Abstract
Gene knockout technology has provided a powerful tool for functional analyses of genes expressed preferentially in a particular tissue. Given marked similarities between human and mouse skin, such studies with epidermally expressed genes have often provided valuable insights into human genetic skin disorders. Efficient silencing of a specified gene in a temporally regulated and epidermal-specific fashion could extend functional analyses to broadly expressed genes and increase the categories of human skin disorders to which parallels could be drawn. We have generated transgenic mice expressing Cre and a fusion protein between Cre recombinase and the tamoxifen responsive hormone-binding domain of the estrogen receptor (CreERtam) under the control of the human keratin 14 (K14) promoter. This promoter is strongly active in dividing cells of epidermis and some other stratified squamous epithelia. With K14–Cre, transgenic embryos recombine genetically introduced loxP sequences efficiently and selectively in the genomes of keratinocytes that reside in embryonic day 14.5 skin, tongue, and esophagus. With K14–CreERtam, postnatal transgenic mice show no Cre activity until tamoxifen is administered. If orally administered, tamoxifen activates keratinocyte-specific CreERtam, allowing recombination of loxP sequences in epidermis, tongue, and esophagus. If topically administered, tamoxifen allows recombination in the area of skin where tamoxifen was applied. Finally, we show that epidermal cells harboring a Cre-dependent rearranged genome persist for many months after tamoxifen application, indicating that the epidermal stem cell population has been targeted efficiently. These tools now pave the way for testing the functional role of different somatic mutations that may exist in mosaic disorders of the skin, including squamous and basal cell carcinomas.
The use of transgenic mouse technology, coupled with tissue-specific promoters, has enabled researchers to perturb the normal pattern of gene expression in mammals and to assess the consequences of doing so to genetic disease. This method has been extremely powerful in functional studies of genes that are expressed in epidermis and hair follicles of skin. The human keratin 14 (K14) and K5 promoters have been particularly useful in targeting the expression of transgenes to the mitotically active basal layer of mouse epidermis and to the outer root sheath of the hair follicle, oral epithelia, and esophagus (1–3). Conversely, the K10 and involucrin promoters have enabled targeting of gene expression to the differentiating suprabasal layers of epidermis (4, 5), and a segment of the K1 promoter has been useful in targeting expression to all epidermal layers (6).
The use of transgenic mouse technology with these human epidermal promoters has yielded valuable insights into the changes in skin pathology and physiology caused by altering the expression of various genes that affect epidermal growth and/or differentiation. In many cases, these insights have led to the subsequent elucidation of the genetic basis for a human skin disorder. Thus, for example, by expressing a dominant negative mutant K14 gene in transgenic mice, keratin filament assembly was grossly perturbed in the epidermis, leading to basal cell degeneration and skin blistering on mild mechanical stress (7). These features were characteristic of epidermolysis bullosa simplex, a blistering skin disorder in humans (8), and it was verified subsequently that most epidermolysis bullosa simplex cases in humans are caused by mutations in the K14 and K5 genes (9–11). Similarly, correlations between oncogene expression and human skin cancers have been drawn from analyzing transgenic mice expressing Ha-ras in a keratinocyte-specific fashion (4, 6, 12). K14-promoter-driven expression of sonic hedgehog (Shh) in mice led to a basal cell carcinoma-like phenotype, and the characterization of this phenotype in turn led to the identification of Shh mutations in human sporadic basal cell carcinomas (13). A final example is that transgenic mice expressing a K14-promoter-driven, activated form of β-catenin produced pilomatricoma-like tumors in mice (14), and recently, it was discovered that this common skin tumor in humans arises largely from activating mutations in the β-catenin gene (15). Taken together, these studies underscore the importance of a transgenic approach for generating important animal models for the study of human skin diseases and also for guiding scientists to the genetic bases of autosomal dominant human skin disorders of unknown etiology.
Homologous recombination in embryonic stem cells has provided a method to target and mutate a specified gene in the germ line of mice, thereby allowing scientists to assess the relation between loss of gene function and genetic disease (16, 17). This approach has also been employed widely in epidermal biology and has afforded researchers the means to evaluate possible links between the absence or reduction of a particular epidermally expressed protein and recessive disorders of the skin. Examples where this approach has been used successfully include (i) junctional epidermolysis bullosa, a severe skin-peeling disorder caused by the loss-of-function mutations in the genes encoding the α6β4 integrin heterodimer or its adhesive ligand, laminin 5 (18–21); (ii) epidermolysis bullosa simplex with muscular dystrophy, caused by lesions in the plectin gene, encoding a cytoskeletal linker protein expressed abundantly in epidermis and muscle (22); and (iii) lamellar ichthyosis, a severe scaling disorder caused by a loss of transglutaminase 1, an enzyme required for crosslinking epidermal proteins into a cornified envelope in the late stages of terminal differentiation (23).
Although the use of transgenic mouse and gene knockout technology has greatly advanced our understanding of epidermal protein function and of the genetic bases of autosomal dominant and recessive disorders of the skin, there are a number of serious limitations to these strategies. In some cases, for instance, a gene may perform a function that is key to epidermal growth and/or differentiation, but germ-line mutations of the gene may cause early embryonic lethality, thus barring analysis of its function in somatic tissues. A germ-line mutation in a more broadly expressed gene could also cause pleiotropic effects, making it difficult to assess to what extent pathology detected in the skin is attributable directly to loss of expression of the targeted gene in the skin. Finally, a particular gene might function both early and late in epidermal development and/or differentiation, but germ-line mutations would restrict analysis to the earliest essential role of the protein.
In light of these caveats, we have developed a system for conditionally knocking-out genes in developing embryonic epidermis and for inducing the specific knockout of genes in postnatal skin at a selected time and at a selected site. To ablate genes in embryonic epidermis, we have generated transgenic mice expressing K14 Cre, encoding the bacterial Cre recombinase (24, 25) under the control of the well established human K14 promoter. The expression pattern of this promoter has been determined thoroughly and has been used widely in epidermal transgenic technology (1, 3). To ablate genes in postnatal skin, we have generated mice containing a K14–CreERtam transgene, encoding a fusion protein between Cre recombinase and the tamoxifen responsive hormone-binding domain of the estrogen receptor. We show that, in epidermis of postnatal animals, CreERtam transgene expression does not yield active Cre recombinase. However, Cre can be activated by either orally or topically administering tamoxifen. Taken together, these tools now make it possible to conduct functional studies of broadly expressed epidermal genes and to relate loss-of-function mutations to mosaic disorders of the skin, including skin cancer.
MATERIALS AND METHODS
Preparation of Transgene Constructs and Generation of Transgenic Mice.
The expression vector used for generation of Cre transgenic mice contains a 2,100-bp AvaI fragment encompassing the human K14 promoter/enhancer (1). For stabilization of the transcript, the vector contains a rabbit β-globin 5′ untranslated region (UTR) and an intronic sequence 5′ to a BamHI site, as well as the K14 3′ UTR and polyadenylation signal 3′ to this site. The cDNA for Cre recombinase (26) was then subcloned into the BamHI site of the K14 expression vector (3).
For tamoxifen-inducible Cre recombinase, we generated a Cre fusion protein cDNA by using an altered hormone-binding domain of the mouse estrogen receptor, ERtam, which fails to bind estrogen but instead responds only to tamoxifen (27).
Transgenic mice were generated by linearizing the plasmids with KpnI and SphI, injecting DNA into the male pronucleus of fertilized single cell mouse embryos, and implanting embryos into the oviducts of pseudopregnant CD1 female mice as described (1).
Reverse Transcriptase–PCR (RT-PCR).
Tissues and organs were obtained from transgenic mice as described (1). Separation of epidermis and dermis was achieved by using dispase enzyme treatment at 37°C for 30 min to 1 h (3). Note that this procedure leaves some hair follicle contamination in the dermal fraction but results in quite pure epidermal tissue. The tissue specificity of Cre expression was determined by isolating RNAs from different mouse tissues and testing them for Cre sequences by using RT-PCR, the RNA preamplification kit (GIBCO/BRL), and two primers from Cre cDNA: 5′-TGCTGTTTCACTGGTTATGCGG-3′ and 5′-TTGCCCCTGTTTCACTATCCAG-3′.
Histology and Cre Activity Assays.
For routine histology, tissues were fixed in Bouin’s fixative, processed, and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin, examined, and photographed by using an Axiophot microscope (Zeiss). To test for Cre activity in vivo, double-positive pups were analyzed for β-galactosidase activity as described (3). To assay for Cre-mediated recombination events in the Cre–MATE mouse, PCR was carried out on organ tissue DNAs. The following oligonucleotides were used: 5′-GCATTAATAAACTTGAGCAGACTTCAG-3′ and 5′-GCAAAATGATCCAGCGTCCTGGG-3′ for the recombination event; and 5′-TCCACCGCGGTGGCGGCCGCTCTAG-3′ and 5′-GCAAAATGATCCAGCGTCCTGGG-3′ for the unrearranged gene sequence.
RESULTS
Generation of Transgenic Mice Expressing K14–Cre Recombinase at Sufficient Levels to Ablate the Expression of Floxed Genes Quantitatively.
Several other laboratories have used conditional knockout approaches that have resulted in targeted genome rearrangement in the skin. In one study, Feil et al. (28) generated a transgenic mouse driving the expression of the Cre-ERtam-inducible fusion protein under the control of the broadly expressed cytomegalovirus promoter. On systemic injection of tamoxifen, approximately 40% of skin cells showed tamoxifen-induced excision of floxed sequences (29). More recently, Tarutani et al. (30) showed that basal epidermal promoters could be used successfully to knockout genes conditionally in the developing skin of mice.
Although these earlier results are promising, to be broadly applicable for functional studies of many different genes that are expressed in the stratified epithelia of skin, it is essential that Cre recombinase mice are generated in which gene expression is specifically and efficiently targeted to the epidermis in a temporally defined and controllable fashion. To achieve these goals, we began by using an approach similar to the one used by Tarutani et al. (30) and engineered transgenic mice to express Cre recombinase under the control of the extensively characterized human K14 promoter (3). The activity of this promoter is strongly up-regulated at embryonic day 14.5 (E14.5), and in postnatal animals, it is predominantly restricted to the basal, i.e., mitotically active, layer of epidermis, the outer root sheath of hair follicle, and oral epithelium (3).
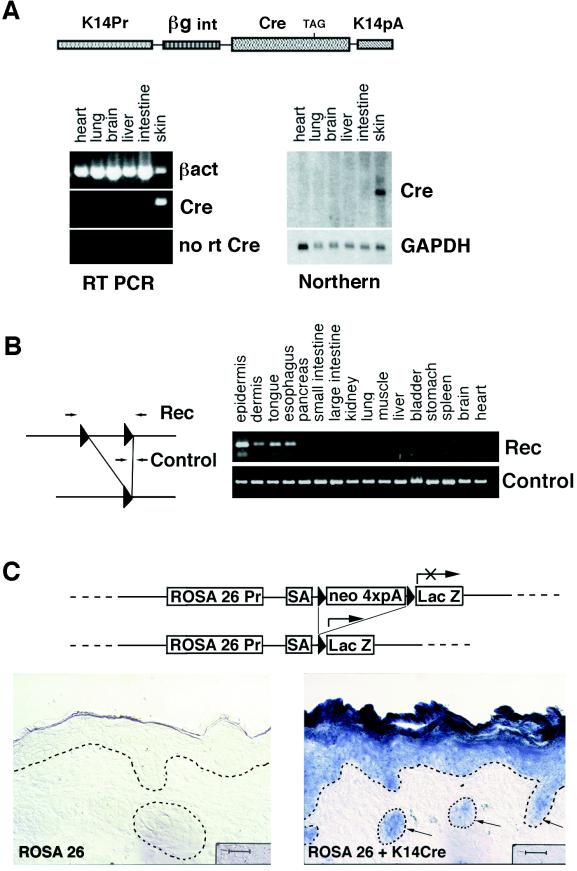
To optimize mRNA expression levels, we used an intron-containing 5′ UTR from the human β-globin gene and a 3′ UTR and polyadenylation signal from the human K14 gene (Fig. 1A). Of four independent lines, the one chosen for this study was that which expressed the highest level of skin-specific Cre mRNA as judged by Northern blot analysis and RT-PCR (Fig. 1A).
Figure 1.
Generation and analysis of transgenic mouse expressing Cre recombinase in skin epithelia. (A) Schematic representation of transgene, as well as RT-PCR and Northern analysis of Cre mRNA expression in organs taken from our K14–Cre mouse line. RNAs isolated from transgenic organs were subjected to RT-PCR with oligonucleotides specific for β-actin or Cre mRNA. Reverse transcriptase was omitted in control reactions for Cre (No RT Cre). For Northern blot hybridization, Cre cDNA was used as a test probe, and a fragment of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was used as a control. K14Pr, the human K14 promoter; βgint, the 5′ untranslated sequences encompassing an intron from the human β-globin gene; Cre, a full-length cDNA encoding bacterial Cre recombinase; K14pA, 3′ UTR and polyadenylation signal from human K14 mRNA. (B) K14–Cre PCR analysis. DNA was extracted from organs of newborn Cre–MATE/K14–Cre double-positive pups. PCR was carried out with oligonucleotides recognizing either the Cre-driven recombination event (Rec) or the nonrecombined allele (Control). (C) Immunohistochemical assay for Cre activity. The ROSA26 Cre recombinase test allele contains a floxed neo gene separating the ROSA26 promoter (ROSA26 Pr) from the bacterial lacZ gene; under these circumstances, β-galactosidase is expressed only in Cre-active cells that have recombined out the floxed neo gene (31). Tissues from ROSA26 (Left) and from ROSA26/K14–Cre double-positive mice (Right) were stained for β-galactosidase activity. Shown are the results from skins. Triangles, loxP sequences; SA, splice acceptor; neo 4xpA, the neo gene with four polyadenylation signals to ensure transcript termination (31); arrows denote hair follicles. Dotted lines indicate the demarcation between epithelium and mesenchyme. The dark patch to the left of each bar is a microscopy shadow. (Bars = 50 μm.)
To test for Cre activity, we first bred our K14–Cre transgenic mice to a Cre test mouse line, Cre–MATE, that has, in its genome, one allele that contains an insertion of two loxP sites, in head-to-tail orientation, separated by 3 kilobases of genomic sequence (Fig. 1B). Different organs were then taken from double-positive animals, and after extraction of genomic DNAs, the allele was then analyzed by PCR for the Cre-dependent recombination event that resulted in the deletion of the 3-kilobase sequence internal to the loxP sites. Of the tissues examined, recombination was detected only in those that normally express K14 gene, including the epidermis of skin, esophagus, and tongue (Fig. 1B). Based on our previous extensive study of K14 promoter activity, Cre recombinase activity is also likely in cornea, vaginal epithelium, salivary glands, and mammary epithelium (3).
A small level of recombination was detected consistently in the dermal sample. As judged by Western blot analysis with an antibody against keratinocyte-specific K14 (not shown), this recombination seems to be attributable to contamination of the dermal fraction with keratinocytes. Most likely the contaminating keratinocytes come from hair follicles, which are not removed easily by the dispase treatment. Further evidence of keratinocyte specificity of Cre is provided below.
For the studies described here, both the efficiency and the specificity of Cre-mediated recombination were crucial. To test for these features in detail, we bred our K14–Cre transgenic animals to the ROSA26 Cre reporter mouse. This mouse has a ROSA26 locus genetically manipulated such that just downstream from the ROSA26 promoter is a neo gene flanked by lox sequences, followed by an adjacent lacZ coding sequence (31). Under these circumstances, even though the ROSA26 promoter is broadly active, β-galactosidase activity will be detected only when the floxed neo gene has been removed by Cre recombinase action (31). As shown in Fig. 1C, epidermal-specific and hair follicle-specific β-galactosidase activity was detected only when the ROSA26 mice were bred onto the K14–Cre background. β-Galactosidase activity was also detected in the tongue and esophagus but not in organs and tissues where the K14 promoter is silent (data not shown). Importantly, most, if not all, the K14-expressing cells within the skin section scored positive for Cre recombinase activity. The persistent blue staining in suprabasal epidermal layers was expected, given that these cells are derived from the basal layer. Taken together, our analyses established that our K14–Cre mouse is able to recombine floxed sequences in a highly efficient and keratinocyte-specific fashion.
Inducing Cre Recombinase Activity and Targeted Genome Rearrangement in the Stratified Epithelial Tissues of Postnatal Animals.
To regulate the activity of Cre in vivo, we modified the K14–Cre construct and replaced Cre cDNA with an in-frame fusion of the coding sequences of the complete Cre cDNA, coupled to the sequences encompassing an altered estrogen receptor hormone-binding site and regulatory domain (Fig. 2A). The hormone-binding site harbors a point mutation that makes the site unresponsive to the natural ligand, 17β-estradiol, but responsive to estrogen antagonists such as tamoxifen or 4-hydroxytamoxifen (27). Similar fusion proteins have been found to be regulated tightly in vivo when they are expressed under the control of the cytomegalovirus or Wnt1 promoters (28, 29, 32).
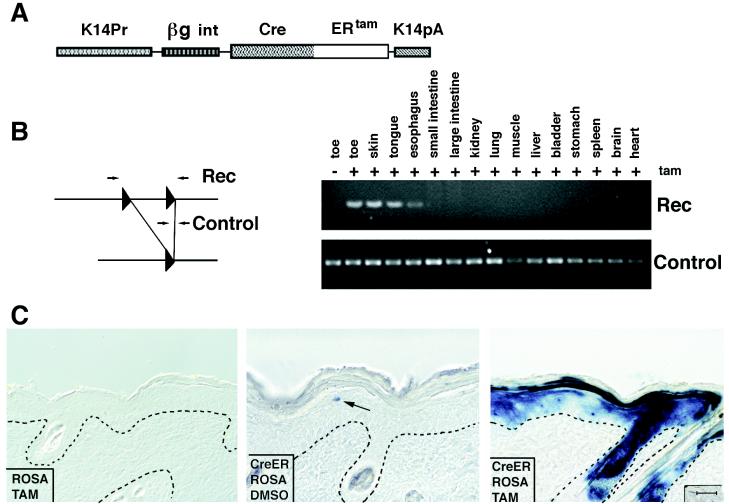
Figure 2.
Generation and analysis of transgenic mice expressing the CreERtam fusion protein in skin epithelia. (A) Schematic representation of the transgene. (B) PCR analysis of the DNA extracted from different organs of K14–CreERtam/Cre–MATE double-positive animals before and after oral administration of tamoxifen. PCR was conducted as described in the legend for Fig. 1. (C) Immunohistochemical assay for Cre activity in transgenic mouse skin. Double-positive K14–CreERtam/ROSA26 Cre test or single-positive ROSA26 Cre test transgenic mice were used in this experiment. Tamoxifen in DMSO or DMSO alone was applied to the skin once a day for 5 days. A day after the last application, skin biopsies were taken and sections were stained for β-galactosidase activity. Shown are the results from ROSA26 Cre test mouse treated with tamoxifen (Left); double-positive ROSA26 Cre test/K14–CreERtam mice treated with DMSO (Center); and double-positive mice treated with tamoxifen (Right). Triangles, loxP sequences; the arrow denotes a single cell positive for β-galactosidase in the skin from double-positive ROSA26 Cre test/ K14–CreERtam mouse treated with DMSO. (Bar = 50 μm.)
Transgenic mice were again engineered, this time harboring the K14–CreERtam transgene. Two transgenic lines were obtained, and of these, the highest-expressing animal was chosen for further study. To analyze the activity of CreERtam, we used the same approach as in the case of regular Cre. We first bred the K14–CreERtam mouse with our Cre–MATE test mouse, and the resulting double-positive mice were used for induction experiments.
To induce Cre activity in all stratified squamous epithelia known to have K14 promoter activity, we administered tamoxifen orally to the K14–CreERtam mice. Tamoxifen (1 mg per mouse per day) was given for 5 consecutive days. A day after the last dose was administered, mice were killed, and genomic DNAs were extracted from different organs. PCR analysis specific for the Cre recombinase-dependent recombination event identified recombination in DNA samples from toe, back skin, and tongue but not from nonstratified epithelial tissues (Fig. 2B). Importantly, no recombination was detected before tamoxifen administration (see Fig. 2B, toe samples ± tam treatment).
In the course of these experiments, we noticed that the oral doses of tamoxifen that we used (1 mg per mouse) were quite toxic to the animals. This dose, also used by other researchers (29, 32), is close to the lethal dose for mice, which is about 2 mg per mouse. In rodents, tamoxifen is converted to α-hydroxytamoxifen in the liver, where it can cause damage and even hepatocarcinomas if administered chronically (33).
The period of drug administration was relatively short for the purposes of inducible gene targeting, and the toxicity we observed seemed to be reversible, as long as only a single dose of the drug was administered. However, these effects could complicate the interpretation of a gene knockout phenotype produced by internal tamoxifen administration if the phenotype is analyzed before the recovery period, which seems to be approximately 1 week after administration of the drug.
Inducing Cre Recombinase Activity and Homologous Recombination in a Specific Patch of Skin on Postnatal Animals.
We next turned to establishing the technology for regulating the activity of Cre in a highly restricted fashion in a patch of skin on a postnatal animal. For this purpose, we used our K14–CreERtam/ROSA26 double-positive animals and determined whether we could induce Cre activity by topical application of a saturating solution of tamoxifen in DMSO. In this case, we applied 100 μl of 200 mg/ml tamoxifen solution once a day for 5 days to a small, approximately 1.5 × 1.5-cm patch of shaved skin on the back of test animals. As a control, an equivalent patch received DMSO alone.
After the experiment, the skins were processed for PCR analysis of genomic DNAs and for β-galactosidase activity assays. As judged by PCR, CreERtam recombinase had been activated specifically in skin that had been topically treated with tamoxifen but not in DMSO-treated skin (data not shown). As judged by β-galactosidase activity assays on sections taken from treated patches of ROSA26/K14–CreERtam skin, Cre recombinase had been activated in many of the cells of the epidermis and outer root sheath of hair follicles (Fig. 2C Right). In contrast, β-galactosidase activity was not detected in skin cells of ROSA26 single-positive mice that were treated with tamoxifen (Fig. 2C Left). Occasionally, we detected a blue-stained cell in the epidermis of K14–CreERtam/ROSA26 double-positive mice that had been treated with DMSO alone (Fig. 2C, arrow in Center). These cells were rare and seem to be indicative of a very low background level of CreERtam activity that can occur in the absence of tamoxifen induction.
Tamoxifen-induced recombinations were detected in all layers of skin epidermis, consistent with the fact that K14 promoter activity exists in the basal layer of epidermis, from which all differentiating layers are derived. Approximately 50–60% of the cells in the epidermis showed evidence of Cre-induced recombination. This efficiency was significantly better than the 25% observed in Wnt1-promoter driven CreERtam transgenic mice and than the systemic administration of tamoxifen used to activate cytomegalovirus–CreERtam (29, 32). Moreover, not only did topical application of tamoxifen prove to be a more efficient method for activating CreERtam in postnatal animals, but in addition, the precision and selectivity of activation was improved significantly by using this method. Importantly, animals topically treated with tamoxifen remained physically healthy in appearance, in contrast to those that were fed the drug orally.
Induction of Cre Recombinase in K14–CreERtam Mice Targets Epidermal and Hair Follicle Stem Cells.
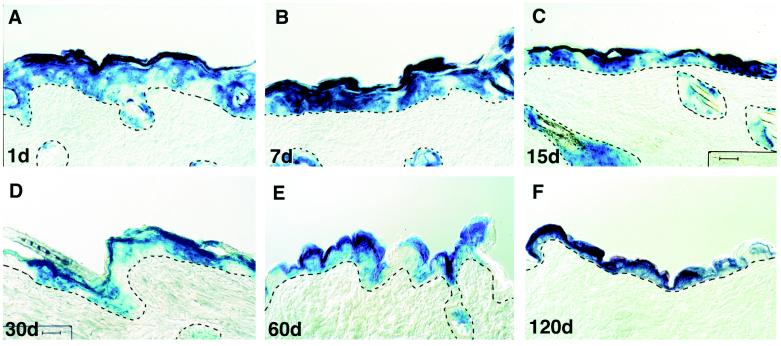
An important issue in inducible knockout technology is the extent to which a targeting event becomes a permanent feature of the animal. To address this issue, we turned to evaluating the persistence of Cre-dependent recombination of the ROSA26 locus in skin. As shown in Fig. 3, blue-staining keratinocytes were still detected in both epidermal and hair follicle cells for more than 4 months after the recombination events had been induced at the ROSA26 locus. Overall, the percentage of the β-galactosidase-positive cells was not significantly different between skin examined at 1 week after the initial application and that examined at 4 months after application. Given that the rate of the renewal of mouse skin epidermis is about 1 week, the persistence of blue cells is reflective of successful targeting of epidermal and hair follicle stem cells.
Figure 3.
Persistence of cells with a Cre-activated, recombined genome: evidence for targeting epidermal stem cells. Double-positive ROSA26 Cre test/K14–CreERtam transgenic mice were topically treated with tamoxifen once a day for 5 days. Skin biopsies were taken 1 day after the last tamoxifen application (A); 7 days (B); 15 days (C); 30 days (D); 2 months (E); or 4 months (F). Biopsies were sectioned and stained for β-galactosidase activity. The dotted lines denote the demarcation between epithelim and mesenchyme. (Bar = 50 μm.)
Although our results with topical applications of tamoxifen were encouraging for efficient, postnatal induction of Cre-mediated homologous recombination in the skin, more extensive testing of our older animals indicated the presence of blue-stained keratinocytes at other skin body sites extending beyond the sites where tamoxifen was applied (data not shown). If the mice had licked the tamoxifen-treated body site, it is possible that the drug could have been transferred to these additional sites. Alternatively, systemic entry through adsorption of the drug into the skin could have led to a distribution of tamoxifen to other skin sites. Future studies will be needed to explore these possibilities in greater detail and to optimize the dose and means of topical tamoxifen administration so as to minimize its distribution to other keratinocytes.
DISCUSSION
Our findings provide a study of the efficacy of keratinocyte-specific promoters in targeting the ablation of genes in mice in a developmental-specific, differentiation-specific, and inducible-specific fashion. Our findings support and extend the earlier studies of Tarutani et al. (30) and Brocard et al. (29), who obtained partially specific ablation of genes in skin epidermis. In our study, we show that targeting with the K14 promoter occurs efficiently in epidermis and hair follicles in mice aged from E14.5 to adulthood and that efficient activation of Cre recombinase also occurs in other K14-promoter active tissues, including tongue, esophagus, cornea, and oral epithelia.
The epidermis develops in a patterned fashion, and the basal epidermal keratin promoters are activated in a similar manner (2, 3). Thus, it is important to note that although E14.5 is the first day at which a high level of K14 promoter activity occurs over the entire body surface of the mouse embryo, activity actually occurs in a few select regions of the skin as early as E9.5. Thus, for a single mouse embryo, if the appropriate body sites are sampled, the consequences of a particular gene-targeting event can be examined over a broad range of developmental stages. An additional feature is that, because K14–Cre is expressed in the basal epidermal layer, the entire epidermis becomes targeted for the desired recombination event shortly after Cre activation.
A myriad of potential uses exists for keratinocyte-specific knockouts in mice. Most importantly, this technology will enable researchers to examine the consequences of targeting gene ablation to stratified squamous epithelia under conditions where the gene need not be expressed uniquely in keratinocytes. One interesting group of genes are those encoding intercellular adhesion proteins, such as α-catenin. Full-scale ablation causes loss of epithelial adherens junctions in preimplantation embryos (34). α-Catenin also has been implicated in epithelial polarity in somatic tissues, and its absence has been correlated with metastatic cancer progression in adult tissues (35). Analysis of its role in postnatal epithelia will be key to evaluating the extent to which such correlations are functionally relevant.
Another prime candidate for keratinocyte-specific targeting is the gene encoding β-catenin, a protein involved not only in adherens junctions, but also Wnt signaling (36). Although β-catenin ablation results in gastrulation defects in mice (37), it has also been implicated in hair follicle formation and epidermal stem cell fate determination (14, 38), neither of which occur until well after gastrulation. Other genes of interest for conditional targeting in skin include cell-cycle-regulated genes, such as c-myc, known to play an important role in epidermal growth and differentiation (39), and TGFβs and their receptors, which influence proliferation in many tissues, including epidermis (ref. 40 and references therein). For these and many other uses, this knockout technology will be useful in enhancing our general understanding of the biology of epithelial tissue and function, ranging from epithelial polarity to balancing proliferation and differentiation to epithelial–mesenchymal interactions.
Inducible knockout technology is an additional step toward a more complete understanding of epithelial biology, in that we can now target specific genes for ablation in adult skin and in patches of skin. This technology opens the door for understanding the consequences of ablating specific genes in the epidermal and hair follicle stem cells of adult animals. Such research is expected to be invaluable not only for advancing our knowledge of the genetic bases for mosaic skin disorders, but also for unraveling the pathways involved in squamous and basal cell carcinomas of the skin. Finally, although the study reported here has concentrated on the use of our K14–CreERtam animals for topically inducing Cre-mediated recombination in the skin, it may be possible to develop methods to localize tamoxifen in specific tissues where K14 promoter activity is high, thereby enabling induced ablation of genes specifically in, for example, cornea or oral epithelia. Such technology could be valuable for generating mouse models for various types of human epithelial cancers and genetic disorders.
Acknowledgments
We are grateful to Wenyu Bai for her help in generating the transgenic mice presented in this study, to Phil Soriano (Fred Hutchinson Cancer Center) for kindly providing the ROSA26 mice before publication, and for the help and support received from the University of Chicago Cancer Research Center Transgenic Animal Facility. This work was supported by grants from the National Institutes of Health and the National Cancer Institute. E.F. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- Kn
keratin n
- CreERtam
fusion protein between Cre recombinase and the tamoxifen responsive hormone-binding domain of the estrogen receptor
- UTR
untranslated region
- K14–Cre
transgene containing the Cre coding sequence 3′ from the human K14 promoter/enhancer
- RT-PCR
reverse transcriptase–PCR
- En
embryonic day n
References
- 1.Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E. Proc Natl Acad Sci USA. 1989;86:1563–1567. doi: 10.1073/pnas.86.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne C, Tainsky M, Fuchs E. Development (Cambridge, UK) 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Zinkel S, Polansky K, Fuchs E. Proc Natl Acad Sci USA. 1997;94:219–226. doi: 10.1073/pnas.94.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailleul B, Surani M A, White S, Barton S C, Brown K, Blessing M, Jorcano J, Balmain A. Cell. 1990;62:697–708. doi: 10.1016/0092-8674(90)90115-u. [DOI] [PubMed] [Google Scholar]
- 5.Carroll J M, Albers K M, Garlick J A, Harrington R, Taichman L B. Proc Natl Acad Sci USA. 1993;90:10270–10274. doi: 10.1073/pnas.90.21.10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenhalgh D A, Rothnagel J A, Quintanilla M I, Orengo C C, Gagne T A, Bundman D S, Longley M A, Roop D R. Mol Carcinog. 1993;7:99–110. doi: 10.1002/mc.2940070208. [DOI] [PubMed] [Google Scholar]
- 7.Vassar R, Coulombe P A, Degenstein L, Albers K, Fuchs E. Cell. 1991;64:365–380. doi: 10.1016/0092-8674(91)90645-f. [DOI] [PubMed] [Google Scholar]
- 8.Fitzpatrick T B, Freedberg I M, Eisen A Z, Wolff K, Austen K F, Goldsmith L A, Katz S I. Fitzpatrick’s Dermatology in General Medicine. 1–2. New York: McGraw–Hill; 1999. [Google Scholar]
- 9.Coulombe P A, Hutton M E, Letai A, Hebert A, Paller A S, Fuchs E. Cell. 1991;66:1301–1311. doi: 10.1016/0092-8674(91)90051-y. [DOI] [PubMed] [Google Scholar]
- 10.Bonifas J M, Rothman A L, Epstein E H. Science. 1991;254:1202–1205. doi: 10.1126/science.1720261. [DOI] [PubMed] [Google Scholar]
- 11.Lane E B, Rugg E L, Navsaria H, Leigh I M, Heagerty A H M, Ishida-Yamamoto A, Eady R A J. Nature (London) 1992;356:244–246. doi: 10.1038/356244a0. [DOI] [PubMed] [Google Scholar]
- 12.Wang X J, Greenhalgh D A, Lu X R, Bickenbach J R, Roop D R. Oncogene. 1995;10:279–289. [PubMed] [Google Scholar]
- 13.Oro A E, Higgins K M, Hu Z, Bonifas J M, Epstein E H, Scott M P. Science. 1997;276:817–821. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- 14.Gat U, DasGupta R, Degenstein L, Fuchs E. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 15.Chan E F, Gat U, McNiff J M, Fuchs E. Nat Genet. 1999;21:410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 16.Capecchi M R. Trends Genet. 1989;5:70–76. doi: 10.1016/0168-9525(89)90029-2. [DOI] [PubMed] [Google Scholar]
- 17.Pirity M, Hadjantonakis A K, Nagy A. Methods Cell Biol. 1998;57:279–293. doi: 10.1016/s0091-679x(08)61585-x. [DOI] [PubMed] [Google Scholar]
- 18.Dowling J, Yu Q-C, Fuchs E. J Cell Biol. 1996;134:559–572. doi: 10.1083/jcb.134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georges-Labouesse E, Messaddeq N, Cadalbert L, Dierich A, Le Meur M. Nat Genet. 1996;13:370–373. doi: 10.1038/ng0796-370. [DOI] [PubMed] [Google Scholar]
- 20.van der Neut R, Krimpenfort P, Calafat J, Niessen C M, Sonnenberg A. Nat Genet. 1996;13:366–369. doi: 10.1038/ng0796-366. [DOI] [PubMed] [Google Scholar]
- 21.Christiano A M, Uitto J. Exp Dermatol. 1996;5:1–11. doi: 10.1111/j.1600-0625.1996.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 22.Andra K, Lassmann H, Bittner R, Shorny S, Fassler R, Propst F, Wiche G. Genes Dev. 1997;11:3143–3156. doi: 10.1101/gad.11.23.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuki M, Yamashita F, Ishida-Yamamoto A, Yamada K, Kinoshita C, Fushiki S, Ueda E, Morishima Y, Tabata K, Yasuno H, et al. Proc Natl Acad Sci USA. 1998;95:1044–1049. doi: 10.1073/pnas.95.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter A. Trends Genet. 1998;14:73–79. doi: 10.1016/s0168-9525(97)01326-7. [DOI] [PubMed] [Google Scholar]
- 25.Rossant J, McMahon A. Genes Dev. 1999;13:142–145. doi: 10.1101/gad.13.2.142. [DOI] [PubMed] [Google Scholar]
- 26.O’Gorman S, Wahl G M. Science. 1997;277:1025. doi: 10.1126/science.277.5329.1021c. [DOI] [PubMed] [Google Scholar]
- 27.Littlewood T D, Hancock D C, Danielian P S, Parker M G, Evan G I. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Proc Natl Acad Sci USA. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brocard J, Warot X, Wendling O, Messaddeq N, Vonesch J L, Chambon P, Metzger D. Proc Natl Acad Sci USA. 1997;94:14559–14563. doi: 10.1073/pnas.94.26.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarutani M, Itami S, Okabe M, Ikawa M, Tezuka T, Yoshikawa K, Kinoshita T, Takeda J. Proc Natl Acad Sci USA. 1997;94:7400–7405. doi: 10.1073/pnas.94.14.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 32.Danielian P S, Muccino D, Rowitch D H, Michael S K, McMahon A P. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 33.Wogan G N. Semin Oncol. 1997;24:S1-87–S1-97. [PubMed] [Google Scholar]
- 34.Torres M, Stoykova A, Huber O, Chowdhury K, Bonaldo P, Mansouri A, Butz S, Kemler R, Gruss P. Proc Natl Acad Sci USA. 1997;94:901–906. doi: 10.1073/pnas.94.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barth A I, Nathke I S, Nelson W J. Curr Opin Cell Biol. 1997;9:683–690. doi: 10.1016/s0955-0674(97)80122-6. [DOI] [PubMed] [Google Scholar]
- 36.Wodarz A, Nusse R. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 37.Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Development (Cambridge, UK) 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 38.Jian-Zhu A, Watt F M. Development (Cambridge, UK) 1999;126:2285–2298. doi: 10.1242/dev.126.10.2285. [DOI] [PubMed] [Google Scholar]
- 39.Gandarillas A, Watt F M. Genes Dev. 1997;11:2869–2882. doi: 10.1101/gad.11.21.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui W, Fowlis D J, Cousins F M, Duffie E, Bryson S, Balmain A, Akhurst R J. Genes Dev. 1995;9:945–955. doi: 10.1101/gad.9.8.945. [DOI] [PubMed] [Google Scholar]