Abstract
Objective: To investigate whether a correlation between gene methylation and expression of aggrecan in chondrocytes exists.
Methods: The human aggrecan promoter region was analysed computationally for CpG-rich regions. These were investigated for the methylation of C residues in normal (aged) and osteoarthritic chondrocytes by the bisulphite method for modifying DNA as well as sequence analysis using DNA directly extracted from normal and osteoarthritic cartilage tissue. Additionally, chondrocytic cell lines were investigated for methylation within the aggrecan promoter region.
Results: The CpG-rich promoter region of the human aggrecan gene contains a 0.6 kb region that meets the criteria of a CpG island as defined by prediction programmes. A significant correlation of aggrecan mRNA expression levels and methylation status in normal (aged) and osteoarthritic chondrocytes as well as in different chondrocytic cell lines was not found.
Conclusions: Expression of aggrecan in normal cartilage and diseased states is not modulated by gross changes of CpG methylation of its promoter region. CpG methylation does not have a central role in the switch off of aggrecan promoter activity in human adult articular chondrocytes.
Full Text
The Full Text of this article is available as a PDF (353.6 KB).
Figure 1.
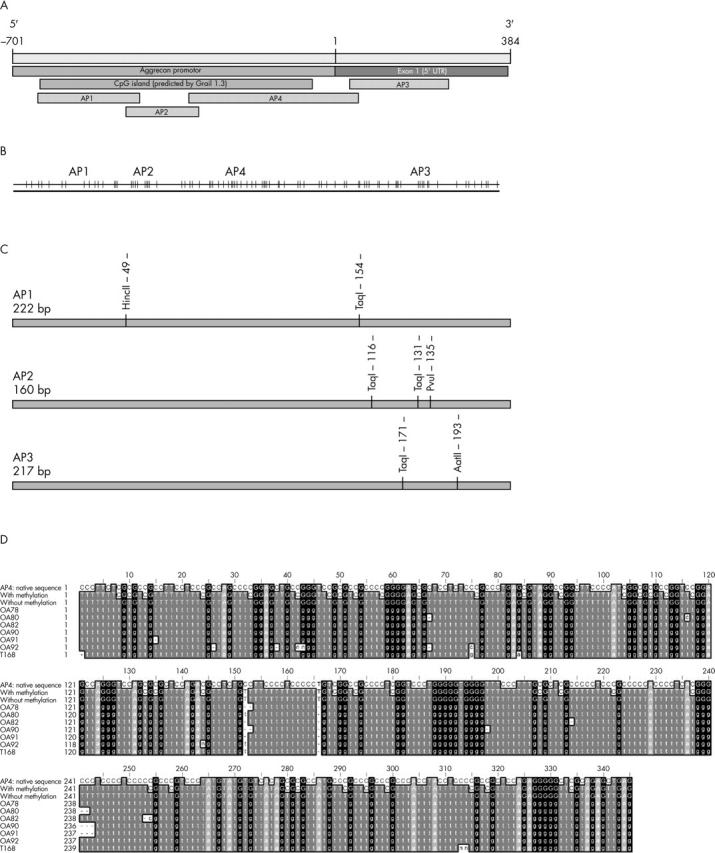
Genomic organisation of the human aggrecan promoter region. (A) Promoter region and exon 1 (GB: AF031586) include a predicted CpG island (Grail 1.3, CpG Island Searcher). The regions AP1-AP4 were analysed for their methylation status by PCR amplification after bisulphite treatment of genomic DNA. (B) Distribution of CpG sites (vertical lines) in the promoter region. (C) Map of new restriction sites within the fragments AP1-AP3, generated by bisulphite treatment of CpG methylated sites. (D) Sequence analyses of cloned amplification products of AP4 after bisulphite treatment of genomic DNA. The unconverted sequence of the region AP4 (lane 1), the converted sequences after bisulphite treatment assuming complete CpG methylation (lane 2) or without methylation (lane 3) are compared with the sequences of six OA samples (lanes 4–9) as well as one normal case (lane 10) after bisulphite treatment, amplification, and cloning. The OA samples showed no indication of CpG methylation, as all cytosines were converted to thymines as seen with the non-methylated sequences (lane 3).
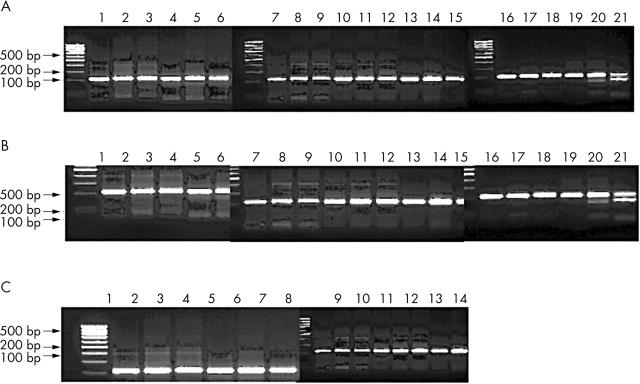
Figure 2.
Detection of new restriction sites in the regions AP1-AP3 after bisulphite treatment. The fragments AP1-AP3 were amplified after bisulphite treatment and potential new restriction sites were detected by restriction digestions with the indicated enzymes (see fig 1B). (A) Region AP1 from three normal samples (1–3, 4–6, 7–9), three osteoarthritic (OA) samples (10–12, 13–15, 16–18), and control DNA treated with SssI methylase (19–21). Fragments were either untreated (1, 4, 7, 10, 13, 16, 19), restricted with HincII (2, 5, 8, 11, 14, 17, 20), or restricted with TaqI (3, 6, 9, 12, 15, 18, 21). The appearance of new restriction sites is only apparent in SssI treated control DNA. (B) Region AP2 was analysed as in (A) but restrictions were done with TaqI and PvuI, respectively. (C) Region AP3 was analysed from three OA samples (1–2, 3–4, 5–6), three normal samples (7–8, 9–10, 11–12), and a positive control (13–14).