cells were subjected to analysis. (A) ASK1-transfected cells show nuclear morphology characteristic of apoptosis, as determined by DAPI staining and fluorescence microscopy. Representative cells with apoptotic nuclei are indicated by arrows. (B) ASK1-induced apoptosis is modulated by 14-3-3 binding. Top shows the quantitation of apoptotic cell death by using nuclear morphology. The ratio of apoptotic transfected cells to total counted transfected cells is indicated above each bar. Middle shows expression levels of HA–ASK1 and Flag-tagged 14-3-3ζ proteins as determined by immunoblot. Bottom depicts the DNA integrity of transfected cells. Total DNA from the transfected samples was isolated, and an equal amount of DNA from each was separated on a 1.5% agarose gel.
Overexpression of 14-3-3ζ Suppresses ASK1-Induced Apoptosis.
To directly test whether the 14-3-3 expression level can regulate the proapoptotic activity of ASK1, we monitored ASK1-induced cell death in the presence of transiently expressed 14-3-3ζ. Indeed, overexpression of 14-3-3ζ blocked ASK1wt-induced apoptosis, with transfected cells showing normal nuclear morphology and intact genomic DNA (Fig. 4B). This 14-3-3 effect is specific for ASK1 because overexpression of 14-3-3ζ had no effect on ASK1S967A-induced cell death. Strikingly, 14-3-3ζ K49E, a ligand binding-defective mutant, exhibited a dominant-negative effect, giving rise to accelerated cell death on ASK1 coexpression. The level of cell death induced by coexpression of 14-3-3ζ K49E with ASK1wt is comparable to that induced by the ASK1 mutant, ASK1S967A. Thus, the above data confirm a critical role of 14-3-3 in controlling the death-promoting activity of ASK1.
It has been suggested that active suppression of cell death is required to maintain cell survival (44). We show here that the death-promoting activity of ASK1 is antagonized by its binding to 14-3-3 proteins. This finding identifies a novel mechanism for suppression of apoptosis through intercepting the ASK1-mediated proapoptotic process.
It has been demonstrated that ASK1 kinase activity is required for its death-promoting function (14, 17). 14-3-3 may suppress ASK1-induced cell death by directly inhibiting the catalytic activity of ASK1. However, we found that neither the autokinase activity nor the trans-kinase activity of ASK1 was altered by the addition of purified 14-3-3ζ protein to ASK1 IP (data not shown). Also, the 14-3-3-binding-defective ASK1S967A IP exhibited a time-dependent kinase activity indistinguishable from that of ASK1wt (data not shown). These data suggest that 14-3-3 may regulate ASK1 function through a mechanism independent of ASK1 kinase activity. However, an unequivocal conclusion requires further examination of ASK1 kinase activity under physiological conditions.
It is possible that 14-3-3 binding controls the interaction of ASK1 with its death effectors. ASK1 has been shown to be a mitogen-activated protein kinase kinase kinase that leads to the activation of the c-Jun N-terminal kinase (JNK, also known as stress-activated protein kinase, SAPK) and p38 mitogen-activated protein kinase pathways (13, 14, 18). Binding of 14-3-3 to ASK1 may inhibit the ability of ASK1 to activate JNK and p38 pathways or other potential effectors directly involved in apoptosis. Alternatively, 14-3-3 may determine the localization of ASK1 in cells, sequestering it in a compartment distinct from ASK1’s death effectors. 14-3-3 is known to bind many proteins involved in promoting cell survival and proliferation, such as Raf-1 (29). Thus, it is conceivable that 14-3-3 binding recruits ASK1 to phosphorylate substrates critical for cell survival. Our results also suggest that 14-3-3-mediated suppression of cell death is linked to the action of an upstream kinase that phosphorylates Ser-967 of ASK1. Such an event would place ASK1 under the control of a signal-transduction pathway responsive to cell survival stimuli. The ASK1–14-3-3 interaction is reminiscent of the Akt-induced Bad–14-3-3 interaction and suppression of Bad-induced cell death (refs. 10, 11, and 45; S. C. Masters, S. R. Datta, M. E. Greenberg, and H.F., unpublished data). It is possible that 14-3-3 targets and suppresses the proapoptotic activity of multiple death-promoting proteins in response to cell survival signals, thus acting as a general mediator of cell survival.
Acknowledgments
We thank H. Ichijo for generously providing pcDNA3–HA–ASK1, G. Pavlath for advice in using the fluorescence microscope, J. Barbieri, J. Pohl, R. Kahn, A. Boman, T. J. Murphy, H. Wang, H. Zhong, C. Zhang, J. Kuai, and R. Subramanian for reagents and assistance, and S. C. Masters and B. A. Wilson for critical reading of the manuscript and helpful comments. We also thank members of the Fu lab for assistance and enlightening discussions. This work was supported in part by National Institutes of Health Grant GM53165. H.F. is a recipient of the Burroughs Wellcome Fund New Investigator Award.
ABBREVIATIONS
- ASK1
apoptosis signal-regulating kinase 1
- β-gal
β-galactosidase
- DAPI
4′,6-diamidino-2-phenylindole
- eGFP
enhanced green fluorescent protein, ExoS, exoenzyme S
- HA
hemagglutinin
- His
hexahistidine
- IP
immunoprecipitate
- TNF
tumor necrosis factor
- wt
wild type
References
- 1.Jacobson M D, Weil M, Raff M C. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 2.Rudin C M, Thompson C B. Annu Rev Med. 1997;48:267–281. doi: 10.1146/annurev.med.48.1.267. [DOI] [PubMed] [Google Scholar]
- 3.Ellis R E, Yuan J Y, Horvitz H R. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- 4.Cryns V, Yuan J. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 5.Ashkenazi A, Dixit V M. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T I, Jones D P, Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 7.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 8.Pan G, O’Rourke K, Dixit V M. J Biol Chem. 1998;273:5841–5845. doi: 10.1074/jbc.273.10.5841. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Benedict M A, Wu D, Inohara N, Nunez G. Proc Natl Acad Sci USA. 1998;95:4386–4391. doi: 10.1073/pnas.95.8.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 11.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 12.Cardone M H, Roy N, Stennicke H R, Salvesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 13.Wang X S, Diener K, Jannuzzi D, Trollinger D, Tan T H, Lichenstein H, Zukowski M, Yao Z. J Biol Chem. 1996;271:31607–31611. doi: 10.1074/jbc.271.49.31607. [DOI] [PubMed] [Google Scholar]
- 14.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 15.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang T H, Wang H S, Ichijo H, Giannakakou P, Foster J S, Fojo T, Wimalasena J. J Biol Chem. 1998;273:4928–4936. doi: 10.1074/jbc.273.9.4928. [DOI] [PubMed] [Google Scholar]
- 17.Chang H Y, Nishitoh H, Yang X, Ichijo H, Baltimore D. Science. 1998;281:1860–1863. doi: 10.1126/science.281.5384.1860. [DOI] [PubMed] [Google Scholar]
- 18.Nishitoh H, Saitoh M, Mochida Y, Takeda K, Nakano H, Rothe M, Miyazono K, Ichijo H. Mol Cell. 1998;2:389–395. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- 19.Gotoh Y, Cooper J A. J Biol Chem. 1998;273:17477–17482. doi: 10.1074/jbc.273.28.17477. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Seimiya H, Naito M, Mashima T, Kizaki A, Dan S, Imaizumi M, Ichijo H, Miyazono K, Tsuruo T. Oncogene. 1999;18:173–180. doi: 10.1038/sj.onc.1202276. [DOI] [PubMed] [Google Scholar]
- 21.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu H, Coburn J, Collier R J. Proc Natl Acad Sci USA. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulich S M, Frank D W, Barbieri J T. Infect Immun. 1993;61:307–313. doi: 10.1128/iai.61.1.307-313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Wang H, Liu D, Liddington R, Fu H. J Biol Chem. 1997;272:13717–13724. doi: 10.1074/jbc.272.21.13717. [DOI] [PubMed] [Google Scholar]
- 25.Muslin A J, Tanner J W, Allen P M, Shaw A S. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 26.Yaffe M B, Rittinger K, Volinia S, Caron P R, Aitken A, Leffers H, Gamblin S J, Smerdon S J, Cantley L C. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 27.Aitken A. Trends Cell Biol. 1996;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- 28.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 29.Morrison D K, Cutler R E. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 30.Campbell S L, Khosravi-Far R, Rossman K L, Clark G J, Der C J. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 31.Tobiume K, Inage T, Takeda K, Enomoto S, Miyazono K, Ichijo H. Biochem Biophys Res Commun. 1997;239:905–910. doi: 10.1006/bbrc.1997.7580. [DOI] [PubMed] [Google Scholar]
- 32.Pallas D C, Fu H, Haehnel L C, Weller W, Collier R J, Roberts T M. Science. 1994;265:535–537. doi: 10.1126/science.8036498. [DOI] [PubMed] [Google Scholar]
- 33.Fu H, Xia K, Pallas D C, Cui C, Conroy K, Narsimhan R P, Mamon H, Collier R J, Roberts T M. Science. 1994;266:126–129. doi: 10.1126/science.7939632. [DOI] [PubMed] [Google Scholar]
- 34.Cavenagh M M, Whitney J A, Carroll K, Zhang C, Boman A L, Rosenwald A G, Mellman I, Kahn R A. J Biol Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- 35.Liu D, Bienkowska J, Petosa C, Collier R J, Fu H, Liddington R. Nature (London) 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- 36.Xiao B, Smerdon S J, Jones D H, Dodson G G, Soneji Y, Aitken A, Gamblin S J. Nature (London) 1995;376:188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Zhang L, Liddington R, Fu H. J Biol Chem. 1998;273:16297–16304. doi: 10.1074/jbc.273.26.16297. [DOI] [PubMed] [Google Scholar]
- 38.Petosa C, Masters S C, Bankston L A, Pohl J, Wang B, Fu H, Liddington R C. J Biol Chem. 1998;273:16305–16310. doi: 10.1074/jbc.273.26.16305. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe M, Isobe T, Ichimura T, Kuwano R, Takahashi Y, Kondo H, Inoue Y. Brain Res Mol Brain Res. 1994;25:113–121. doi: 10.1016/0169-328x(94)90285-2. [DOI] [PubMed] [Google Scholar]
- 40.Masters S C, Pederson K J, Zhang L, Barbieri J T, Fu H. Biochemistry. 1999;38:5216–5221. doi: 10.1021/bi982492m. [DOI] [PubMed] [Google Scholar]
- 41.Furukawa Y, Ikuta N, Omata S, Yamauchi T, Isobe T, Ichimura T. Biochem Biophys Res Commun. 1993;194:144–149. doi: 10.1006/bbrc.1993.1796. [DOI] [PubMed] [Google Scholar]
- 42.Michaud N R, Fabian J R, Mathes K D, Morrison D K. Mol Cell Biol. 1995;15:3390–3397. doi: 10.1128/mcb.15.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denizot F, Lang R. J Immunol Methods. 1986;89:271–277. doi: 10.1016/0022-1759(86)90368-6. [DOI] [PubMed] [Google Scholar]
- 44.Raff M C. Nature (London) 1992;356:397–400. doi: 10.1038/356397a0. [DOI] [PubMed] [Google Scholar]
- 45.Hsu S Y, Kaipia A, Zhu L, Hsueh A J. Mol Endocrinol. 1997;11:1858–1867. doi: 10.1210/mend.11.12.0023. [DOI] [PubMed] [Google Scholar]