Abstract
The US2 and US11 glycoproteins of human cytomegalovirus facilitate destruction of MHC class I heavy chains by proteasomal proteolysis through acceleration of endoplasmic reticulum-to-cytosol dislocation. Modification of the class I heavy chain was used to probe the structural requirements for this sequence of reactions. The cytosolic domain of the class I heavy chain is required for dislocation to the cytosol and for its subsequent destruction. However, interactions between US2 or US11 and the heavy chain are maintained in the absence of the class I cytosolic domain, as shown by chemical crosslinking in vivo and coprecipitation when translated in vitro. Thus, substrate recognition and accelerated destruction of the heavy chain, as facilitated by US2 or US11, are separable events.
The central role of MHC class I products in an antiviral immune response is well established. A number of viruses encode proteins that can inhibit or abolish surface display of MHC class I on infected cells, and the viral products responsible act at different points in the MHC class I biosynthetic pathway (1). Human cytomegalovirus encodes two endoplasmic reticulum (ER)-resident glycoproteins, US2 and US11, either of which is sufficient to induce the rapid dislocation of newly synthesized MHC class I glycoproteins from the ER to cytosol, where the class I heavy chains are degraded by the proteasome (2–4).
The detailed mechanism of dislocation is not understood, although some clues have emerged. First, when proteasomal proteolysis is inhibited, a characteristic deglycosylated intermediate is observed, which is the product of N-glycanase attack on the class I heavy chain. This intermediate accumulates in the cytosol. Second, this process does not seem to be limited to the rapid (t1/2 < 5 min) reaction catalyzed by US2 or US11—other polypeptides that fail to fold properly are also degraded in a proteasome-dependent manner. Examples include yeast secretory proteins prepro-α factor and improperly folded carboxypeptidase Y (5, 6) as well as the mammalian polytopic CFTR protein (7–9), apolipoprotein B (10), and α1-antitrypsin (11). The T cell receptor α chain, unstable when expressed without its folding partners, was also shown to be degraded by the proteasome (12). For T cell receptor α chain and some of the above cases, deglycosylated cytosolic intermediates are seen when proteasome is inhibited. Also, class I heavy chains synthesized in the absence of β2-microglobulin (β2m, light chain) fail to fold properly, and characteristic dislocation intermediates can be detected in these cells on inhibition of proteasomal proteolysis (13, 14). Third, the translocon (Sec61p complex) may be the conduit through which the polypeptide chain is removed to the cytosol, as shown not only by coimmunoprecipitation experiments in cells actively dislocating MHC class I heavy chains (4) but also by genetic studies in yeast (15, 16). Finally, ER-to-cytosol dislocation is sensitive to agents that modify free thiols or alter the redox state of the cell more generally. Not only did agents such as diamide or N-ethylmaleimide (NEM) inhibit dislocation of the class I heavy chain, but a T cell receptor α chain mutant lacking cysteines was likewise affected, indicating that substrate thiols are not primarily responsible for the NEM block of dislocation (14). The balance between oxidation and reduction within the cell thus contributes not only to protein folding in general, but also to functioning of the dislocation machinery.
Here we examine the substrate requirements for dislocation through modification of the class I heavy chain. We demonstrate that the cytosolic tail of the class I heavy chain is essential for its dislocation from ER to cytosol, but not for binding of US2 or US11. The recognition elements whereby US2 and US11 bind to the class I glycoprotein can be preserved, yet without concomitant dislocation of the substrate.
MATERIALS AND METHODS
Construction of MHC Class I Heavy Chain Mutants.
The HLA-A*0201 heavy chain (HLA-A2) was amplified by PCR from a full length cDNA in which the signal peptide was converted to the mouse H-2 K signal peptide, which enhances in vitro translation in some instances. The primers used were A2NEco (5′-CGGAATTCACCACCATGGTACCGTGCACGCTGCTCCT-3′) and A2TMXho (5′-CCGCTCGAGTCAGCTCTTCCTCCTCCACATCACAG-3′). EcoRI and XhoI cloning sites, as well as a consensus translation start site (17), are encoded in the primers. The second primer introduces a stop codon, resulting in the truncated heavy chain diagrammed in Fig. 1. All constructs were sequenced.
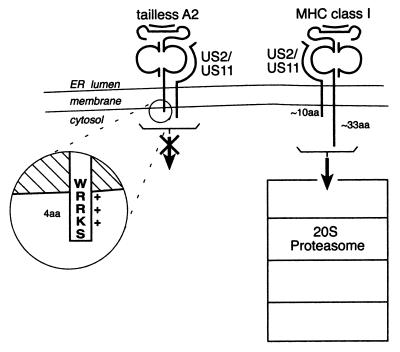
Figure 1.
Schematic representation of mutant and wild-type class I heavy chain. Truncated class I heavy chains (tailless A2) are not dislocated, yet associate with US2 and US11. The truncated heavy chains retain four amino acids that protrude into the cytosol to facilitate proper insertion in the membrane (21). The sizes of the proteasome particle and the class I protein are rendered approximately to scale.
Metabolic Labeling and Immunoprecipitation.
MG373 astrocytoma cells were collected and preincubated in methionine/cysteine-free DMEM with or without proteasome inhibitor. The proteasome inhibitor ZL3VS (50 μm) was added as indicated in the figures (18). Metabolic labeling and immunoprecipitation were performed as described (3, 19). Rabbit anti-class I heavy chain serum (αHC) recognizes free heavy chains; the monoclonal antibody W6/32 recognizes assembled class I molecules (20). The US2 and US11 antisera were generated by immunizing rabbits with peptides and will be described elsewhere. Fractionation experiments were done as described previously (3). Quantitation of autoradiograms was done by using alphaimager software by Alpha Innotech (San Leandro, CA).
Crosslinking.
Astrocytoma cells were incubated for 1 hr with ZL3VS, then metabolically labeled for 30 min. Cells were placed on ice and washed with ice-cold PBS. Dithiobis(succinimidyl propionate) (DSP, 5 mM; Pierce) was added from 30 × stock in DMSO, and crosslinking was allowed to proceed on ice. After 30 min, Tris buffer, pH 7.4 (100 mM), was added to quench for 15 min on ice. Samples were diluted with 10 volumes of Nonidet P-40 lysis buffer and immunoprecipitated with W6/32 antibody. One-fifth of the W6/32 immunoprecipitate was set aside for direct analysis, whereas two-fifths was precipitated with αHC and two-fifths with α-US2 or α-US11, as appropriate.
In Vitro Transcription/Translation.
In vitro transcription was performed as described (12) on 5 μg of HLA-A2, tailless HLA-A2, US11, and US2 cDNAs cloned into the pCDNA3.1 plasmid (Invitrogen) or HLA-DR1α and β2m cDNA in the pSP72 plasmid (Promega). In vitro translations were performed for 2 hr in a total reaction mixture of 30 μl, containing 17.5 μl Flexi Rabbit Recticulocyte Lysate (Promega), 0.8 μl KCl (2.5 M), 0.5 μl amino acid mixture minus methionine (1 mM; Promega), 2.5 μl [35S]methionine (10 mCi/ml, translation grade; NEN), 1 μl canine pancreatic microsomes, and 2 μl mRNA. DTT (1 mM) was added to some translations as indicated in Fig. 5. After translation, for the US2 experiment (Fig. 5a), microsomes were sedimented (15 min, 12,000 × g) and lysed in 1% digitonin (Boehringer Mannheim) in digitonin buffer (DB, 25 mM Hepes, 150 mM KOAc, pH 7.7). Immunoprecipitation was done as above, except washes were in 0.2% digitonin in DB. The US11 translations (Fig. 5b) were lysed in Nonidet P-40 lysis buffer rather than digitonin.
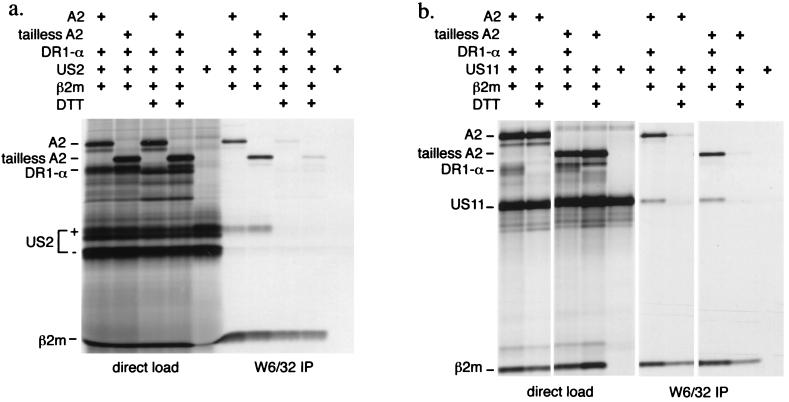
Figure 5.
Tailless HLA-A2 associates with US2 and US11 in vitro. Full length and tailless A2 mRNAs were translated in a microsome-supplemented in vitro translation system together with β2m, HLA-DR1α, and US2 or US11 mRNA in the presence and absence of 1 mM DTT, as indicated. A portion of the total translation mixture was loaded directly (left lanes). The remainder of the microsomes was lysed and subjected to immunoprecipitation with W6/32 antibody (right lanes).
RESULTS
Tailless MHC Class I Heavy Chains Are Stable in the Presence of US2 or US11 Products.
The heavy chain of MHC class I (heavy chain) is rapidly degraded in the presence of either the US2 or US11 glycoprotein encoded by human cytomegalovirus. To investigate the possible involvement of the cytosolic tail of class I heavy chains in their dislocation, a deletion of all but four amino acids of the cytosolic domain of HLA-A2 was generated (Fig. 1). The remaining four residues were retained to allow the class I heavy chain to properly insert in the membrane (21). Others have shown that similarly truncated HLA-A2 molecules behave like the full length heavy chain with respect to folding, expression on the cell surface, and internalization from the plasma membrane (22, 23).
Astrocytoma cell lines (24) expressing US2, US11, or neither (control cells) were transfected with the tailless form of HLA-A2 (tailless A2), and stable clones were selected. The fate of endogenous and tailless heavy chains was examined by metabolic labeling and immunoprecipitation in pulse–chase experiments in the absence of proteasome inhibitor (Fig. 2). The rabbit anti-heavy chain serum (αHC, Top) reacts with free heavy chains, whereas the W6/32 monoclonal antibody (Bottom) reacts with properly folded, β2m-complexed heavy chains. In US2- or US11-expressing cells, in the absence of proteasome inhibitors, the endogenous class I heavy chains (wild-type HC) are mostly degraded by 30 min of chase (Fig. 2 a and b, left lanes). This is in marked contrast with the tailless heavy chains (Fig. 2 a and b, right lanes), which persist in the presence of US2 and US11. Free full length heavy chains in the tailless A2-transfected cells decrease over the chase such that 27% and 45% of the starting material is recovered at 30 min for US2 and US11 cells, respectively. Folded full length heavy chains recovered from these cells also decrease over the chase, to 51% and 42% of starting material for US2 and US11 cells, respectively. In contrast, there is a 10% increase in the recovery of tailless free heavy chains over the chase for both US2 and US11 cells, but more importantly, the recovery of folded (W6/32-reactive) tailless A2 is substantial at the zero time point and increases 30% over the chase in the US2-expressing cells, with a three-fold increase in recovery of folded tailless A2 in the US11-expressing cells. In control cells (Fig. 2c), both the endogenous heavy chains and the tailless A2 molecules are stable, and their folding occurs during the chase period as reflected by a continuous increase in recovery of W6/32-reactive material.
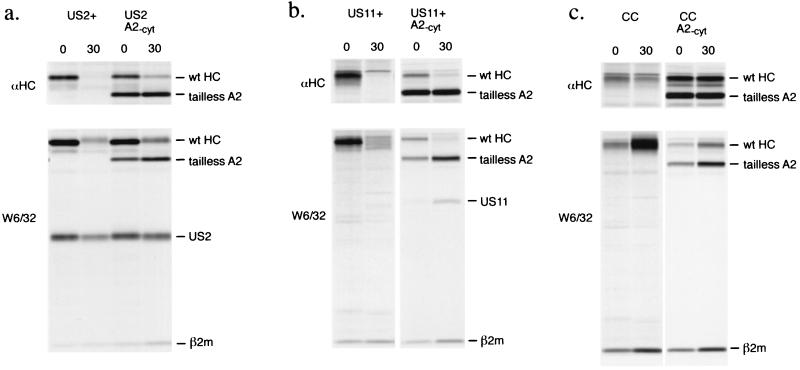
Figure 2.
Tailless A2 is stable in the presence of US2 and US11 while the endogenous class I heavy chains are destroyed. Cells were pulse labeled for 5 min and chased for the indicated times in the absence of proteasome inhibitors. US2-expressing cells (a), US11-expressing cells (b), or control cells (CC; c) transfected with tailless A2 (A2-cyt) were subjected to pulse–chase analysis. Immunoprecipitations were done with antibodies reactive with free heavy chains (αHC) or folded class I products (W6/32). The US2 and US11 molecules that coprecipitate in the W6/32 complex are indicated.
The rate of heavy chain loss is slowed in the presence of tailless A2. The rates of full length heavy chain loss seen for US2 cells (Fig. 2a) and US11 cells (Fig. 2b), respectively, are 2.3 times slower and 4 times slower when tailless A2 is present. Nonetheless, the behavior of the endogenous and tailless heavy chains is clearly distinct. We observe an increase in the recovery of folded W6/32-reactive material over the chase only for the tailless heavy chain, with concomitant loss of folded wild-type heavy chain. We typically observe the glycosylated form of US2 in association with folded heavy chains [W6/32, (4)], and the added presence of tailless A2 does not seem to inhibit this association (Fig. 2a).
Dislocation of Endogenous MHC Class I Heavy Chains Is Inhibited by the Presence of Tailless A2.
The degradation of class I heavy chains targeted by US2 and US11 occurs by their dislocation to the cytosol where they are attacked by the proteasome. The addition of proteasome inhibitors to cells expressing US2 or US11 allows the recovery of heavy chain intermediates that are cytosolic and have lost their N-linked glycan in a peptide N-glycanase-type reaction (3). These deglycosylated intermediates, which accumulate during the chase period, are seen most clearly in cells that do not express tailless A2 (Fig. 3a left lanes; αHC). The deglycosylated intermediate does not accumulate in control cells (Fig. 3b). When tailless A2 is introduced into US2 or US11 cells, we observe a retardation of dislocation of endogenous heavy chains, reflected by decreased recovery of its deglycosylated intermediate and by a slower loss of fully glycosylated heavy chains (similar for US2 cells; data not shown). The half-life of full length glycosylated heavy chains calculated for US11+ cells is approximately 5 min, whereas for the US11+ cells transfected with tailless A2 the recovery of glycosylated full length heavy chains is nearly constant over the course of the experiment. In subcellular fractionation experiments, we do not observe accumulation of glycosylated heavy chains in the cytosol fraction. The absence of deglycosylated intermediate is therefore a valid indicator of a lack of dislocation. One representative fractionation experiment is shown (Fig. 3c). Deglycosylated full length heavy chains are recovered quantitatively in the cytosol, whereas no tailless heavy chains are found in the cytosol.
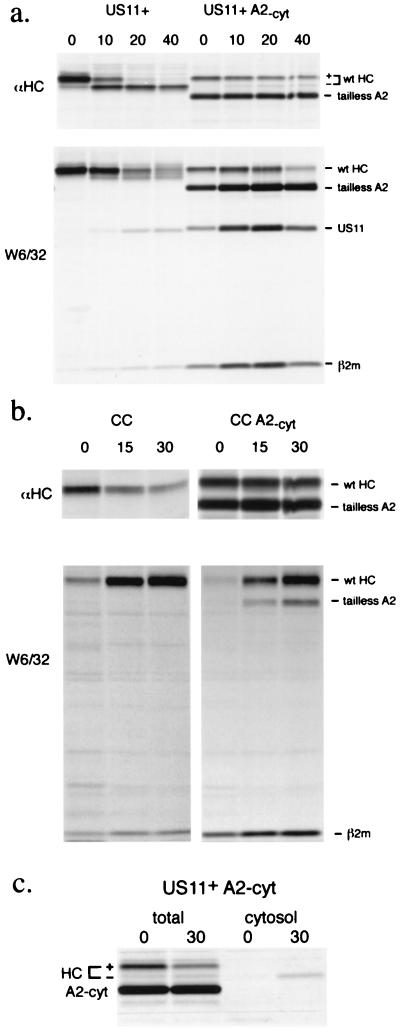
Figure 3.
Presence of tailless A2 inhibits dislocation of endogenous MHC class I heavy chains. Cells were pulse labeled for 5 min and chased for the times shown, all in the presence of ZL3VS. The bracket with + and - denotes the forms of heavy chain with and without glycan, respectively. In US11-expressing cells (a), full length class I heavy chain is lost (a decrease in W6/32-reactive material) or dislocated (an increase in −CHO form). The tailless A2 continues to fold (increasing W6/32) while no deglycosylated tailless A2 accumulates. There is increased recovery of US11 in the cells expressing tailless A2. In control cells (b), endogenous and tailless heavy chains mature at similar rates. Fractionation of US11-expressing cells transfected with tailless A2 (c) shows the presence of deglycosylated full length heavy chains in the cytosol and no accumulation of tailless A2 in the cytosol.
Though the retardation in heavy chain dislocation is more clearly seen in the presence of proteasome inhibitors, this result agrees with our observations in untreated cells (Fig. 2) and suggests that the additional heavy chains are competing for some aspect of the dislocation machinery. The association of US11 in the W6/32 complex is readily observed (Fig. 3a). Note that the recovery of US11 is significantly enhanced when tailless A2 is present and when proteasomes are inhibited (compare Figs. 2b and 3a). In the presence of proteasome inhibitors, the endogenous heavy chains in US11 cells decrease during the chase. The tailless heavy chains, in contrast, are stable and continue to fold during the chase. The expression of epitope-tagged dislocation-competent heavy chains in US2 or US11 cells also retards the dislocation of endogenous heavy chains, but to a lesser extent (C.M.S., data not shown).
These experiments do not allow us to determine whether US2 or US11 associates with tailless A2 or full length heavy chains, especially because protein synthesis was not inhibited during the chase. Because dislocation of class I heavy chains is slowed in these cells, US2 or US11 may have more time to associate with both tailless and full length class I molecules in the ER. For US2, we now have biochemical evidence using recombinant protein that suggests US2 and class I molecules associate in a one-to-one stoichiometry (B. Gewurz, personal communication). We do not observe either US2 or US11 in association with free heavy chains (data not shown). The decreased rate of dislocation of endogenous heavy chain in the presence of tailless A2 would be expected if the tailless heavy chains were to compete for binding to US2 and US11. The possibility of direct binding of both full length and tailless MHC class I heavy chains to US2 and US11 is examined below.
US2 and US11 Bind Tailless A2 in Vivo and in Vitro.
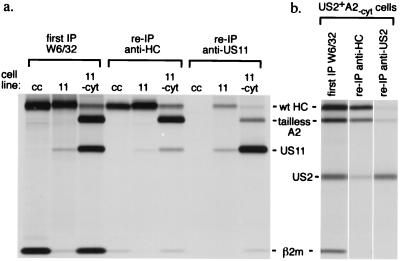
We examined the capacity of US11 and US2 to interact with the tailless class I heavy chain more directly by chemical crosslinking. Metabolically labeled cells were crosslinked with the cell-permeable thiol-cleavable crosslinker DSP. Folded heavy chain complexes were recovered first with W6/32. A portion of this immunoprecipitate was set aside for direct analysis. The remainder was denatured by boiling in SDS in the absence of reducing agent to maintain the crosslinked condition. Reimmunoprecipitation of crosslinked material was then performed with αHC and α-US2 or US11 antibodies, as indicated, followed by analysis on reducing SDS/PAGE. After crosslinking, tailless heavy chains are recovered with both α-US2 or α-US11 (Fig. 4), indicating their association in the W6/32-reactive complex. Also, US2 and US11 are recovered with the αHC serum. The recovery of tailless heavy chain by reprecipitation with α-US2 or α-US11 is direct evidence that US2 and US11 form complexes with tailless HLA-A2. The presence of DSP was necessary to observe these effects (data not shown).
Figure 4.
DSP crosslinking of cells expressing tailless A2 reveals an association between US11 or US2 and tailless A2. Cells exposed to the crosslinker DSP (see Materials and Methods) were lysed in Nonidet P-40 buffer. Folded MHC class I products were recovered by immunoprecipitation with the W6/32 antibody. A portion of this immunoprecipitate was loaded directly (first IP). The remainder was denatured in SDS without reductant and reprecipitated with αHC and either α-US2 or α-US11 antibodies. Reimmunoprecipitation with α-US11 antibody recovers tailless A2, indicative of an association between them (a, right lane). Likewise, US2 antibodies recover crosslinked tailless A2 (b, right lane). Reduction of the thiol-cleavable crosslinker before SDS/PAGE analysis dissociates the crosslinked molecules and yields the constituent polypeptides that then migrate at their characteristic molecular weights.
Physical association between either US2 or US11 and tailless heavy chains can also be demonstrated in vitro (Fig. 5). Wild-type or tailless A2 mRNAs were translated in vitro together with β2m and either US2 or US11 mRNA. Inclusion of canine microsomes allows the formation of the class I MHC heterodimer, as judged by accumulation of a W6/32-reactive complex. HLA-DR1α mRNA was included in translation reactions as a polypeptide that should not be recovered in the W6/32-reactive complex and thus served as an internal negative control. A portion of the microsomes was then analyzed directly by SDS/PAGE, while the remainder was lysed in detergent and immunoprecipitated with W6/32. The lysates were immunoprecipitated with αHC and α-US2 or α-US11 to confirm the presence of comparable amounts of the relevant polypeptides in each translation (not shown). Translation of all transcripts can be seen in the samples analyzed directly. Immunoprecipitation with W6/32 recovers both full length A2 and tailless A2 associated with β2m, together with US2 (Fig. 5a) and US11 (Fig. 5b). DTT strongly inhibits protein folding, as it blocks proper intrachain disulfide bond formation in the translated polypeptides. Inclusion of DTT inhibited the coprecipitation of US2 or US11 with W6/32 and also inhibited heavy chain association with β2m, as evidenced by little recovery of W6/32-reactive material in the DTT samples. We consistently observe considerable recovery of radiolabeled β2m in the DTT-containing samples immunoprecipitated with W6/32. One explanation for this result is that the in vitro-translated β2m continues to fold in the presence of this modest concentration of DTT and can also associate with preexisting unlabeled canine class I heavy chains that reside in the microsomes used for translation. Indeed, interspecies exchange of β2m is a common occurrence (25, 26). At no time does HLA-DR1α coprecipitate with class I molecules or US2/US11. Thus, US2 and US11 are recovered by virtue of their specific association with the class I complex. Furthermore, the W6/32 antibody does not precipitate US2 or US11 when these products are translated in the absence of heavy chain. Our ability to recover specific complexes between class I and US2 or US11, both in vitro and by chemical crosslinking in vivo, shows that the association between these partners does not require the cytosolic tail of the class I heavy chain.
DISCUSSION
Cells that express US2 or US11 provide a system to study the process of ER-to-cytosol dislocation, an important route of disposal for improperly folded or incorrectly multimerized proteins. This pathway has been coopted by the human cytomegalovirus to target MHC class I glycoproteins for destruction. In one and the same US2- or US11-expressing cell, the fate of endogenous heavy chains can be compared with that of altered forms of heavy chain introduced by transfection. We show that MHC class I heavy chains lacking their cytosolic tail are refractory to US2- and US11-mediated attack, while the endogenous heavy chains continue to be destroyed.
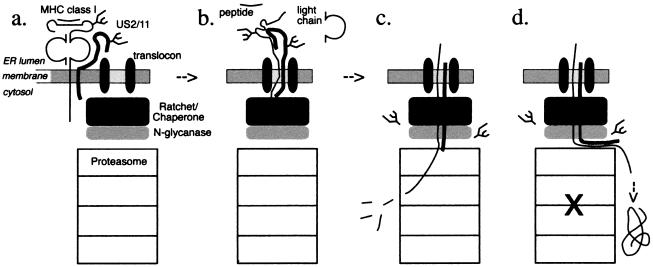
For dislocation of heavy chains, the cytosolic domain may provide a point of contact for machinery in the cytosol to extract the MHC class I heavy chain from the ER membrane. The following model is consistent with our observations of heavy chain dislocation (Fig. 6): US2 and US11 bind to class I heavy chain within the ER lumen and deliver it to the site of dislocation (a). US2 and US11 may interact with the translocon, perhaps modifying it in such a way that the heavy chain may reenter the translocon, in what is a reversal of the normal process of release from the Sec61p channel into the lipid environment of the membrane. The heavy chain is then dislocated by extraction from the ER on its way to proteolysis in the cytosol (b). The cytosolic tail of class I heavy chain is in fact required for extraction. Therefore, cytosolic factors (black box) such as heat-shock proteins of the Hsp70 family could be involved in this extraction, perhaps providing energy for the reaction or acting as a ratchet to prevent heavy chain from sliding back into the ER lumen. Such a mechanism would be akin to what has been proposed as the ratchet role for Hsp70 family members in protein import into the ER in yeast (reviewed in ref. 27) and the mitochondrion (28). Because the cytosolic deglycosylated breakdown intermediates of the class I heavy chain are not observed in the absence of proteasomal inhibitors, even when using pulse-labeling times of 1 min or less, dislocation and degradation by the proteasome are tightly coupled (c). When proteasomal activity is compromised, the bulk of the dislocating substrate is deflected into the cytosol and accumulates as a deglycosylated polypeptide (d). The N-glycanase activity likely acts immediately on removal of the polypeptide from the ER environment, because glycosylated heavy chains are not observed in the cytosol in the course of dislocation when proteasome function is preserved. For entry of the class I heavy chain into the proteasome, the glycan could pose steric constraints and would have to be removed for the heavy chain to reach the active sites within the proteasome. In cells that express tailless heavy chains, the heavy chains may engage the dislocation machinery, but the process cannot be completed for lack of the cytosolic portion required for extraction from the membrane. The tailless class I heavy chain should thus compete with endogenous class I heavy chains and, at high levels of expression, competitively inhibit dislocation of endogenous heavy chains. Such competition is in fact observed (Figs. 2 and 3).
Figure 6.
Model for dislocation of class I heavy chains by US2 and US11. (a) The dislocation reaction is initiated by binding of US2 or US11 to the heavy chain with most intermolecular contacts within the ER lumen. (b) Once in the translocon, the class I heavy chain unfolds. The light chain of class I and MHC-bound peptide may then dissociate within the ER. Cytosolic factors (black box) bind to the cytosolic tail of the class I heavy chain and aid in its extraction. (c) N-glycanase (gray box) attacks the glycan when it emerges into the cytosol. Proteasome cleaves the class I heavy chain. (d) In the presence of proteasomal inhibitors, a portion of the heavy chain is deflected into the cytosol directly and accumulates as a deglycosylated intermediate.
The cytosolic domain of HLA-A2 contains three lysines in or near its cytosolic domain. Mutagenesis of these lysines to arginine does not prevent dislocation by US2 and US11 (C. Shamu, C.M.S., T. Rapoport, H.L.P., unpublished observation). Thus, we may rule out ubiquitin conjugation of the cytosolic tail as a requirement for the occurrence of heavy chain dislocation. This proposal is at odds with several reports in the literature in which ubiquitination of substrate was required for substrate degradation and suggests that several distinct modes of protein removal from the ER may be defined based on this criterion alone (7, 9, 29, 30).
Endocytosis of growth hormone receptor (31) and yeast alpha factor receptor (32) involve an intact ubiquitin conjugation apparatus. Because ubiquitination of the cytosolic tail is not required for the dislocation of MHC class I heavy chains, and because a major fraction of cytosolic heavy chains that accumulate in the presence of proteasome inhibitors appears not to be ubiquitinated, we suggest that for dislocation of class I heavy chain, the putative factors involved are likely distinct from the ubiquitin-conjugation machinery. However, we cannot exclude the possibility that these factors could be involved in both processes.
Candidates for cytosolic factors involved in extracting substrates from the translocon include chaperones and also the PA700/19S regulatory complex of the proteasome. Cytosolic Hsp70, together with J-domain-containing proteins, is known to bind extended stretches of polypeptide that would be exposed during dislocation (33, 34). The degradation of apolipoprotein B involves dislocation to the cytosol and is influenced by levels of cytosolic Hsp70 in HepG2 cells (35). Also, Hsp70 is modified by thiol alkylating agents like NEM (36), consistent with our observation that modest levels of NEM block dislocation of MHC class I heavy chains and of a T cell receptor α dislocation substrate lacking cysteines (14).
The 20S proteasome core particle combines in the cytosol with a large regulatory particle, the PA700/19S complex, which is thought to unfold polypeptides before they can enter the proteasome, as well as to remove ubiquitin chains (37). Where ubiquitination of the substrate is required for dislocation, the proteasome itself may be involved in unfolding and removal of the substrate from the ER environment. This process could be enhanced by the proteasome’s recognition of ubiquitin moieties. In yeast, where proteasome mutant strains are available, the rate of membrane extraction of a short-lived ER membrane protein depends on the functionality of the proteasome, suggesting a role of the proteasome in extracting the substrate (38). We have also observed that when the proteasome is blocked in US2 or US11 cells, the rate of dislocation of MHC class I molecules is slowed (C.M.S., unpublished observations). The proteasome could be the factor that binds the cytosolic tail of class I heavy chain and extracts MHC class I heavy chains from the ER membrane. The PA700/19S complex is likely to possess chaperone-like properties that allow it to interact with substrates that are not ubiquitinated.
The notion that the US2 and US11 glycoproteins themselves contain all the elements required for dislocation, as long as binding to MHC products is established, is ruled out by our experiments with truncated heavy chains. Because binding between US2 or US11 and tailless class I is preserved, yet no dislocation of this substrate is observed, we conclude that US2 and US11 alone do not have the capability to remove heavy chains from the ER. Our experiments do not rule out the possibility that the binding between US2 or US11 and class I heavy chains is subtly altered, nor can we exclude the possibility that class I heavy chain’s cytosolic tail is involved in an essential interaction with the translocon. However, we believe our interpretation is the simplest one based on topological considerations. The identification of elements in the carboxyl terminus of the heavy chain that are essential for its dislocation is therefore an important experimental goal.
Acknowledgments
The authors thank Tom Rapoport and Caroline Shamu for a critical reading of the manuscript. This work was supported by the National Institutes of Health (grant 5R37-A133456), Boehringer Ingelheim, and a postdoctoral fellowship from the Cancer Research Institute of New York (C.M.S.).
ABBREVIATIONS
- ER
endoplasmic reticulum
- NEM
N-ethylmaleimide
- αHC
anti-class I heavy chain serum
- DSP
dithiobis(succinimidyl propionate)
- heavy chain
heavy chain of MHC class I
- HLA-A2
HLA-A*0201 heavy chain
- β2m
β2-microglobulin
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Ploegh H L. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- 2.Jones T R, Hanson L K, Sun L, Slater J S, Stenberg R M, Campbell A E. J Virol. 1995;69:4830–4841. doi: 10.1128/jvi.69.8.4830-4841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiertz E J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 4.Wiertz E J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Nature (London) 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 5.Hiller M M, Finger A, Schweiger M, Wolf D H. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 6.McCracken A A, Brodsky J L. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 8.Bebok Z, Mazzochi C, King S A, Hong J S, Sorscher E J. J Biol Chem. 1998;273:29873–29878. doi: 10.1074/jbc.273.45.29873. [DOI] [PubMed] [Google Scholar]
- 9.Xiong X, Chong E, Skach W R. J Biol Chem. 1999;274:2616–2624. doi: 10.1074/jbc.274.5.2616. [DOI] [PubMed] [Google Scholar]
- 10.Zhou M, Fisher E A, Ginsberg H N. J Biol Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
- 11.Qu D, Teckman J H, Omura S, Perlmutter D H. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- 12.Huppa J B, Ploegh H L. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- 13.Hughes E A, Hammond C, Cresswell P. Proc Natl Acad Sci USA. 1997;94:1896–1901. doi: 10.1073/pnas.94.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tortorella D, Story C M, Huppa J B, Wiertz E J, Jones T R, Ploegh H L. J Cell Biol. 1998;142:365–376. doi: 10.1083/jcb.142.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilon M, Schekman R, Romisch K. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plemper R K, Bohmler S, Bordallo J, Sommer T, Wolf D H. Nature (London) 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 18.Bogyo M, McMaster J S, Gaczynska M, Tortorella D, Goldberg A L, Ploegh H. Proc Natl Acad Sci USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beersma M F, Bijlmakers M J, Ploegh H L. J Immunol. 1993;151:4455–4464. [PubMed] [Google Scholar]
- 20.Parham P, Barnstable C J, Bodmer W F. J Immunol. 1979;123:342–349. [PubMed] [Google Scholar]
- 21.von Heijne G. Nature (London) 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 22.Vega M A, Strominger J L. Proc Natl Acad Sci USA. 1989;86:2688–2692. doi: 10.1073/pnas.86.8.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gur H, Geppert T D, Lipsky P E. Mol Immunol. 1997;34:125–132. doi: 10.1016/s0161-5890(97)00007-2. [DOI] [PubMed] [Google Scholar]
- 24.Jones T R, Hanson L K, Sun L, Slater J S, Stenberg R M, Campbell A E. J Virol. 1995;69:4830–4841. doi: 10.1128/jvi.69.8.4830-4841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernabeu C, van de Rijn M, Lerch P G, Terhorst C P. Nature (London) 1984;308:642–645. doi: 10.1038/308642a0. [DOI] [PubMed] [Google Scholar]
- 26.Jefferies W A, MacPherson G G. Eur J Immunol. 1987;17:1257–1263. doi: 10.1002/eji.1830170907. [DOI] [PubMed] [Google Scholar]
- 27.Simon S M, Peskin C S, Oster G F. Proc Natl Acad Sci USA. 1992;89:3770–3774. doi: 10.1073/pnas.89.9.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider H C, Berthold J, Bauer M F, Dietmeier K, Guiard B, Brunner M, Neupert W. Nature (London) 1994;371:768–774. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- 29.Jeffers M, Taylor G A, Weidner K M, Omura S, Vande Woude G F. Mol Cell Biol. 1997;17:799–808. doi: 10.1128/mcb.17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Virgilio M, Weninger H, Ivessa N E. J Biol Chem. 1998;273:9734–9743. doi: 10.1074/jbc.273.16.9734. [DOI] [PubMed] [Google Scholar]
- 31.Govers R, ten Broeke T, van Kerkhof P, Schwartz A L, Strous G J. EMBO J. 1999;18:28–36. doi: 10.1093/emboj/18.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hicke L, Riezman H. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 33.Flynn G C, Pohl J, Flocco M T, Rothman J E. Nature (London) 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 34.Misselwitz B, Staeck O, Rapoport T A. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 35.Fisher E A, Zhou M, Mitchell D M, Wu X, Omura S, Wang H, Goldberg A L, Ginsberg H N. J Biol Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- 36.Liu Q, Levy E J, Chirico W J. J Biol Chem. 1996;271:29937–29944. doi: 10.1074/jbc.271.47.29937. [DOI] [PubMed] [Google Scholar]
- 37.Glickman M H, Rubin D M, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V A, Finley D. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 38.Mayer T U, Braun T, Jentsch S. EMBO J. 1998;17:3251–3257. doi: 10.1093/emboj/17.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]