Abstract
Dipeptidyl peptidase I (DPPI) is a lysosomal cysteine protease that has been implicated in the processing of granzymes, which are neutral serine proteases exclusively expressed in the granules of activated cytotoxic lymphocytes. In this report, we show that cytotoxic lymphocytes derived from DPPI−/− mice contain normal amounts of granzymes A and B, but these molecules retain their prodipeptide domains and are inactive. Cytotoxic assays with DPPI−/− effector cells reveal severe defects in the induction of target cell apoptosis (as measured by [125I]UdR release) at both early and late time points; this defect is comparable to that detected in perforin−/− or granzyme A−/− × B−/− cytotoxic lymphocytes. DPPI therefore plays an essential role in the in vivo processing and activation of granzymes A and B, which are required for cytotoxic lymphocyte granule-mediated apoptosis.
One of the major contact-dependent mechanisms used by cytotoxic lymphocytes to induce target-cell death is the granule exocytosis pathway (1). In this pathway, perforin facilitates the target cell entry and/or trafficking of granzymes (2–8), which then deliver the lethal hits that cause DNA fragmentation, the hallmark of apoptosis (9). Cytotoxic lymphocytes derived from perforin−/− mice have a profound defect in their ability to induce DNA fragmentation in vitro and in vivo (10–14). Granzyme B−/− cytotoxic lymphocytes have a severe defect in the early induction of apoptosis which is slowly—but almost completely—corrected by prolonged incubation with target cells (15, 16). We and others have recently shown that this late, perforin-dependent, granzyme B-independent activity is mediated by granzyme A (17, 18). When the rapid granzyme B pathway and the slow granzyme A pathway are both disarmed (as in granzyme A × B-deficient mice) the cytotoxic defect seen in vitro and in vivo is as severe as that detected in perforin-deficient mice. This result suggests that a single mechanism of inhibiting the activities of both granzymes A and B could provide a powerful way to regulate the ability of immune effector cells to kill their targets.
Granzymes belong to a family of highly related neutral serine proteases that are expressed exclusively in the granules of activated cytotoxic lymphocytes. They are synthesized as preproenzymes with an 18- to 26-residue leader sequence that is cleaved and removed, leaving a prodipeptide (usually GlyGlu or GluGlu) at the N terminus of the enzyme (19). Activation of the granzymes requires the cleavage of the prodipeptide, which then presumably allows the mature enzyme to fold into a catalytically active conformation (20). Several recent studies have suggested that DPPI is capable of performing this processing event in vitro (21–25).
Dipeptidyl peptidase I (DPPI, also known as cathepsin C) is a lysosomal cysteine protease that is expressed in most tissues (26, 27). In cytotoxic lymphocytes and in myeloid cells, DPPI is found in the secretory granule compartment (28). DPPI belongs to the papain superfamily of proteases and shares a number of similarities with other lysosomal cysteine proteases, such as cathepsins B, H, and L. We recently cloned and characterized the murine DPPI gene (27). Southern blot analysis and chromosomal analysis by fluorescence in situ hybridization (FISH) revealed that DPPI represents a single locus on mouse chromosome 7; no highly related genes were identified on two independent bacterial artificial chromosome clones that contained at least 150 kilobase (kb) of DNA from the DPPI locus (27). Genes encoding additional mouse lysosomal cysteine proteases (cathepsins B, H, L, and S) have been mapped to other chromosomes (29). Cathepsins L and S have been shown to play an important role in the degradation of the invariant chain of major histocompatibility class II complexes and in CD4 T cell selection (30–32), but none of the related cathepsins is a diaminopeptidase, and none is known to affect the function of either CD8+ T cells or natural killer cells.
To determine whether DPPI is specifically required for the processing of granzymes in vivo and to further define its physiologic functions, we generated mutant mice that are deficient in DPPI. Cytotoxic lymphocytes derived from these mice contain normal amounts of granzymes A and B, but these enzymes retain their prodomains and are essentially inactive. These data show that DPPI is required for the processing of the prodomains of granzymes A and B and demonstrate that alternative dipeptidyl proteases cannot substitute for its function in vivo.
MATERIALS AND METHODS
Construction of the DPPI Targeting Vector.
A 6.2-kb EcoRI–HindIII fragment containing exons 1 and 2 derived from bacterial artificial chromosome 7930 (Genome Systems, St. Louis) was used to develop the targeting vector. The 1.2-kb 5′ targeting arm was created by PCR amplification of DNA derived from bacterial artificial chromosome 7930 by using the primer set 5′-GACAGAAGTGGTTTTGTTCTTC-3′ (forward) and 5′-CTGACGGCGGCCGCGGAG-3′ (reverse), and the product was subcloned into pCR2.1 (Invitrogen). The reverse primer was engineered to introduce a NotI site, which facilitated subsequent subcloning. The 5-kb HindIII fragment used for the 3′ targeting arm was directly subcloned from bacterial artificial chromosome 7930 into pBluescript KS(−) (Stratagene). To assemble the targeting vector, a 3-kb BamHI–EcoRI LacZ reporter gene (33) was subcloned into pBluescript KS(−). The entire 1.8-kb LoxP/PGK-Neo cassette(34) was subcloned into the EcoRI site downstream from the LacZ reporter gene in the same transcriptional orientation. The left arm of the targeting vector was excised from pCR2.1 as a NotI fragment and subcloned into the NotI site upstream from the LacZ gene. The right arm was excised as a HindIII fragment and subcloned into the HindIII site downstream of the PGK-Neo gene. The targeting vector was linearized with SalI and gel-purified before electroporation into embryonic stem cells.
Generation of DPPI−/− Mice.
DPPI+/− embryonic stem cells were generated as described (15). Neomycin-resistant clones were expanded and homologous recombination events confirmed by Southern blot analysis by using external probe A (Fig. 1A) after PstI digestion. Cells from clones 100 and 102 were injected into C57BL/6 blastocysts. Chimeric males were bred with either C57BL/6 or 129/SvJ mice. Germ-line transmission was confirmed by using Southern blot analysis. Heterozygous animals were intercrossed to generate homozygous mutant mice. All assays were performed with mice from both lines.
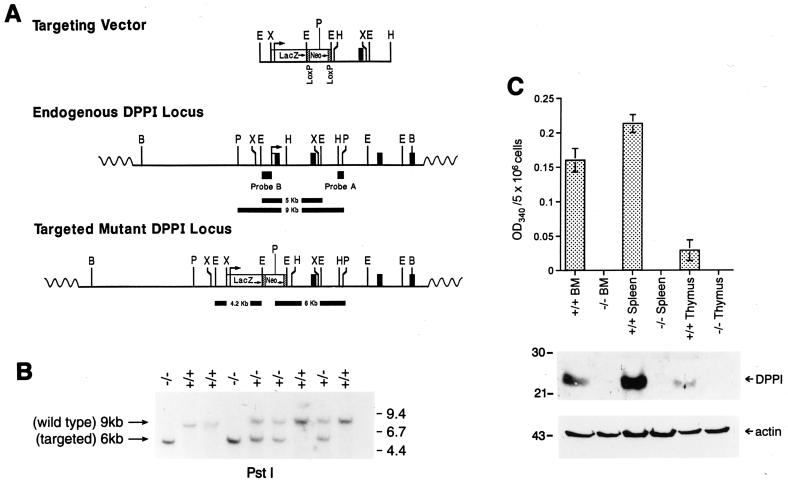
Figure 1.
Targeting of the DPPI gene. (A) Creation of the targeting vector. The DPPI locus is shown in the Middle and the targeting construct is depicted at Top. The locations and transcriptional orientations of the β-galactosidase (LacZ) and PGK-Neo cassettes are indicated. The targeting construct was designed to replace exon 1 and part of intron 1 with the selectable marker cassette. The locations of the external (probe A) and internal (probe B) probes used to detect homologous recombination are shown. (B) Southern blot analysis of tail DNA from wild-type, DPPI+/−, and DPPI−/− animals. DNA was digested with PstI and hybridized with probe A. The wild-type allele is found within a 9-kb fragment; the targeted allele is reduced to 6 kb because of an internal PstI site in the PGK-Neo cassette. (C) DPPI activity in wild-type and DPPI−/− tissues. Freshly isolated bone marrow cells, splenocytes, and thymocytes were lysed and assayed for DPPI activity as determined by the hydrolysis of Gly-Phe-β-naphthylamide. Activity was defined as the OD at 405 nm for 5 × 106 cells. Results represent the mean of duplicate determinations. Equal amounts of total lysates were also fractionated on 10% SDS/PAGE gels and immunoblotted with a specific rabbit anti-mouse DPPI antibody. A β-actin antibody was used to control for protein content and loading.
Protease Activity Assays.
DPPI activity was assayed by the hydrolysis of Gly-Phe-β-naphthylamide (Sigma) as described (23). Controls included buffer blanks and duplicate samples that received no substrate (background absorbance). The results were expressed as OD at 340 nm per 5 × 106 cells. Tryptase activity was determined by the hydrolysis of N-α-benzyloxycarbonyl-l-lysine thiobenzyl ester (BLT), and aspase activity was determined by the hydrolysis of N-t-butyloxycarbonyl-l-Ala-Ala-Asp-thiobenzyl ester (BAADT), as described (25). Controls included duplicate samples that contained 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) without substrate.
Western Blot Analysis.
Tissue and cell lysates were prepared and analyzed by using standard Western blotting techniques as described (25). Primary antibodies included rabbit anti-mouse DPPI antiserum (25), rabbit anti-mouse granzyme B antiserum (15), rabbit anti-mouse granzyme A antiserum (18), rat anti-mouse perforin mAb (clone P1–8, Kamiya Biomedical, Thousand Oaks, CA), and mouse β-actin antibody (Sigma).
In Vitro Lytic Assays.
Primary mixed lymphocyte reaction cultures were obtained by culturing splenocytes (H-2b) from 8- to 12-week old wild-type (129/SvJ), DPPI−/− (129/SvJ), granzyme A−/− × B−/− (129/SvJ), and perforin−/− (129/SvJ × C57BL/6) mice with irradiated splenocytes from BALB/c mice (H-2d), as described (15). Lymphokine-activated killer (LAK) cells were obtained by culturing splenocytes in the presence of high-dose rhIL-2 (1,000 units/ml) for 6–7 days as described (16). Lytic assays were performed by using [125I]deooxyuridine ([125I]UdR) (ICN) radiolabeled YAC-1, TA3, and P815 target cells as described (16).
Microsequencing.
LAK cells from wild-type and DPPI−/− splenocytes were harvested and purified by using Ficoll 1077 (Sigma) gradients. Cells were resuspended in lysis buffer (1 M NaCl/50 mM Tris, pH 8.0/0.2% Triton X-100 containing 1 mM phenylmethylsulfonyl fluoride/5 μg/ml pepstatin A/5 μg/ml leupeptin/5 μg/ml antipain, all obtained from Sigma) and sonicated, and cellular debris was removed by centrifuging at 10,000 × g at 4°C for 10 min. The cleared lysates were loaded onto an Accell Plus cation exchange column (Millipore), and proteins were eluted by using a linear gradient of 0.25–2 M NaCl in 50 mM Mes, 0.1 mM EGTA, and 10% betaine (all obtained from Sigma). Granzymes A, B, and C eluted between 0.6 and 0.8 M NaCl. Fractions were collected and proteins were separated by using denaturing, nonreducing SDS/PAGE, transferred to poly(vinylidene difluoride) membranes (Bio-Rad) in 10 mM 3-[cyclohexylamino]-1-propanesulfonic acid (CAPS) buffer containing 10% methanol for 2 hr at 4°C, and stained briefly with Coomassie blue (Promega). Bands at Mr 26,000, 29,000, and 60,000 were excised and subjected to Edman degradation (8–10 cycles) to define the N terminus. Amino acid sequence analysis was performed by the Protein Chemistry Laboratory at Washington University (St. Louis).
RESULTS AND DISCUSSION
To generate DPPI−/− embryonic stem cells, a targeting vector that contains a PGK-Neo cassette was designed to replace exon 1 and part of intron 1 of the DPPI gene (Fig. 1A). Because DPPI is ubiquitously expressed and thought to be involved in the processing of many enzymes and hormones (26, 35–38), we predicted that a null mutation would result in embryonic lethality. We therefore inserted a β-galactosidase cDNA just downstream from the DPPI transcription initiation site to allow us to track DPPI expression during development. The linearized targeting construct was electroporated into RW-4 (129/SvJ) embryonic stem cells, and G418-resistant clones were selected and examined by Southern blotting analysis. Five independent homologously recombined clones were obtained from 247 G418-resistant colonies, and two were subsequently injected into C57BL/6 blastocysts to generate chimeras. Two chimeras from each clone were bred to C57BL/6 or 129/SvJ mice. All four chimeras successfully contributed to germ-line transmission. DPPI heterozygotes were intercrossed to produce homozygous mutant mice. Genotyping of the first 108 animals derived from the breeding of heterozygous mice produced 30 wild-type animals (28%), 53 heterozygotes (49%), and 25 homozygous mutants (23%). Southern blotting analysis of genomic tail DNA confirmed the presence of the mutation (Fig. 1B). DPPI mRNA (data not shown) and protein could not be detected in mouse spleen, bone marrow, or thymus (Fig. 1C). DPPI activity, as measured by the hydrolysis of Gly-Phe-β-naphthylamide, was absent in DPPI−/− mice, confirming that the mutation produces a null phenotype. Expression of the β-galactosidase could not be detected in DPPI+/− or −/− splenocytes, either because this cDNA did not contain a local polyadenylation site, or because the retained PGK-Neo cassette reduced the output of the DPPI gene itself (data not shown). DPPI-deficient mice developed normally with equal representation of both sexes, appeared healthy, and were fertile.
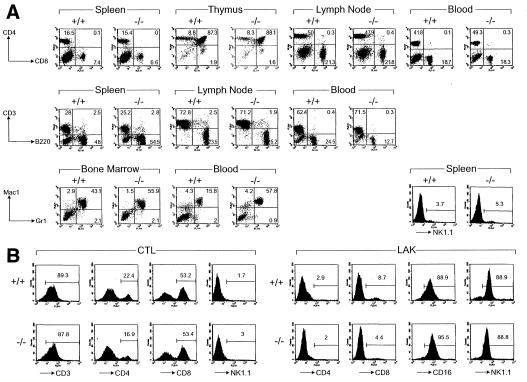
Histopathological evaluation of all major organs including spleen, liver, lung, heart, kidney, thymus, brain, and bone marrow showed no apparent abnormalities (data not shown). Flow cytometric analyses of various T cell subsets, B cells, or NK cells derived from the spleen, thymus, lymph nodes, or blood of young mice revealed no significant differences between wild-type and mutant animals (Fig. 2A). Hematopoiesis was also normal, except for a 3- to 4-fold increase in granulocytes (Mac-1+/Gr-1+) in the peripheral blood (Fig. 2A) of 3 of 11 animals tested. The significance of this finding is not clear; even though these mice have defects in cell-mediated cytotoxicity and/or granulocyte function (see below), they displayed no serologic evidence of infection with any common mouse pathogens (data not shown; all mice were maintained in a specific-pathogen-free barrier facility). Splenocytes from DPPI−/− mice can be stimulated with allogeneic splenocytes in primary, one-way mixed lymphocyte reactions to generate cytotoxic T lymphocytes (CTL), or with high-dose IL-2 to generate LAK cells (Fig. 2B). DPPI protein and activity in these cytotoxic lymphocytes were absent (data not shown), indicating that there is no alternative, inducible form of this enzyme produced on lymphocyte activation.
Figure 2.
Normal immune system development and cytotoxic lymphocyte activation in DPPI mutant mice. (A) Flow cytometric analyses of lymphoid and myeloid cells in wild-type and DPPI−/− mice. Cells harvested from spleen, thymus, mesenteric lymph nodes, and peripheral blood were analyzed for the expression of the indicated surface markers. The percentages of cells expressing CD3, CD4, CD8, B220, and NK1.1 were similar for all mice. Note that there was a moderate increase in the Mac-1+/Gr-1+ in the example shown here; 3 of 11 tested mice had a similar finding. (B) Normal proliferation and activation of DPPI−/− splenocytes in primary MLR and LAK activation. Splenocytes from wild-type and DPPI−/− animals were activated in a 5-day mixed lymphocyte reaction culture or in the presence of high-dose IL-2 to generate LAK cells. The expansion of the CD8+ CTL and NK1.1+ LAK cell populations were essentially equivalent in DPPI+/+ and −/− splenocytes.
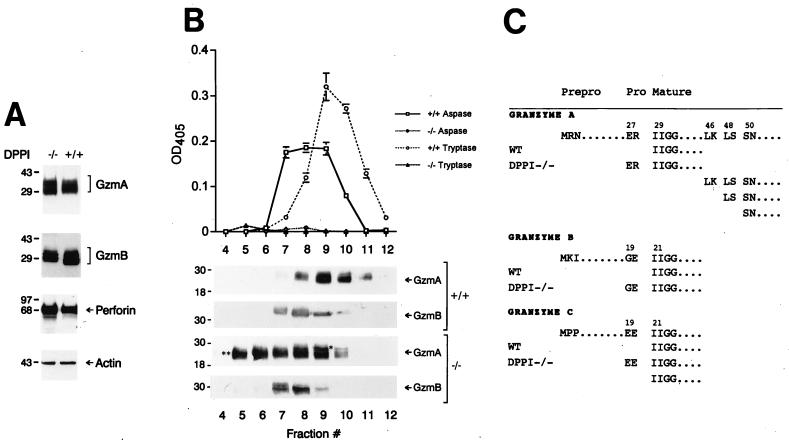
Western analyses with specific anti-granzyme A, anti-granzyme B, or anti-granzyme C (data not shown) antisera revealed a normal abundance of these proteins in DPPI−/− LAK cells (Fig. 3A) and CTL (data not shown). Granzyme A and B activities from total LAK cell lysates or from cationic exchange column fractions were assayed by using peptide-based chromogenic substrates (to date, there is no known substrate nor function for granzyme C). Granzyme A-mediated tryptase activity (as assayed by the hydrolysis of the BLT substrate) from DPPI−/− LAK lysates was virtually identical to that detected in granzyme A−/− LAK lysates (data not shown). In addition, column fractions that contained abundant immunoreactive granzyme A had no detectable tryptase activity (Fig. 3B). However, we detected a small amount of residual aspase activity (2–5% that of wild-type levels, as assayed by the hydrolysis of the BAADT substrate) in crude DPPI−/− CTL/LAK cell lysates (data not shown) and in the column fractions that contained immunoreactive granzyme B (Fig. 3B). DPPI−/− LAK and CTL lysates were unable to process in vitro-translated caspase 3 (a known substrate of granzyme B both in vitro and in vivo) (38–40) to its active p20 form (data not shown). These results demonstrate that both granzymes A and B are nearly completely inactive in CTL/LAK cells derived from DPPI−/− mice. In preliminary studies, two other related serine proteases that also contain similar prodomains (cathepsin G and neutrophil elastase, found specifically in neutrophils) were also nearly completely inactive (data not shown).
Figure 3.
Lack of granzyme processing and activity in LAK cells derived from DPPI−/− mice. (A) Granzyme A and B levels are normal in DPPI−/− LAK cells. Equal amounts of protein derived from wild type DPPI−/− LAK cells were analyzed by Western blotting by using polyclonal antibodies directed against mouse granzymes A and B and mAbs against mouse perforin and β-actin. The specificities of these antibodies have been determined previously (see Materials And Methods). (B) Granzyme A and B activities in LAK cell lysates. Wild-type and DPPI−/− LAK cell lysates were fractionated on a cationic exchange column by using a linear 0.25–2 M NaCl gradient, and each fraction was analyzed for aspase and tryptase activity. These same fractions were separated by SDS/PAGE, transferred to nitrocellulose, and incubated with anti-granzyme A and B antibodies to confirm the abundance and sizes of the proteins. (C) Granzymes are abnormally processed in DPPI−/− LAK cells. Column fractions from B were fractionated by using nonreducing SDS/10% PAGE and transferred to poly(vinylidene difluoride) membranes; specific bands were excised, eluted, and subjected to microsequencing to determine the N termini. These results were confirmed in two independent experiments.
Granzymes A and B from DPPI−/− LAK lysates migrate slightly differently on SDS/PAGE gels compared with granzymes from wild-type LAK cells (Fig. 3A), suggesting that these proteins are abnormally processed. To define the N termini of the granzymes, LAK cell lysates were fractionated on cation exchange columns and granzymes A, B, and C were isolated and subjected to microsequencing. We confirmed that granzymes derived from wild-type LAK cells start with the prerequisite isoleucine at position 29 for granzyme A and position 21 for both granzymes B and C (Fig. 3C). However, the only species of granzyme B recovered from fractions 7 and 8 derived from DPPI−/− LAK cells (Fig. 3C) was found to be in the proform (i.e., with the prodipeptide still attached). DPPI is therefore required for the processing of granzyme B in vivo. Because granzyme A eluted from the column in a broad peak in the DPPI−/− LAK lysate, we sequenced granzyme A derived from fractions 5 and 9 to define the N termini of all of the different isoforms of granzyme A. The upper band marked ∗ in fraction 9 (Fig. 3B) contains granzyme A in the proform (with the GluArg prodipeptide still attached). Sequencing of the more rapidly migrating bands in both fractions 9 and 5 (Fig. 3B, ∗∗) revealed a mixture with equal proportions of three different cleavage products of granzyme A, starting with Leu (46), Leu (48), or Ser (50). These shorter products of granzyme A can dimerize (data not shown), but they are unable to hydrolyze the granzyme A-specific peptide substrate (Fig. 3B). Even though all in vitro manipulations were carried out in a mixture of protease inhibitors (see Materials and Methods), we cannot completely rule out random degradation of granzyme A after the cells were lysed. Alternatively, progranzyme A may be in a vulnerable conformation that allows it to be processed by intracellular exo- or endopeptidases. Surprisingly, 50% of the granzyme C from DPPI−/− LAK cells was correctly processed to the mature form (Fig. 3C); these results were repeated and confirmed in a separate experiment. The reason for this finding is unclear, but it suggests that the GluGlu prodipeptide of granzyme C can be processed by another protease. One possible candidate is dipeptidyl peptidase II (DPPII), another ubiquitously expressed lysosomal serine protease with a slightly different aminopeptidase activity (41, 42). Finally, subcellular fractionation studies confirmed that the proforms of granzymes A and B can be efficiently packaged into lysosomal granules along with perforin (data not shown). Thus, DPPI is not required for signal peptide removal, nor is it required for the subsequent transport of the granzymes into lysosomes.
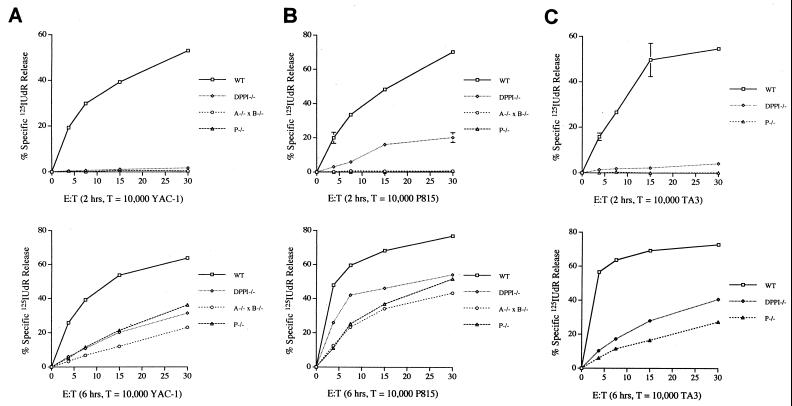
To examine the ability of DPPI−/− cytotoxic lymphocytes to induce apoptosis in target cells, we performed standard cytolytic assays in which [125I]UdR release was measured at increasing effector/target ratios. We compared DPPI−/− effectors to granzyme A−/− × B−/− and perforin−/− effector cells; the [125I]UdR release defect seen in perforin−/− effectors is virtually equivalent to that detected in granzyme A−/− × B−/− effectors (17, 18), because perforin is required to effectively deliver granzymes and/or traffic them within target cells (5–8). LAK cells were generated by culturing splenocytes in the presence of high-dose IL-2. Flow cytometric studies revealed that these populations contained nearly equivalent fractions of CD16+ and NK1.1+ cells (Fig. 2B). Western analyses of wild-type and DPPI−/− LAK lysates revealed equivalent amounts of granzymes A, B, and perforin, indicating that these cells were similarly activated (data not shown). LAK cells from wild-type, DPPI−/−, granzyme A−/− × B−/−, and perforin−/− splenocytes were incubated with YAC-1 target cells for two hours (early) or six hours (late). The [125I]UdR release assays revealed that DPPI−/− LAK cells have a severe defect both at early and late time points and that this defect is comparable to the defect seen in granzyme A−/− × B−/− and perforin−/− LAK cells (Fig. 4A). These results were highly reproducible in three separate experiments. Mixed lymphocyte reaction cultures from wild-type, DPPI−/−, or perforin−/− splenocytes also contained equivalent proportions of various T cells subsets (Fig. 2B) and equal amounts of granzymes A and B (data not shown). These were tested against two different allogeneic target cell lines (TA3 and P815). After 2 hours of incubation, perforin −/− CTL induced no [125I]UdR release from either target cell line, because perforin−/− effectors cannot deliver and/or traffic granzyme B (15). DPPI−/− CTL can mediate low levels of specific [125I]UdR release (Fig. 4B Upper), but this defect varies slightly among target cell lines and experiments; this variability may reflect the small but variable amount of residual aspase activity (<5% that of wild-type amounts) in DPPI−/− CTL. After 6 hr of incubation, DPPI−/− CTL have a persistent defect in [125I]UdR release (Fig. 4 B and C). These results confirm that DPPI−/− cytotoxic lymphocytes have a severe defect in their ability to cause DNA fragmentation that is similar to the defect detected in LAK cells and CTL derived from mice deficient for both granzymes A and B or for perforin. Because the mutant DPPI locus contains a PGK-Neo cassette, the observed phenotype could be influenced by the down-regulation of tightly linked genes because of a “neighborhood” effect (43), but no highly related genes have been localized to the same region of mouse chromosome 7.
Figure 4.
Defective [125I]UdR release exhibited by DPPI−/− CTL and LAK cells. Standard cytotoxicity assays of wild-type (□), DPPI−/− (◊), granzyme A−/− × B−/− (○), and perforin−/− (▵) LAK cells tested against YAC-1 target cells at increasing effector/target (E:T) ratios (Left); CTL were tested against TA3 and P815 target cells (Center and Right). Release of [125I]UdR is measured after 2 (Upper) and 6 (Lower) hr. The results are represented as mean ± SEM of duplicate samples. This experiment was repeated three times with similar results. Note that DPPI−/− LAK and CTL have an early defect in [125I]UdR release that is due to the loss of active granzyme B, and a persistent defect at 6 hr that is caused by the loss of active granzyme A.
In summary, we have shown that DPPI is essential for the processing and activation of granzymes A and B in vivo. This observation was further supported by the finding of a severe and persistent defect in cytotoxicity displayed by DPPI−/− CTL and LAK cells. Our findings support a rationale for developing specific inhibitors of DPPI as a potential means of disabling both granzymes A and B in immune effector cells. However, we have only begun to characterize the defects seen in the DPPI-deficient mice. Neutrophils and mast cells, which contain related serine proteases such as cathepsin G, neutrophil elastase, and mast cell chymases, should also be affected by this mutation. Because some of these related proteases serve important biological functions in vivo (44, 45), an effective DPPI inhibitor will probably need to be specifically targeted to lymphocytes to avoid untoward effects in these other cellular compartments.
Acknowledgments
The authors thank Eileen Grass, Rick Goforth, Tim Corbin, and Pam Goda for performing the embryonic stem cell work and the microinjections. We thank Kelly Schrimpf for excellent mouse husbandry. We thank April Adkison and Jamie Mercer for outstanding technical assistance. We also thank Nancy Reidelberger for her expert assistance in preparing the manuscript. We owe a debt of gratitude to Dr. Don Payan, who encouraged us to pursue this project. This work was supported by National Institutes of Health Grants KO8-HL03774 (C.T.N.P.) and DK49786 and CA49712 (T.J.L.).
ABBREVIATIONS
- DPPI
dipeptidyl peptidase I
- kb
kilobase
- CTL
cytotoxic T lymphocytes
- LAK
lymphokine-activated killer
Footnotes
A Commentary on this article begins on page 8312.
References
- 1.Henkart P A. Immunity. 1994;1:343–346. doi: 10.1016/1074-7613(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 2.Masson D, Tschopp J. J Biol Chem. 1985;260:9069–9072. [PubMed] [Google Scholar]
- 3.Podack E R, Young J D, Cohn Z A. Proc Natl Acad Sci USA. 1985;82:8629–8633. doi: 10.1073/pnas.82.24.8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C-C, Walsh C M, Young J D. Immunol Today. 1995;16:194–201. doi: 10.1016/0167-5699(95)80121-9. [DOI] [PubMed] [Google Scholar]
- 5.Shi L, Mai S, Israels S, Browne K, Trapani J A, Greenberg A H. J Exp Med. 1997;185:855–866. doi: 10.1084/jem.185.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jans D A, Jans P, Briggs L J, Sulton V, Trapani J A. J Biol Chem. 1996;271:30781–30789. doi: 10.1074/jbc.271.48.30781. [DOI] [PubMed] [Google Scholar]
- 7.Froelich C J, Orth K, Turbov J, Seth P, Gottlieb R, Babior B, Shah G M, Bleackley R C, Dixit V M, Hanna W. J Biol Chem. 1996;271:29073–29079. doi: 10.1074/jbc.271.46.29073. [DOI] [PubMed] [Google Scholar]
- 8.Pinkoski M J, Hobman M, Heibein J A, Tomaselli K, Li F, Seth P, Froelich C J, Bleackley R C. Blood. 1998;92:1044–1054. [PubMed] [Google Scholar]
- 9.Russell J H. Immunol Rev. 1983;72:97–118. doi: 10.1111/j.1600-065x.1983.tb01074.x. [DOI] [PubMed] [Google Scholar]
- 10.Kagi D, Lederman B, Burki K, Seiler P, Odermatt B, Olsen K J, Podack E R, Zinkernagel R M, Hengartner H. Nature (London) 1994;369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 11.Kagi D, Vignaux F, Ledermann B, Burki K, Depraetere V, Nagata S, Hengartner H, Golstein P. Science. 1994;265:5528–5530. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- 12.Lowin B, Hahne M, Tschopp J. Nature (London) 1994;370:650–652. doi: 10.1038/370650a0. [DOI] [PubMed] [Google Scholar]
- 13.Kojima H, Shinohara N, Hanaoka S, Someya-Shirota Y, Takagaki Y, Ohno H, Saito T, Katayama T, Yagita H, Okumura K, et al. Immunity. 1994;1:357–365. doi: 10.1016/1074-7613(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 14.Walsh C M, Matloubian M, Liu C C, Ueda R, Kurahara C G, Christensen J L, Huang H T, Young J D, Ahmed R, Clark W R. Proc Natl Acad Sci USA. 1994;91:10854–10858. doi: 10.1073/pnas.91.23.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heusel J W, Wesselschmidt R, Shresta S, Russell J, Ley T J. Cell. 1994;76:977–987. doi: 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 16.Shresta S, MacIvor D M, Heusel J W, Russell J H, Ley T J. Proc Natl Acad Sci USA. 1995;92:5679–5683. doi: 10.1073/pnas.92.12.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon M M, Hausmann M, Tran T, Ebnet K, Tschopp J, ThaHla R, Mullbacher A. J Exp Med. 1997;186:1781–1786. doi: 10.1084/jem.186.10.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shresta S, Graubert T A, Thomas D A, Raptis S Z, Ley T J. Immunity. 1999;10:595–605. doi: 10.1016/s1074-7613(00)80059-x. [DOI] [PubMed] [Google Scholar]
- 19.Jenne D E, Tschopp J. Immunol Rev. 1988;103:53–57. doi: 10.1111/j.1600-065x.1988.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 20.Caputo A, Garner R S, Winkler U, Hudig D, Bleackley R C. J Biol Chem. 1993;268:2458–2467. [PubMed] [Google Scholar]
- 21.McGuire M J, Lipsky P E, Thiele D L. J Biol Chem. 1993;268:2458–2467. [PubMed] [Google Scholar]
- 22.Thiele D L, McGuire M J, Lipsky P E. J Immunol. 1997;158:5200–5210. [PubMed] [Google Scholar]
- 23.Smyth M J, McGuire M J, Thia K Y. J Immunol. 1995;154:6299–6305. [PubMed] [Google Scholar]
- 24.Kummer J A, Kamp A M, Citarella F, Horrevoets A J, Hack C E. J Biol Chem. 1996;271:9281–9286. doi: 10.1074/jbc.271.16.9281. [DOI] [PubMed] [Google Scholar]
- 25.Pham C T N, Thomas D A, Mercer J D, Ley T J. J Biol Chem. 1998;273:1629–1633. doi: 10.1074/jbc.273.3.1629. [DOI] [PubMed] [Google Scholar]
- 26.Kominami E, Ishido K, Muno D, Sato N. Biol Chem Hoppe-Seyler. 1992;373:367–373. doi: 10.1515/bchm3.1992.373.2.367. [DOI] [PubMed] [Google Scholar]
- 27.Pham C T N, Armstrong R J, Zimonjic D B, Popescu N C, Payan D G, Ley T J. J Biol Chem. 1997;272:10695–10703. doi: 10.1074/jbc.272.16.10695. [DOI] [PubMed] [Google Scholar]
- 28.Brown G R, McGuire M J, Thiele D L. J Immunol. 1993;150:4733–4742. [PubMed] [Google Scholar]
- 29.Deussing J, Roth W, Rommerskirch W, Wiederanders B, von Figura K, Peters C. Mamm Genome. 1997;8:241–245. doi: 10.1007/s003359900401. [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, Villadangos J A, Ploegh H, Peters C, Rudensky A Y. Science. 1998;280:450–453. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- 31.Shi G P, Villadangos J A, Dranoff G, Small C, Gu L, Haley K J, Riese R, Ploegh H L, Chapman H A. Immunity. 1999;10:197–206. doi: 10.1016/s1074-7613(00)80020-5. [DOI] [PubMed] [Google Scholar]
- 32.Nakagawa T Y, Roth W, Wong P, Nelson A, Farr A, Deussing J, Villadangos J A, Ploegh H, Peters C, Rudensky A Y. Immunity. 1999;10:207–217. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- 33.Graubert T A, Hug B A, Wesselschmidt R, Hsieh C-L, Ryan T M, Townes T M, Ley T J. Nucl Acid Res. 1998;26:2849–2858. doi: 10.1093/nar/26.12.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hug B, Wesselschmidt R L, Fiering S, Bender M A, Epner E, Groudine M, Ley T J. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald J K, Callahan P X, Zeitman B B, Ellis S. J Biol Chem. 1969;244:2693–2709. [PubMed] [Google Scholar]
- 36.McDonald J K, Zeitman B B, Callahan P X, Ellis S. J Biol Chem. 1974;249:234–240. [PubMed] [Google Scholar]
- 37.D’Agrosa R M, Callahan J W. Biochem Biophys Res Commun. 1988;157:770–775. doi: 10.1016/s0006-291x(88)80316-4. [DOI] [PubMed] [Google Scholar]
- 38.Darmon A J, Nicholson D W, Bleackley R C. Nature (London) 1995;377:446–448. doi: 10.1038/377446a0. [DOI] [PubMed] [Google Scholar]
- 39.Darmon A J, Ley T J, Nicholson D W, Bleackley R C. J Biol Chem. 1996;271:21709–21712. doi: 10.1074/jbc.271.36.21709. [DOI] [PubMed] [Google Scholar]
- 40.Yang X, Stennickes H R, Wang B, Green D R, Janicke R U, Srinivasan A, Seth P, Salvesen G S, Froelich C J. J Biol Chem. 1998;273:34278–34283. doi: 10.1074/jbc.273.51.34278. [DOI] [PubMed] [Google Scholar]
- 41.Huang K, Takagaki M, Kani K, Ohkubo I. Biochim Biophys Acta. 1996;1290:149–156. doi: 10.1016/0304-4165(96)00013-x. [DOI] [PubMed] [Google Scholar]
- 42.Grossrau R, Lojda Z. Histochemistry. 1980;70:53–76. doi: 10.1007/BF00508846. [DOI] [PubMed] [Google Scholar]
- 43.Pham C T N, MacIvor D M, Hug B A, Heusel J W, Ley T J. Proc Natl Acad Sci USA. 1996;93:13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abbott R E, Corral C J, MacIvor D M, Lin X, Ley T J, Mustoe T A. Arch Surg. 1998;133:1002–1006. doi: 10.1001/archsurg.133.9.1002. [DOI] [PubMed] [Google Scholar]
- 45.Belaaouaj A, McCarthy R, Baumann M, Gao Z, Ley T J, Abraham S N, Shapiro S D. Nat Med. 1998;4:615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]