Abstract
Polycystic ovary syndrome (PCOS) is a common endocrine disorder of women, characterized by hyperandrogenism and chronic anovulation. It is a leading cause of female infertility and is associated with polycystic ovaries, hirsutism, obesity, and insulin resistance. We tested a carefully chosen collection of 37 candidate genes for linkage and association with PCOS or hyperandrogenemia in data from 150 families. The strongest evidence for linkage was with the follistatin gene, for which affected sisters showed increased identity by descent (72%; χ2 = 12.97; nominal P = 3.2 × 10−4). After correction for multiple testing (33 tests), the follistatin findings were still highly significant (Pc = 0.01). Although the linkage results for CYP11A were also nominally significant (P = 0.02), they were no longer significant after correction. In 11 candidate gene regions, at least one allele showed nominally significant evidence for population association with PCOS in the transmission/disequilibrium test (χ2 ≥ 3.84; nominal P < 0.05). The strongest effect in the transmission/disequilibrium test was observed in the INSR region (D19S884; allele 5; χ2 = 8.53) but was not significant after correction. Our study shows how a systematic screen of candidate genes can provide strong evidence for genetic linkage in complex diseases and can identify those genes that should have high (or low) priority for further study.
Polycystic ovary syndrome (PCOS) is a common endocrine disorder that is found in ≈4% of women of reproductive age (1) and results in reduced fertility and a 7-fold increased risk for type 2 diabetes mellitus (2). The syndrome is characterized by hyperandrogenism and chronic anovulation. It is also associated with polycystic ovaries, hirsutism, obesity, and insulin resistance. The observation of familial aggregation of PCOS (3–5) is consistent with a genetic basis for this disorder. However, the mode of inheritance of PCOS has not been firmly established. Although some studies support a single dominant gene with high penetrance (6–8), others do not (9).
Several pathways have been implicated in the etiology of PCOS. These include the metabolic or regulatory pathways of steroid hormone synthesis (10, 11), regulatory pathways of gonadotropin action (12), the insulin-signaling pathway (13–15), and pathways regulating body weight (16). Several genes from these pathways have been tested as candidate genes for PCOS (10, 11, 17–23). In particular, in the insulin receptor gene (INSR), mutations have been identified in several rare syndromes that, like PCOS, are characterized by hyperandrogenism and insulin-resistant diabetes mellitus. These syndromes include leprechaunism, Rabson–Mendenhall syndrome, and type A syndrome (20–23). Although mutation analysis, linkage studies, and case-control association studies have been carried out with these candidate genes, evidence that any of them play a role in PCOS has not been replicated widely and is still inconclusive. These uncertainties are common in “complex” genetic diseases, where identifying the contributing genes is made difficult by likely genetic heterogeneity, environmental contributions, and multiple etiologies.
As an initial step in the identification of genes playing a role in the etiology of PCOS, we carried out a genetic analysis of 37 candidate genes for PCOS. We chose to analyze candidate genes for PCOS, in part because several well characterized metabolic pathways and candidate genes had been implicated in the etiology of PCOS, but also because we have not yet assembled enough families to carry out a complete genome scan. We tested for linkage with the candidate genes by the affected sib-pair (ASP) test (24), and we tested for association between alleles of the candidate gene markers by the transmission/disequilibrium test (TDT; ref. 25). These methods require no assumption about mode of inheritance, and the TDT, unlike case-control studies, is not influenced by population structure or heterogeneity (26).
MATERIALS AND METHODS
Family Ascertainment and Phenotypes.
We studied 150 nuclear families with at least one affected index case. Among the families, 148 were of European origin and 2 were of Caribbean origin. Criteria for diagnosis are described by Legro et al. (6). Briefly, an index case was considered affected if she met the following criteria: chronic menstrual irregularity (amenorrhea or ≤ six menses per year; ref. 27) and hyperandrogenemia (HA), i.e., elevated levels of total testosterone or testosterone not bound to sex hormone-binding globulin. Hormone levels were considered elevated if they were more than two standard deviations above the control mean; in our assay these thresholds were 58 ng/dl and 15 ng/dl for total testosterone and testosterone not bound to sex hormone-binding globulin, respectively. Nonclassical 21-hydroxylase deficiency, hyperprolactinemia, and androgen-secreting tumors were excluded (28). HA is a salient and unambiguous biochemical feature of PCOS and is found in a significant proportion of sisters of patients with PCOS, even in the absence of oligomenorrhea (6, 8, 29). Our previous studies have suggested that HA is the major reproductive endocrine phenotype in our families with PCOS (6). For genetic analysis, therefore, female relatives of index cases were considered affected if they had elevated androgen levels, whether or not they had oligomenorrhea (6), and we used the designation “PCOS/HA” to describe this combined category. Female relatives were not screened for nonovarian causes of HA. Women were considered unaffected if they had normal circulating androgen levels, were not taking any confounding medications (e.g., oral contraceptives or insulin-sensitizing agents), and had regular menstrual cycles (menses every 27–35 days; ref. 6). Women not of reproductive age and those not fulfilling the criteria for affected or unaffected phenotypes were assigned the phenotype “unknown” (6). Because the male phenotype corresponding to PCOS is unclear, all men in the study also were assigned the phenotype “unknown.”
There were 134 sisters of index cases; 39 sisters were affected (PCOS/HA); 46 sisters were unaffected; and for 49 sisters, the phenotype was unknown. Of the 39 affected sisters, 14 had HA but not oligomenorrhea. Among the 28 multiplex families, the number of sibships with two, three, four, or five affected offspring were 21, 4, 2, and 1, respectively. Maximum sample size for TDT was 163 trios (affected daughter and both parents).
Candidate Genes.
We chose 37 candidate genes from four metabolic pathways that have been implicated in the etiology of PCOS (Table 1). These 37 genes map to 33 distinct chromosomal locations. Where possible, we typed polymorphic sites within each candidate gene. For candidate genes without polymorphisms, we chose closely linked short tandem repeat polymorphisms (STRPs). For 28 of the 37 candidate genes, there is at least one polymorphic marker within 1 cM of the candidate gene. For the remaining nine candidate genes, polymorphic markers are 1–4 cM from the candidate gene.
Table 1.
Genotyping panel for 37 PCOS candidate genes
| Marker locus | Gene symbol | Candidate gene | Distance, in centimorgans (cM)* | Chromosomal location |
|---|---|---|---|---|
| Steroid hormone | ||||
| AR | AR | Androgen receptor | 0 | Xq11.2 |
| D15S519 | CYP11A | CYP11A-cytochrome P450 side-chain cleavage enzyme | 0 | 15q23–24 |
| D15S520 | CYP11A | CYP11A-cytochrome P450 side-chain cleavage enzyme | 0 | 15q23–24 |
| D10S192 | CYP17 | CYP17-cytochrome P450 17α-hydroxylase/17,20-desmolase | <1 | 10q24.3 |
| CYP19 | CYP19 | CYP19-cytochrome P450 aromatase | 0 | 15q21 |
| D17S934 | HSD17B1 | 17 β-hydroxysteroid dehydrogenase, type I | <2 | 17q11–21 |
| HSD17B2 | HSD17B2 | 17 β-hydroxysteroid dehydrogenase, type II | 0 | 16q24.2 |
| D9S1809 | HSD17B3 | 17 β-hydroxysteroid dehydrogenase, type III | <1 | 9q22 |
| D1S514 | HSD3B1+2 | 3 β-hydroxysteroid dehydrogenase, type I and II | <1 | 1p31.1 |
| D8S1821 | STAR | Steroidogenic acute regulatory protein | <2 | 8p11.2 |
| Gonadotropin action | ||||
| D12S347 | ACTR1 | Activin receptor 1 | <1 | 12q13.12 |
| D2S2335 | ACTR2A | Activin receptor 2A | <1 | 2q22.2 |
| D3S1298 | ACTR2B | Activin receptor 2B | <1 | 3p22.2 |
| D5S474 | FS | Follistatin | <2 | 5p14 |
| D5S623 | FS | Follistatin | <0.5 | 5p14 |
| D5S822 | FS | Follistatin | <1 | 5p14 |
| D2S163 | INHA | Inhibin A | <1 | 2q33.34 |
| INHBA | INHBA | Inhibin β-A | 0 | 7p13–15 |
| D2S293 | INHBB | Inhibin β-B | 2 | 2cen–2q13 |
| D12S1691 | INHC | Inhibin C | <1 | 12q13 |
| D17S1353 | SHBG | Sex hormone binding globulin | <1 | 17p13.2 |
| D2S1352 | LHCGR | Luteinizing hormone/choriogonadotropin receptor | <2 | 2p21 |
| D2S1352 | FSHR† | Follicle-stimulating hormone receptor | <2 | 2p21 |
| D18S474 | MADH4 | Mothers against decapentaplegic homolog 4 | <1 | 18q21 |
| Obesity and energy regulation | ||||
| D18S64 | MC4R | Melanocortin 4 receptor | <3 | 18q21.32 |
| D7S1875 | OB | Leptin | 0.2 | 7q31.3–32.1 |
| D1S198 | OBR | Leptin receptor | 0.5 | 1p31 |
| D2S131 | POMC | Pro-opiomelanocortin | <1 | 2p23 |
| D11S911 | UCP2+3 | Uncoupling protein 2+3 | <4 | 11q13 |
| Insulin action | ||||
| IGF1 | IGF1 | Insulin-like growth factor I | 0 | 12q22–23 |
| IGF1R | IGF1R | Insulin-like growth factor I receptor | 0 | 15q25–26 |
| D7S519 | IGFBP1+3 | Insulin-like growth factor binding protein 1 + 3 | 1 | 7p13–7p12 |
| HphI site | INS VNTR | Insulin gene VNTR | 0 | 11p15.5 |
| INSR | INSR | Insulin receptor | 0 | 19p13.3 |
| D19S216 | INSR | Insulin receptor | 4.2 | 19p13.3 |
| D19S905 | INSR | Insulin receptor | 0 | 19p13.3 |
| D19S884 | INSR | Insulin receptor | 1.2 | 19p13.3 |
| D19S922 | INSR | Insulin receptor | 1.2 | 19p13.3 |
| D19S391 | INSR | Insulin receptor | 3.6 | 19p13.2 |
| D19S865 | INSR | Insulin receptor | 7.2 | 19p13.2 |
| D19S906 | INSR | Insulin receptor | 11 | 19p13.2 |
| D19S840 | INSR | Insulin receptor | 14 | 19p13.2 |
| D19S212 | INSL3 | Leydig insulin-like protein 3 | <1 | 19p13.1 |
| D19S410 | INSL3 | Leydig insulin-like protein 3 | <1 | 19p13.1 |
| IRS1 | IRS1 | Insulin receptor substrate 1 | 0 | 2q36–37 |
| D3S1263 | PPARG | Peroxisome proliferator-activated receptor-gamma | <0.2 | 3p25–24.2 |
The list contains 45 polymorphic markers closely linked to 37 PCOS candidate genes.
*Distance between polymorphic marker and candidate gene.
D2S1352 was used for the two closely linked genes, LHCGR and FSHR.
Radiation Hybrid (RH) Mapping.
Candidate genes for which accurate mapping information was not available were mapped physically by using the Stanford Human Genome Center (SHGC) medium resolution G3 RH mapping panel (Research Genetics, Huntsville, AL). DNA (40 ng) from each somatic hybrid clone was amplified in a total volume of 8 μl in the presence of 200 μM dNTPs (Amersham Pharmacia), 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 1.0–2.0 mM MgCl2, 0.36 units AmpliTaq polymerase (Roche Molecular Systems, Branchburg, NJ), and 0.5 μM of each primer. The forward primer was labeled with [γ-33P]ATP and samples were electrophoresed on 6% acrylamide, 5 M urea gels at 70 W. Care was taken to choose primers that showed low levels of cross-species homology and, when relevant, low levels of homology to closely related human genes. Genotypes for the RH panel were submitted to a web server (shgc-www.stanford.edu) managed by the SHGC for chromosomal localization. STRP markers were chosen to map as closely as possible to the location determined by RH mapping (see Results).
Genotyping.
Genotypes were determined at 45 polymorphic loci linked to the 37 candidate genes (Table 1), and the 44 STRPs were assayed by denaturing PAGE. Radioactively labeled primers were used to label 2 of the STRPs (at AR and D11S911), whereas the remaining 42 STRPs were visualized by using fluorescently labeled primers and the ABI Sequencing system (PE Applied Biosystems). The HphI site at the insulin gene VNTR is a single nucleotide polymorphism, used as a surrogate for the VNTR itself (30), and was assayed by single-strand conformational polymorphism analysis.
For each fluorescently labeled STRP, 45 ng of genomic DNA was amplified as described for the RH mapping, except that the forward primer was fluorescently labeled. For some markers, it was necessary to add 9% (vol/vol) DMSO to obtain suitable PCR product. The STRPs were grouped in five “panels” of eight or nine markers each. The PCR products of any one panel were pooled to give approximately equal signal intensities. Pooled PCR products were electrophoresed in the presence of an internal size standard (Genescan 500) on 4% acrylamide, 5 M urea denaturing gels by using a 377 DNA sequencer (PE Applied Biosystems, Foster City, CA). Genotypes were determined by using the genescan analysis and genotyper programs (PE Applied Biosystems, Foster City, CA).
The radioactive PCRs were carried out as described for the RH mapping. The PCRs for the HphI polymorphism were carried out as described for the fluorescently labeled primers, in the presence of 1.5 mM MgCl2 and [α-33P]dCTP. Samples were electrophoresed overnight at room temperature at 9 W on an MDE gel (FMC).
Statistical Analysis.
The extent of identity by descent (IBD) in ASPs was used to test for linkage between the candidate gene and PCOS/HA (24). To incorporate sibships with more than two affected sisters, IBD was calculated by using the weighting scheme described by Suarez and Hodge (31). This method takes into account the fact that the sib pairs in larger sibships are not all independent and sometimes results in fractional numbers of transmitted alleles. The conventional χ2 statistic calculated with these data is “conservative”; the true significance levels would be more extreme than those quoted. In the present study of 33 independent regions, the apparent significance of any single test will be exaggerated as a result of the multiple tests. The P value for each single test was, therefore, multiplied by 33, and where appropriate, we also report the resulting corrected value Pc. Haplotypes used in multilocus IBD analysis were generated by the genehunter program (32) when both parents were available. Otherwise haplotypes were reconstructed manually (see below). We tested for association between specific alleles at the candidate gene markers and PCOS/HA by using the TDT (25).
Missing Parental Genotypes.
DNA samples could not be obtained from 20 parents. The analysis of sharing in families with one or two missing parental genotypes was done only if the transmissions to the affected could be determined unambiguously and without bias. Genotypes for missing parents were reconstructed by using genotypes of unaffected siblings or those with unknown phenotype. None of these siblings were included in the statistical analysis. Among the 28 multiplex families, there were 4 with one parent missing and 2 with both missing. For the TDT, when one parent was missing, the available parent’s genotype was used only if the inheritance could be determined unambiguously and without bias in affected individuals (33, 34).
RESULTS
RH Mapping.
RH mapping localized eight candidate genes whose detailed map positions were previously unknown. The results of chromosomal localization, as determined with the SHGC web server, are shown in Table 2. Two-point logarithm of odds scores between the candidate gene and the most closely linked marker ranged from 8.5 (SHBG) to 1,000 (INHA and MADH4), indicating high confidence in the localizations. The markers used for RH mapping were nonpolymorphic expressed sequence tags; using the RH localization, we chose a closely linked highly polymorphic STRP for genotyping. The polymorphic markers used for the genetic analysis are indicated with the approximate map distance between the marker and candidate gene in centimorgans (Table 2).
Table 2.
RH mapping of candidates genes for PCOS
| Gene* | Chromosomal location | Linked marker | Logarithm of odds† | STRP marker for linkage analysis | Distance between STRP and candidate gene, cM |
|---|---|---|---|---|---|
| FS | 5p14 | SHGC-36388 | 13.8 | D5S623 | <0.5 |
| SHBG | 17p13.2 | SHGC-35513 | 8.5 | D17S1353 | <1.0 |
| INHA | 2q36.1 | SHGC-11864 | 1,000 | D2S163 | <1.0 |
| INHC | 12q13 | AFM312XF5 | 11.5 | D12S1691 | <1.0 |
| ACTR1 | 12q13.12 | AFM298ZB1 | 9.4 | D12S347 | <0.8 |
| ACTR2A | 2q22.2 | SHGC-9391 | 10.3 | D2S2335 | <1.0 |
| ACTR2B | 3p22.2 | SHGC-115353 | 9.7 | D3S1298 | <1.0 |
| MADH4 | 18q21 | SHGC-33967 | 1,000 | D18S474 | <1.0 |
*Abbreviations are defined in Table 1.
Two-point maximum logarithm of odds score between candidate gene and most closely linked marker.
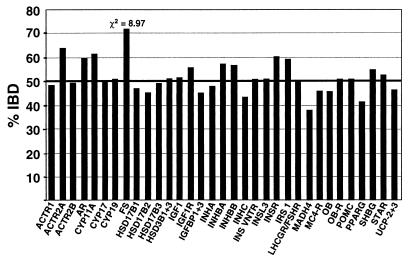
ASP Analysis.
The results of the ASP analysis for all 33 regions are shown in Fig. 1. By far the strongest evidence for linkage was observed for follistatin. The IBD for D5S623, the marker mapping closest to follistatin, was 72% (33.8 of 47 transmissions; χ2 = 8.97; P = 2.7 × 10−3). Haplotypes generated from D5S623 and two flanking STRPs also showed 72% IBD (47.9 of 66.5 transmissions), but the increase in the number of informative transmissions (from 47 to 66.5) resulted in χ2 = 12.91 (P = 3.27 × 10−4). Even after correction for multiple testing, this finding remains statistically significant (Pc = 0.01). The IBD for the 25 ASPs with PCOS (HA and menstrual irregularities) did not differ appreciably from that for the 14 ASPs where the nonindex sister had HA alone (data not shown).
Figure 1.
Summary of ASP analysis. IBD results for the marker with the highest IBD in each candidate gene region are shown. IBD was calculated for 39 ASPs in 28 families at each of 33 candidate gene regions (x axis). The IBD expected under the null hypothesis of no linkage is 50%. The only χ2 value > 3.84 (critical value for P < 0.05) is for follistatin.
We also found a modest increase in sharing at CYP11A. IBD was 62% for each of the two markers tested in this region. Haplotypes generated from these markers elevated the IBD to 67% (χ2 = 5.34). However, after correction for multiple testing, these results were not statistically significant at the P = 0.05 level. For several other markers (ACTR2A, AR, INSR, and IRS1), IBD was ≈60%, but in each case, small sample size (≤36 transmissions) led to nonsignificant results.
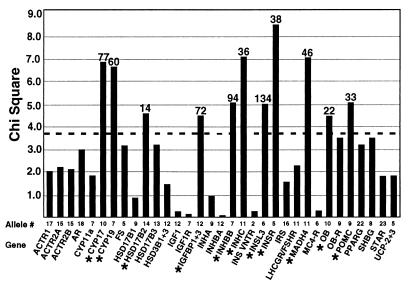
TDT.
The results of the TDT are shown in Fig. 2. Only alleles with at least 10 transmissions from a heterozygous parent to an affected daughter were included in the analysis. There were 349 such alleles. There was evidence for association (χ2 > 3.84; nominal P < 0.05) between at least one allele and PCOS/HA for 14 markers, mapping to 11 candidate genes (CYP17, CYP19, HSD17B2, IGFBP1+3, INHBB, INHC, INSL3, INSR, MADH4, OB, and POMC). The largest TDT was observed in the INSR region with allele 5 of D19S884 (χ2 = 8.53; P = 0.004; see Table 3). After correction for 349 tests, however, no alleles had a significantly elevated TDT.
Figure 2.
Summary of TDT analysis. The dashed line indicates a χ2 value of 3.84 (P = 0.05). The candidate gene regions and the allele with the highest χ2 value for each region are listed on the x axis. Each allele with a nominally elevated χ2 (>3.84) is indicated with an asterisk, and the number of transmissions tested is shown above the bar. A total of 349 alleles at 45 loci were tested.
Table 3.
Results from the present study for linkage and TDT analysis of genes previously tested in other studies
| Gene (ref.) | ASP Analysis
|
TDT
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IBD | Not IBD | Total, n | IBD, % | χ2 | P | Allele | Transmitted | Not transmitted | χ2 | P | |
| INS VNTR (35,36) | Class III | ||||||||||
| 15.2 | 14.8 | 30 | 51 | 0.00 | >0.5 | Total | 50 | 54 | 0.15 | >0.5 | |
| Paternal | 23 | 24 | 0.02 | >0.5 | |||||||
| Maternal | 27 | 30 | 0.16 | >0.5 | |||||||
| CYP11A (10) | |||||||||||
| D15S519 | 22.8 | 14.2 | 37 | 62 | 2.03 | 0.15 | 7 | 73 | 90 | 1.77 | 0.18 |
| D15S520 | 21 | 12.7 | 33.7 | 62 | 2.06 | 0.15 | 5 | 72 | 82 | 0.65 | 0.42 |
| Haplotype* | 30.8 | 15.2 | 46 | 67 | 5.34 | 0.02 | — | — | — | — | — |
| CYP19 (10) | |||||||||||
| 25.9 | 24.7 | 50.7 | 52 | 0.03 | <0.5 | 6 | 56 | 36 | 4.35 | 0.04 | |
| 7 | 20 | 40 | 6.67 | 0.01 | |||||||
| CYP17 (11,37) | |||||||||||
| D10S192 | 28.5 | 29.5 | 58 | 49 | 0.02 | >0.5 | 10 | 27 | 50 | 6.87 | 0.01 |
| INSR (18–20) | |||||||||||
| INSR | 17.5 | 12.5 | 30 | 58 | 0.83 | 0.36 | 13 | 39 | 21 | 5.40 | 0.02 |
| D19S884 | 27.3 | 24.7 | 52 | 53 | 0.14 | >0.5 | 5 | 10 | 28 | 8.53 | 0.004 |
| Haplotype† | 34.7 | 29.9 | 64.7 | 54 | 0.36 | >0.5 | — | — | — | — | — |
*D15S519–D15S520.
pterD19S905–INSR–D19S884–D19S922cen.
Previously Tested Candidate Genes.
We tested five gene regions (INS VNTR, CYP11A, CYP19, CYP17, and INSR) that have been previously tested by others for association or linkage to PCOS. In those studies (10, 11, 35, 36), PCOS was defined by polycystic ovaries (and various associated findings) and premature male pattern baldness (proposed as the male phenotype corresponding to PCOS). Waterworth et al. (36) found evidence for linkage with the insulin gene VNTR polymorphism (nonparametric linkage score = 3.25; P = 0.002). We did not see any significant excess IBD (IBD = 51%) in this region. Our results for this gene and other previously tested genes are shown in Table 3. Waterworth and colleagues (35, 36) also found evidence for association between the insulin VNTR and PCOS but only in the form of preferential transmission of the class III allele of the insulin VNTR from heterozygous fathers (χ2 = 7.54; P = 0.006), but not from mothers, to daughters with PCOS. In contrast, we saw no evidence for association between the class III alleles of the insulin VNTR and PCOS/HA. This finding held for transmissions from both parents to daughters with PCOS/HA or specifically from either fathers or mothers to affected daughters. In fact, there is a nonsignificant excess in the direction opposite to that observed by Waterworth et al (36).
Gharani et al. (10) found evidence for linkage with the cholesterol side-chain cleavage enzyme, CYP11A, (nonparametric linkage score = 3.03; P = 0.003). They allowed for genetic heterogeneity and estimated that ≈60% of their 20 families had the linked form. We analyzed two of the STRPs tested by Gharani et al. (ref. 10; D15S519 and D15S520) and found modest evidence for linkage (see above).
Gharani et al. (10) also found an association with D15S520, which is located in the promoter region of CYP11A. They found that, compared with controls, allele 5 of D15S520 was seen significantly less often in affected women (P = 0.03) and in women with elevated androgen levels alone (P = 0.002). In our families, there was no significant association between PCOS/HA and any alleles at this marker or the closely linked D15S519; allele 5 of D15S520 was transmitted at a slightly reduced frequency (72:82), but the difference was not statistically significant.
Gharani et al. (10) were able to exclude linkage with CYP19. We also found no significant evidence for linkage in this region (IBD = 51%). There were two alleles with elevated TDT (allele 6, χ2 = 4.35; allele 7, χ2 = 6.67), but after correction for multiple testing, these findings were no longer statistically significant.
Like Carey et al. (11), we found no evidence for linkage between CYP17 and PCOS/HA (IBD = 49%). Carey et al. (11) did find evidence for association with a variant nucleotide in the CYP17 promoter region, although these findings did not remain significant when more patients were added to the analysis (37). We found that one allele (allele 10 of D10S192) in the CYP17 region does have a somewhat elevated TDT (χ2 = 6.87), but after correcting for multiple testing, this finding was not statistically significant.
Several studies have sought, but failed to find, mutations in the INSR coding region of patients with PCOS (14, 17, 19, 21–23). Our findings in this region are consistent with previous studies in that we also do not find evidence for linkage between PCOS and the INSR. IBD for the INSR region ranges from 53% at D19S884 to 61% at D19S922; neither is statistically significant, and IBD for the much more informative 1.2-cM haplotype for this region (65 transmissions) is only 54% (χ2 = 0.36). We did, however, find evidence for association (elevated TDT) in the INSR region. The strongest evidence for association is with allele 5 of D19S884; however, this finding is not statistically significant after correction.
DISCUSSION
We tested for linkage and association between 37 candidate genes and PCOS/HA in data from 150 families, including 39 affected sister pairs. The phenotype PCOS/HA was defined by HA and oligomenorrhea in index cases and HA with or without oligomenorrhea in affected sisters. We found evidence for linkage with two genes: follistatin and CYP11A. Only the linkage with follistatin remains significant after correction for multiple testing.
Both of these regions are worthy of follow up studies. The cholesterol side-chain cleavage enzyme CYP11A converts cholesterol to pregnenolone, a rate-limiting step of steroidogenesis. A mutation that causes up-regulation of CYP11A activity could therefore result in an increase in androgen levels, one of the criteria used to define affected status in this study (6). The evidence for linkage with CYP11A (Table 3) was not very strong when each marker was considered separately, but when we assessed IBD by considering sharing of the haplotype defined by D15S519–D15S520 (a span of <1 kb), the IBD was 67% of 46 transmissions, and the corresponding χ2 was 5.34 (nominal P = 0.02). However, these results are no longer significant after correction for multiple testing (multiplying the P value by 33, the number of regions tested). Because Gharani et al. (10) also found evidence for linkage with CYP11A, our findings are, to some extent, a confirmation. In this situation, multiplying by the full 33 tests probably provides a correction that is too stringent, but it is not known what correction should be used instead.
By far the most convincing evidence for linkage was found with follistatin. Follistatin, an activin-binding protein, neutralizes the biological activity of activin in vitro and in vivo (38, 39). Activin, a member of the transforming growth factor-β superfamily, and follistatin are expressed in numerous tissues, including the ovary, pituitary, adrenal cortex, and pancreas. Activin promotes ovarian follicular development, inhibits thecal-cell androgen production, and increases pituitary follicle-stimulating hormone secretion and insulin secretion by pancreatic β-cells (39, 40). An increase in level or in functional activity of follistatin might, therefore, be expected to arrest follicular development, increase ovarian androgen production, reduce levels of circulating follicle-stimulating hormone, and impair insulin release. These changes are all characteristic features of PCOS (3, 41). Indeed, overexpression of follistatin in transgenic mice results in suppression of serum levels of follicle-stimulating hormone and arrested ovarian folliculogenesis (38).
With 66 transmissions of informative haplotypes at the follistatin locus, the finding of 72% IBD is highly significant, even after correction for 33 tests (Pc = 0.01). Although, in principle, some gene other than follistatin could give rise to the evidence for linkage of PCOS/HA with this region, we have focused on follistatin, because it is the candidate gene that led us to study this region.
We also tested for association in the follistatin and CYP11A regions. No allele of any marker in these regions showed significant evidence for allelic association. Although an allelic association detected by the TDT would have provided support for linkage, the absence of association is not inconsistent with linkage, because the effect detected by the TDT requires linkage disequilibrium in addition to linkage. It follows that genetic markers may reveal linkage without showing allelic association with the disease, especially if, as in the case of follistatin, the marker is not extremely tightly linked (Table 1).
We carried out a very complete analysis of association by the TDT for all markers, because this type of analysis is a particularly appropriate test for a possible role of a candidate gene. However, the very large number of alleles tested (349) makes it difficult to interpret nominally significant results. Furthermore, 6 of the 11 nominally significant tests shown in Fig. 2 are based on a relatively small number of transmissions (<50). Even for the larger samples with χ2 values close to 7 (CYP17 and CYP19), we are uncertain about the ultimate significance of the associations we observed. The strongest evidence for association was seen with allele 5 of D19S884 (χ2 = 8.53; P = 0.004; not significant after correction). D19S884 was chosen as a marker for the insulin receptor, considered a candidate gene on the basis of several previous studies (13, 21, 41). Nevertheless, the results are not conclusive, in part because of the modest sample size, and larger independent samples will be needed for a convincing replication of these findings.
In the present study, we have carried out analyses of genetic linkage and population association for a set of candidate genes for PCOS. We show how these genetic analyses can be used to screen a large number of candidate genes, without testing each gene for mutation(s). These approaches identify the candidate genes with the strongest evidence for genetic linkage and suggest which genes make minimal contributions to the etiology of the disease. The alternative procedure of screening one candidate gene at a time for mutations contributing to such diseases would be very inefficient, because variants that predispose to disease are heterogeneous and common in complex diseases such as PCOS. On the other hand, combined analysis of linkage and association can provide evidence that one (or several) candidate genes contribute to susceptibility, even though the precise genetic variant is not known. Such genetic evidence can then be used to guide further studies of those candidate genes. Our results suggest that variation at or near the follistatin gene contributes to the HA of PCOS.
Acknowledgments
We thank all the patients and their families for participating in this study. We also thank the study coordinators (S. Ward, S. Strong, J. Carroll, E. DeFrancesco, and L. Philips) and M. Kahsar-Miller for help with the family studies. We thank K. Vickery, S. Patton, and L. Haig-Ladewig for technical help, L. Demers for hormone assays, and K. Ewens and V. G. Cheung for comments. This work was supported by National Institutes of Health Grants U54 HD34449 (to J.F.S., A.D., and R.S.S.), R01DK40605 (to A.D.), K08HD0118 (to R.S.L.), R01DK47481 (to R.S.S), RR02635 (to Brigham and Women’s Hospital General Clinical Research Center), RR10732 (to Pennsylvania State University General Clinical Research Center), KO8-DK02315 (to D.A.E.), and 5T32DK07314 (to M.U.).
ABBREVIATIONS
- PCOS
polycystic ovary syndrome
- ASP
affected sib pair
- HA
hyperandrogenemia
- RH
radiation hybrid
- STRP
short tandem repeat polymorphisms
- cM
centimorgan
- TDT
transmission/disequilibrium test
- SHGC
Stanford Human Genome Center
- IBD
identity by descent
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
A Commentary on this article begins on page 8315.
References
- 1.Knochenhauer E S, Key T J, Kahsar-Miller M, Waggoner W, Boots L R, Azziz R. J Clin Endocrinol Metab. 1998;83:3078–3082. doi: 10.1210/jcem.83.9.5090. [DOI] [PubMed] [Google Scholar]
- 2.Legro R S, Kunselman A, Dodson W C, Dunaif A. J Clin Endocrinol Metab. 1999;84:165–169. doi: 10.1210/jcem.84.1.5393. [DOI] [PubMed] [Google Scholar]
- 3.Legro R S, Spielman R, Urbanek M, Driscoll D, Strauss J F, Dunaif A. Recent Prog Horm Res. 1998;53:217–256. [PubMed] [Google Scholar]
- 4.Cooper H E, Spellacy W N, Prem K A, Cohen W D. Am J Obstet Gynecol. 1968;100:371–387. doi: 10.1016/s0002-9378(15)33704-2. [DOI] [PubMed] [Google Scholar]
- 5.Givens J R. Endocrinol Metab Clin North Am. 1988;17:771–783. [PubMed] [Google Scholar]
- 6.Legro R S, Driscoll D, Strauss J F, Fox J, Dunaif A. Proc Natl Acad Sci USA. 1998;95:14956–14960. doi: 10.1073/pnas.95.25.14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carey A H, Chan K I, Short F, Williamson R, Franks S. Clin Endocrinol. 1993;38:653–658. doi: 10.1111/j.1365-2265.1993.tb02150.x. [DOI] [PubMed] [Google Scholar]
- 8.Govind A, Obhrai M, Clayton R. J Clin Endocrinol Metab. 1999;84:38–43. doi: 10.1210/jcem.84.1.5382. [DOI] [PubMed] [Google Scholar]
- 9.Jahanfar S, Eden J A, Warren P, Seppala M, Nguyen T V. Fertil Steril. 1995;63:478–486. [PubMed] [Google Scholar]
- 10.Gharani N, Waterworth D M, Batty S, White D, Gilling-Smith C, Conway G S, McCarthy M, Franks S, Williamson R. Hum Mol Genet. 1997;6:397–402. doi: 10.1093/hmg/6.3.397. [DOI] [PubMed] [Google Scholar]
- 11.Carey A H, Waterworth D, Patel K, White D, Little J, Novelli P, Franks S, Williamson R. Hum Mol Genet. 1994;3:1873–1876. doi: 10.1093/hmg/3.10.1873. [DOI] [PubMed] [Google Scholar]
- 12.Franks S. N Engl J Med. 1995;333:853–861. doi: 10.1056/NEJM199509283331307. [DOI] [PubMed] [Google Scholar]
- 13.Dunaif A, Segal K R, Shelley D R, Green G, Dobrjansky A, Licholai T. Diabetes. 1992;41:1257–1266. doi: 10.2337/diab.41.10.1257. [DOI] [PubMed] [Google Scholar]
- 14.Dunaif A, Xia J, Book C B, Schenker E, Tang Z. J Clin Invest. 1995;96:801–810. doi: 10.1172/JCI118126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciaraldi T P, el-Roeiy A, Madar Z, Reichart D, Olefsky J M, Yen S S. J Clin Endocrinol Metab. 1992;75:577–583. doi: 10.1210/jcem.75.2.1322430. [DOI] [PubMed] [Google Scholar]
- 16.Kiddy D S, Hamilton-Fairley D, Bush A, Short F, Anyaoku V, Reed M J, Franks S. Clin Endocrinol. 1992;36:105–111. doi: 10.1111/j.1365-2265.1992.tb02909.x. [DOI] [PubMed] [Google Scholar]
- 17.Conway G S, Avey C, Rumsby G. Hum Reprod. 1994;9:1681–1683. doi: 10.1093/oxfordjournals.humrep.a138773. [DOI] [PubMed] [Google Scholar]
- 18.Talbot J A, Bicknell E J, Rajkhowa M, Krook A, O’Rahilly S, Clayton R N. J Clin Endocrinol Metab. 1996;81:1979–1983. doi: 10.1210/jcem.81.5.8626868. [DOI] [PubMed] [Google Scholar]
- 19.Sorbara L R, Tang Z, Cama A, Xia J, Schenker E, Kohanski R A, Poretsky L, Koller E, Taylor S I, Dunaif A. Metabolism. 1994;43:1568–1574. doi: 10.1016/0026-0495(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 20.Taylor S I, Cama A, Accili D, Barbetti F, Quon M J, de la Luz Sierra M, Suzuki Y, Koller E, Levy-Toledano R, Wertheimer E, et al. Endocr Rev. 1992;13:566–595. doi: 10.1210/edrv-13-3-566. [DOI] [PubMed] [Google Scholar]
- 21.Krook A, Kumar S, Laing I, Boulton A J, Wass J A, O’Rahilly S. Diabetes. 1994;43:357–368. doi: 10.2337/diab.43.3.357. [DOI] [PubMed] [Google Scholar]
- 22.Krook A, O’Rahilly S. Baillieres Clin Endocrinol Metab. 1996;10:97–122. doi: 10.1016/s0950-351x(96)80330-2. [DOI] [PubMed] [Google Scholar]
- 23.O’Rahilly S, Choi W H, Patel P, Turner R C, Flier J S, Moller D E. Diabetes. 1991;40:777–782. doi: 10.2337/diab.40.6.777. [DOI] [PubMed] [Google Scholar]
- 24.Lander E S, Schork N J. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 25.Spielman R S, McGinnis R E, Ewens W J. Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 26.Ewens W J, Spielman R S. Am J Hum Genet. 1995;57:455–464. [PMC free article] [PubMed] [Google Scholar]
- 27.Zawadski J, Dunaif A. In: The Polycystic Ovary Syndrome. Givens J, Haseltine F, Merriam G, editors. Cambridge, MA: Blackwell Scientific; 1992. pp. 377–384. [Google Scholar]
- 28.Dunaif A, Scott D, Finegood D, Quintana B, Whitcomb R. J Clin Endocrinol Metab. 1996;81:3299–3306. doi: 10.1210/jcem.81.9.8784087. [DOI] [PubMed] [Google Scholar]
- 29.Carmina E, Lobo R A. Fertil Steril. 1999;71:319–322. doi: 10.1016/s0015-0282(98)00455-5. [DOI] [PubMed] [Google Scholar]
- 30.Bennett S T, Lucassen A M, Gough S C, Powell E E, Undlien D E, Pritchard L E, Merriman M E, Kawaguchi Y, Dronsfield M J, Pociot F, et al. Nat Genet. 1995;9:284–292. doi: 10.1038/ng0395-284. [DOI] [PubMed] [Google Scholar]
- 31.Suarez B K, Hodge S E. Clin Genet. 1979;15:126–136. doi: 10.1111/j.1399-0004.1979.tb01751.x. [DOI] [PubMed] [Google Scholar]
- 32.Kruglyak L, Daly M J, Reeve-Daly M P, Lander E S. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 33.Curtis D, Sham P. Am J Hum Genet. 1995;56:811–812. [PMC free article] [PubMed] [Google Scholar]
- 34.Spielman R S, Ewens W J. Am J Hum Genet. 1999;64:668–669. doi: 10.1086/302265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett S T, Todd J A, Waterworth D M, Franks S, McCarthy M I. Lancet. 1997;349:1771–1772. doi: 10.1016/S0140-6736(96)08368-7. [DOI] [PubMed] [Google Scholar]
- 36.Waterworth D M, Bennett S T, Gharani N, McCarthy M I, Hague S, Batty S, Conway G S, White D, Todd J A, Franks S, et al. Lancet. 1997;349:986–990. doi: 10.1016/S0140-6736(96)08368-7. [DOI] [PubMed] [Google Scholar]
- 37.Gharani N, Waterworth D M, Williamson R, Franks S. J Clin Endocrinol Metab. 1996;81:4174. doi: 10.1210/jcem.81.11.8923880. [DOI] [PubMed] [Google Scholar]
- 38.Guo Q, Kuma T R, Woodruff T, Hadsell L A, DeMayo F J, Matzuk M M. Mol Endocrinol. 1998;12:96–106. doi: 10.1210/mend.12.1.0053. [DOI] [PubMed] [Google Scholar]
- 39.Mather J P, Moore A, Li R H. Proc Soc Exp Biol Med. 1997;215:209–222. doi: 10.3181/00379727-215-44130. [DOI] [PubMed] [Google Scholar]
- 40.Shibata H, Kanzaki M, Takeuchi T, Miyazaki J, Kojima I. J Mol Endocrinol. 1996;16:249–258. doi: 10.1677/jme.0.0160249. [DOI] [PubMed] [Google Scholar]
- 41.Dunaif A. Endocr Rev. 1997;18:774–800. doi: 10.1210/edrv.18.6.0318. [DOI] [PubMed] [Google Scholar]