Abstract
Aberrant methylation of promoter region CpG islands is associated with transcriptional inactivation of tumor-suppressor genes in neoplasia. To understand global patterns of CpG island methylation in colorectal cancer, we have used a recently developed technique called methylated CpG island amplification to examine 30 newly cloned differentially methylated DNA sequences. Of these 30 clones, 19 (63%) were progressively methylated in an age-dependent manner in normal colon, 7 (23%) were methylated in a cancer-specific manner, and 4 (13%) were methylated only in cell lines. Thus, a majority of CpG islands methylated in colon cancer are also methylated in a subset of normal colonic cells during the process of aging. In contrast, methylation of the cancer-specific clones was found exclusively in a subset of colorectal cancers, which appear to display a CpG island methylator phenotype (CIMP). CIMP+ tumors also have a high incidence of p16 and THBS1 methylation, and they include the majority of sporadic colorectal cancers with microsatellite instability related to hMLH1 methylation. We thus define a pathway in colorectal cancer that appears to be responsible for the majority of sporadic tumors with mismatch repair deficiency.
In the development of colorectal cancers (CRCs), a series of tumor-suppressor genes such as APC, p53, and genes on chromosome 18q (DCC, SMAD2, and DPC4/SMAD4) are inactivated by mutations and chromosomal deletions (1, 2). A subset of CRCs also show a characteristic mutator phenotype which causes microsatellite instability (MSI) and mutations at other gene loci such as TGFβRII (3) and BAX (4). This phenotype usually results from inactivation of mismatch repair (MMR) genes such as hMSH2 and hMLH1 (5). Another molecular defect described in CRC is CpG island (CGI) methylation. CGIs are short sequences rich in the CpG dinucleotide and can be found in the 5′ region of about half of all human genes (6). Methylation of cytosine within 5′ CGIs is associated with loss of gene expression and has been seen in physiological conditions such as X chromosome inactivation and genomic imprinting (7). Aberrant methylation of CGIs has been detected in genetic diseases such as the fragile-X syndrome (8), in aging cells (9), and in neoplasia (10, 11). About half of the tumor-suppressor genes that have been shown to be mutated in the germ line of patients with familial cancer syndromes have also been shown to be aberrantly methylated in some proportion of sporadic cancers, including Rb, VHL, p16, hMLH1, and BRCA1 (10, 11). Tumor-suppressor gene methylation in cancer is usually associated with (i) lack of gene transcription and (ii) absence of coding region mutation. Thus it has been proposed that CGI methylation serves as an alternative mechanism of gene inactivation in cancer.
The causes and global patterns of CGI methylation in human cancers remain poorly defined. We have previously reported that aging could play a factor in this process because methylation of several CGIs could be detected in an age-related manner in normal colon mucosa as well as in CRC (9). In addition, aberrant methylation of CGIs has been associated with the MSI phenotype in CRC (12) as well as specific carcinogen exposures (13). However, an understanding of aberrant methylation in CRC has been somewhat limited by the small number of CGIs analyzed to date. We have used a recently developed PCR-based methylation screening technique, methylated CpG island amplification (MCA), to determine the methylation status of multiple CGIs in a relatively large number of samples. In this report, we analyzed the methylation status of 30 new loci and 3 known tumor-suppressor genes in a panel of 50 primary CRCs and 15 colonic adenomas. We find that (i) the majority of CGI methylation events in CRCs are related to incremental hypermethylation in normal colon as an age-related phenomenon; and (ii) virtually all the other methylation events occur in a distinct subset of CRCs and adenomas which appear therefore to have a new phenotype, which we termed CpG island methylator phenotype (CIMP). These data shed additional light on the global patterns of CGI methylation in human cancer and delineate a distinct pathway involving tumor-suppressor gene hypermethylation in the evolution of CRC.
MATERIALS AND METHODS
Samples and Cell Lines.
Fifty CRCs and 15 colorectal adenoma samples were obtained from The Johns Hopkins Hospital. All patients gave informed consent prior to specimen collection according to institutional guidelines. CRCs used in this study were characterized previously for MSI status (12). Presence or absence of MSI was determined according to strict criteria, requiring band shifts at both dinucleotide and mononucleotide tracts. Of these, 43 cancers were randomly selected and 7 cancers had previously been shown to display MSI. Nine of 43 (21%) randomly selected samples showed MSI. Therefore, in total, 16 samples were MSI positive and 34 samples were MSI negative.
Genes Examined.
In this study, we examined 30 of 33 differentially methylated CpG islands cloned by MCA as described (MINT1–33; ref. 14). The remaining three clones were not examined because of high background or size too small. In addition, we examined the methylation status of p16, THBS1, and hMLH1 by MCA (p16) or bisulfite-PCR (THBS1 and hMLH1).
Detection of Aberrant Methylation by MCA.
MCA was performed essentially as described (14). A detailed protocol is available at http://www.med.jhu.edu/methylation/MCA.html. Presence or absence of methylation in cancers was determined by comparing the signals in the tumor vs. normal lanes. Each sample was blotted in duplicate. Each filter included mixtures of positive control (Caco2) and a negative control (normal colon mucosa from an 18-year-old individual). All 30 MINT clones, as well as p16 (15) were examined by MCA.
Bisulfite-PCR.
Bisulfite treatment and PCR reactions were performed as described previously (16, 17). A detailed protocol is available at http://www.med.jhu.edu/methylation/protocols.html. In total, 12 MINT clones (MINT 1, 2, 4, 6, 7, 11, 12, 23, 25, 29, 31, 32), as well as THBS1, and hMLH1 were examined for methylation status in this way. Primer sequences, conditions for PCR, and restriction enzymes used are available at http://www.med.jhu.edu/methylation/primers.htm.
Southern Blot Analysis.
Five micrograms of DNA was digested with 20–100 units of restriction enzymes as specified by the manufacturer (New England Biolabs). DNA fragments were separated by agarose gel electrophoresis and transferred to a nylon membrane (Zeta-probe, Bio-Rad). Filters were hybridized with 32P-labeled probes and washed at 65°C with 2× SSC/0.1% SDS for 10 min twice, and 0.1× SSC/0.1% SDS for 20 min. Filters were then exposed to a phosphor screen for 24–72 hr and analyzed by using a PhosphorImager (Molecular Dynamics).
RESULTS
Methylation Analysis of Multiple CpG Islands by Using MCA and Bisulfite-PCR.
We have recently developed a PCR-based method termed MCA (14). In MCA, the methylation status of CRC and normal colon mucosa samples can be determined by the presence or absence of a hybridization signal. Using MCA coupled with representational difference analysis, we have isolated 33 sequences (MINT1–33) differentially methylated in Caco2, a CRC cell line, and have confirmed the MCA results by Southern blot analysis. These included 29 CGIs, and several fragments were identical to known gene sequences. Thus, they appear to be fairly representative of hypermethylation events in cancer. The cloning of a large number of CGIs from a CRC cell line allowed us to study the global patterns of hypermethylation in this neoplasm.
To determine the methylation status of these clones in primary tissues, we have used MCA for 30 of the 33 clones (3 clones could not be accurately studied because of high background (MINT19 and MINT29) or small size (MINT33). As shown in Fig. 1, MCA provides semiquantitative methylation data and yields results that are essentially identical to Southern blot analysis or bisulfite-PCR. In 12 cases (MINT1, 2, 4, 6, 7, 11, 12, 23, 25, 29, 31, 32), MCA results were confirmed by bisulfite-PCR for all samples, and the two techniques were concordant in 98% of the cases (examples in Figs. 1 and 2).
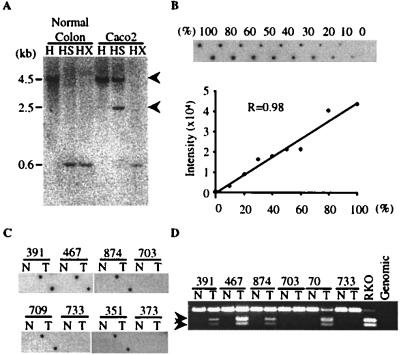
Figure 1.
Comparison of detection of aberrant methylation by using Southern blotting, MCA, or bisulfite-PCR analysis. (A) Methylation of MINT2 detected by Southern blot analysis. Genomic DNA from normal colon mucosa of an 18-year-old person and the CRC cell line Caco2 was digested with restriction endonucleases (H, HindIII; HS, HindIII + SmaI; HX, HindIII + XmaI), electrophoresed, blotted, and hybridized with MINT2 probe. Caco2 DNA fails to digest completely with SmaI, indicating methylation of one or both SmaI sites, but cuts down with XmaI, ruling out polymorphisms at the SmaI sites. (B) Semiquantitative feature of MCA. DNAs from the Caco2 cell line and normal colon were mixed in various proportions before MCA, and methylation of MINT2 was analyzed by dot-blot hybridization (Upper). % refers to the relative proportion of Caco2 DNA. The signal intensity of each dot was determined by PhosphorImager densitometry, and it increased linearly relative to the proportion of tumor cells in the mix. (C) Detection of MINT2 methylation by MCA. We blotted 0.1 μg of MCA products from colorectal tumors and corresponding normal colon mucosa in duplicate onto a nylon filter and hybridized them with a MINT2 probe. Tumors 391, 467, 874, 709, and 351 are methylated at this site. Sample numbers are shown above each lane. N, normal colon; T, colon tumor. (D) Detection of methylation by bisulfite-PCR. Genomic DNA was treated with bisulfite and amplified with primers specific to MINT2. Twenty percent of the PCR products were digested with BstUI and electrophoresed in 3% agarose gels. Arrows indicate bands reflecting methylation of the BstUI site. BstUI cleaves only the methylated alleles, yielding 115- and 88-bp bands (seen in a CRC cell line, RKO, and tumors 391, 467, 874, and 709). The coexistence of methylated and unmethylated bands reflects the fact that all these tumors are not microdissected and contain various amounts of contaminating normal tissues. Sample numbers are shown above. N, normal colon; T, colon tumor.
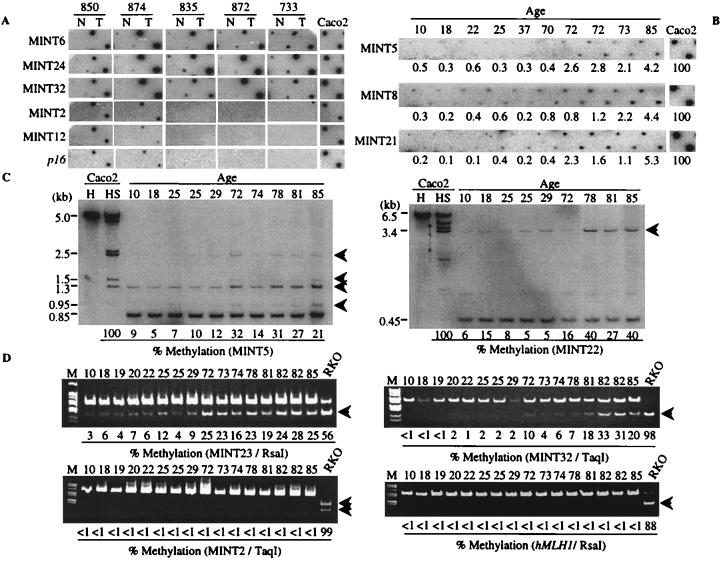
Figure 2.
Methylation of the MINT clones in CRC and mucosa. (A) Examples of MINT clone methylation in colorectal cancer. MCA products from colon tumors (T) and corresponding normal colon mucosa (N) were blotted on a nylon membrane and hybridized with one of the MINT clones (indicated on the left), as well as a p16 probe. MINT6, 24, and 32 are examples of type A methylation, whereas MINT2, 12, and p16 are examples of type C methylation. Tumors 850 and 874 are CIMP+, whereas tumors 835, 872, and 733 are CIMP− (see text). Also shown is the Caco2 cell line (right). (B) Examples of age-related methylation in normal colon as detected by MCA using MINT5, 8, and 21 as probes. MCA products from normal colon mucosa from patients of various ages (indicated at the top in years) were hybridized with the probe indicated on the left. The signal intensity for each sample was determined by densitometry, and a ratio of mucosa/Caco2 (right) is indicated below each sample as a percentage. In each case, methylation is more prominent in DNA from older individuals. (C) Southern blot analysis of clones showing type A methylation in normal colon. DNA from normal colon mucosa from patients of various ages (top) was digested with HindIII and the methylation-sensitive enzyme SmaI and probed with MINT 5 (Left) and MINT22 (Right). Arrows indicate the methylated alleles. The relative proportion of methylated alleles was determined by densitometry and is indicated below each lane. (D) Bisulfite-PCR analysis of clones showing type A and type C methylation in normal colon. DNA from normal colon mucosa from patients of various ages (top) was bisulfite treated, amplified by PCR, and digested with restriction enzymes specific for sites that are created after bisulfite treatment (if the CpG sites are methylated). The arrows indicate the methylated alleles. Percentage methylation was determined by densitometry and is indicated below each lane. In type A loci, the percentage of methylated alleles from older people (>70) was significantly higher than that of younger people (<30) (6.4 ± 1.1 vs. 22.9 ± 1.2, P < 0.000001 for MINT23; 1.3 ± 0.3 vs. 16.1 ± 4.0, P < 0.01 for MINT32). In type C loci no methylation was detected in normal colon mucosa. The loci analyzed and restriction enzymes used are indicated below each gel.
Of the 30 clones analyzed, 26 (87%) were also found to be methylated in some primary CRCs (examples in Figs. 1–3). The four clones methylated only in the cell line Caco2 were (i) MINT14, a LINE element; (ii) MINT14 and MINT18, sequences that had a very low CpG frequency and did not qualify as CpG Islands; and (iii) MINT16 (intron 1 of α-Tubulin). The other 26 clones all qualified as CpG islands on the basis of the criteria >200 bp, G+C content >50%, and CpG/GpC > 0.6 (18). Thus, almost all (26 of 27) nonrepetitive CpG islands recovered from the Caco2 cell line were also methylated in some primary tumors.
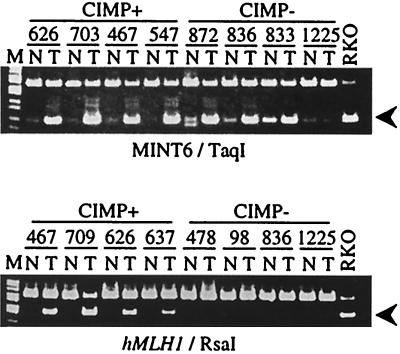
Figure 3.
Methylation analysis of CpG islands in CRC by using bisulfite-PCR. Bisulfite-treated DNA from CRC with (Left) or without (Right) CIMP and paired normal colon mucosa were amplified and digested with restriction enzymes that cleave only the methylated CpG sites. The arrows indicate the methylated alleles. The loci analyzed and restriction enzymes used are indicated below each gel. MINT6 is an example of type A methylation, whereas hMLH1 exemplifies type C methylation.
Two Types of Methylation in CRC.
By examining the methylation status of several known genes in colorectal tumors, we have previously demonstrated that some genes tend to be methylated in an age-dependent manner in normal colon (9). In the present study of 26 CpG islands, we find that hypermethylation patterns in CRC fell into two distinct categories. A majority of the clones (19/26, 73%) were found to be frequently methylated (average 75%, ranging from 30% to 100% of the tumors) in the tumors tested, and a slight amount of methylation was also detected in normal colon mucosa (Fig. 2A). For all of these clones, the normal colon mucosa obtained from young patients showed less methylation compared with the normal mucosa from older patients (Fig. 2 B–D). For this age-related methylation, identical results were obtained by MCA, Southern blot analysis, and bisulfite-PCR analysis, suggesting that it is not related to the technique used to study methylation. Thus, the majority of CGIs hypermethylated in CRC are methylated in normal colon mucosa as well, in an age-related manner. We propose to name this methylation type A for aging-specific methylation.
The remaining 7 clones were methylated exclusively in CRC: no methylation was observed in normal colon by any technique (see Figs. 1 and 2), and their frequency of methylation was significantly lower than type A methylation (ranging from 10% to 50%, see below). We propose to name this type of methylation type C for cancer-specific. There was no significant difference in the G+C content between type A and type C clones (average, 57.1% for type A vs. 58.3% for type C, P = 0.36).
CIMP in CRC.
We next analyzed in detail the methylation status of the type A and type C MINT clones in a panel of 50 primary colorectal cancers and 15 adenomatous polyps. The data obtained by MCA were verified by bisulfite-PCR for 12 clones, and the results obtained with the two techniques were 98% concordant. As mentioned above, all 19 type A clones were frequently methylated in cancers (average 75%, ranging from 30% to 100% of the tumors, see examples in Figs. 2A and 3A and summary in Fig. 4). These frequencies are similar to what we had previously observed for ER (9), MyoD, and N33 (19). When we considered the methylation status of the 7 type C clones however, a remarkable pattern emerged (examples in Figs. 1, 2A, and 3B, summarized in Fig. 4). The 50 cancers fell into two distinct groups: (i) a group with a high level of type C methylation, whereby all the tumors had methylation of 3 or more loci simultaneously (average 5.1 loci per tumor) and (ii) a group where methylation of any type C clone is extremely rare (average 0.3 locus per tumor). In sharp contrast, type A methylation was not significantly different between these two groups of tumors (Figs. 2A, 3A, and 4). Thus, the first group of tumors appears to display a novel process, which we propose to call CpG island methylator phenotype (CIMP). These tumors would then be prone to transcriptional silencing linked to promoter methylation, and they would have the potential to inactivate multiple tumor-suppressor genes simultaneously.
Figure 4.
Methylation profile of colorectal cancer. The methylation status of all type C MINT clones, as well as p16, THBS1, and hMLH1, was determined in a panel of primary CRC and adjacent mucosa. None of the normal tissues studied showed any degree of methylation for these genes. Also shown are representative type A clones (Right). Each column represents a separate gene locus (indicated on top). Each row is a primary CRC (samples above the bold solid line) or adenomatous polyp (below the bold solid line). Black rectangles, methylated tumor; white rectangles, unmethylated tumor. MSI denotes the presence of microsatellite instability and was not determined for the adenomas. In the Center are known genes methylated in CRC (p16, THBS1, hMLH1).
We next studied the relationship of CIMP to known clinicopathological factors in the progression of colorectal neoplasia. In this limited series, CIMP did not appear to correlate with age, gender, or stage of the CRC. However a majority of proximal (cecum, ascending colon) cancers were affected by this pathway (18/22, 82%), which was significantly different from distal (descending, recto-sigmoid) cancers (10/27, 37%; P = 0.003, Fisher’s exact t test). Furthermore, CIMP was also found in preneoplastic adenomatous polyps. Overall, 15 adenomas were studied, and 7/15 were CIMP+. In 6 cases, both an adenoma and a cancer from the same patient were examined. In 1 of these, CIMP was detected in both the adenoma and the cancer; in 3 cases, CIMP was detected in the cancer but not in the adenoma; in 2 cases, CIMP was detected in neither the adenoma nor the cancer. These results suggested that CIMP is an early event in colorectal carcinogenesis.
To determine whether CIMP indeed affects the methylation status of known tumor-suppressor genes important to tumor progression, we studied the p16 gene (20), which is one of the most frequently altered genes in human neoplasms, and the THBS1 gene, which encodes for an angiogenesis-inhibitor with tumor-suppressor properties (21, 22). p16 was studied by both MCA and Southern blot analysis, and THBS1 was studied by Southern blotting and bisulfite-PCR. There was an excellent concordance between methylation of both genes and the presence of CIMP: all 20 cancers and 3 adenomatous polyps methylated at the p16 CpG island had been classified as CIMP+ by using our type C MINT clones (Fig. 4). Similarly, all 9 tumors hypermethylated at THBS1 also belonged to the CIMP+ group (Fig. 4). Thus, CIMP is not limited to the clones recovered by MCA/RDA, but truly reflects a methylator phenotype in these tumors.
Microsatellite Instability Is Linked to CIMP in CRC.
In a previous study (12), we reported a link between microsatellite instability and a hypermethylator phenotype in sporadic CRC. Relatively few mutations in mismatch repair genes have been reported in sporadic MSI+ cancers, but hMLH1 methylation has recently been observed in some cases (23–25) and was suggested to be a primary cause of the MSI phenotype in sporadic cancers. To determine the relation between CIMP and MSI in colorectal cancer, we measured hMLH1 methylation by using bisulfite/PCR in our panel of CRC, which had also been previously typed for the presence of MSI (Fig. 3). hMLH1 was studied by bisulfite-PCR only because it does not have 2 SmaI sites in its CGI. Overall, 16 of 50 (32%) cancers had evidence of MSI. Among the 29 CIMP+ cases, 12 had evidence of hMLH1 methylation, suggesting that hMLH1 is one of the targets of global hypermethylation in CRC. All of these 12 tumors had MSI. By contrast, hMLH1 methylation was detected in only one of the 21 CIMP− cases. These data establish a strong link between the CIMP phenotype, hMLH1 methylation, and MSI in CRC. Two lines of evidence suggest that MSI may follow, and be caused by, CIMP and hMLH1 methylation. First, CIMP is detectable in about half of colonic adenomas, but none of these tumors have hMLH1 methylation, and MSI is rare in this preneoplastic lesion (26, 27). Second, CIMP is not simply caused by mismatch repair defects because, MSI is absent in more than half of the CIMP+ cases, and CIMP was absent in 4 of the 16 cancers with MSI. Overall, these data suggest that, in sporadic CRC, the majority (12/16, 75%) of cases with MSI may be caused by CIMP followed by hMLH1 methylation, loss of hMLH1 expression, and resultant mismatch repair deficiency (24). Interestingly, mismatch repair deficiency in colorectal cancer is also clustered in proximal tumors, similar to CIMP.
DISCUSSION
Recently, several reports have suggested that aberrant methylation of CGIs may play an important role in cancer development (10, 11). However, there is little integrated information on aberrant CGI methylation in cancer at multiple loci, probably because of the lack of a method to detect methylation in a large number of samples for unselected CGIs throughout the genome. Furthermore, it has been shown that cultured cell lines have a high degree of CGI methylation (28) but it was not known to what extent this reflects methylation in primary cancers. In the present study, we have been able to address some of these issues by studying 30 differentially methylated loci in a panel of colorectal cancers, a study facilitated by the relatively quantitative and high-throughput features of MCA.
Despite the fact that all sequences were initially recovered from a colon cancer cell line, 26/27 nonrepeated CpG islands proved to be methylated in some primary colon cancers. Therefore, there appears to be relatively little “artifactual” methylation in cell lines. Nevertheless, cell lines often have more frequent hypermethylation of selected loci than do primary tumors, and it appears probable that this reflects continuous selection in culture.
Analysis of the 26 clones methylated in primary tumors revealed two distinct types of hypermethylation in cancer (type A for aging-specific and type C for cancer-specific), which may have distinct causes, and different roles in cancer development. Type A methylation was seen in the majority of these clones: 19 of 26 (73%) clones were methylated in an age-related manner in normal colon, and hypermethylated at high frequency in colorectal cancer, as we have shown for the ER gene (9) and others (19, 29). These results suggest that a large number of CGIs in the human genome are incrementally methylated during the aging process and, for many genes, this methylation correlates with reduced gene expression as shown for ER (9) and Versican (14). Although we do not know the mechanism of type A methylation, it likely results from physiological processes rather than a genetic alteration because (i) it is very frequent and affects large numbers of cells; (ii) it is present in all individuals, not just patients with cancer; and (iii) this process is gene and tissue specific (19). Because the methylation status at a given CGI is thought to be related to positive (methylator) factors (30–33) and negative (protector) factors (34–37), it is possible that, for some genes, this balance slightly favors de novo methylation, and that this is reflected by progressive hypermethylation after repeated cell divisions.
In contrast to type A methylation, type C methylation is relatively infrequent in primary colorectal cancer, and is never observed in normal colon mucosa. Furthermore, detailed analysis of type C methylation in CRC revealed a striking pattern, suggesting the presence of a hypermethylator phenotype in a subset of these tumors: CIMP+ cases are characterized by frequent concordant methylation of the type C clones examined. By contrast, type C methylation is virtually nonexistent in tumors without CIMP. This concordance cannot be due to simple experimental variation or artifacts because (i) methylation was verified by using separate methods (MCA, bisulfite-PCR, and Southern blotting); (ii) the concordance was not limited to the MINT clones, since it also affected the p16, THBS1, and hMLH1 genes; and (iii) there was no significant difference in type A methylation between CIMP+ and CIMP− tumors. Through its ability to silence multiple genes simultaneously, CIMP would then be functionally equivalent to genetic instability, resulting in the rapid accumulation of molecular alterations with a potential to accelerate the neoplastic process. Because many genes are potential candidates for inactivation through promoter methylation (10, 11), CIMP may have profound pathophysiological consequences in neoplasia through inactivation of tumor-suppressor genes, metastasis-suppressor genes, angiogenesis inhibitors, and others. In fact, our data suggest that CIMP could also result in mismatch repair deficiency through methylation and inactivation of the hMLH1 promoter, and it may explain up to 75% of cases of sporadic CRC with MSI.
The causes of type A and type C methylation are probably different because the latter is detected only in a limited number of cases and the genes affected are distinct. This defect could be either aberrant de novo methylation (through a mutation in DNA-methyltransferase for example), or loss of protection against de novo methylation, through the loss of a trans-activating factor (34, 35). Because DNA-methyltransferase activity is similar in the two groups (data not shown), we favor the latter hypothesis. Thus, at least in CRC, it appears likely that type C methylation (an epigenetic error) is actually caused by a genetic event that results in an increased chance of methylating a subset of CGIs. Ironically, this epigenetic defect may then result in additional genetic lesions through the induction of mismatch-repair deficiency.
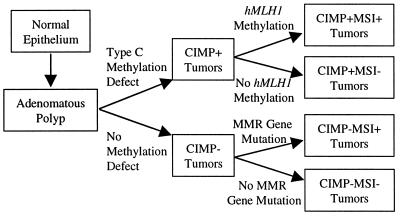
On the basis of these data, we propose the following speculative model integrating CGI methylation into CRC development (Fig. 5). In this model, CGI methylation plays two distinct roles, and it appears to arise through distinct mechanisms. Initially, type A methylation arises as a function of age in normal colorectal epithelial cells. By potentially affecting genes that regulate the growth and/or differentiation of these cells, such methylation could account, in part, for the hyperproliferative state that is thought to precede tumor formation in the colon (38). Such hyperproliferation is known to arise with age in colorectal epithelium (39, 40) and to be marked in patients with colorectal cancer. Furthermore, modulation of type A methylation may provide one possible explanation for the reduction in CRC tumorigenesis by reducing levels of DNA-methyltransferase (41). A second major role for CGI methylation appears later, possibly at the transition between small and large adenomas in the colon (on the basis of 15 adenomas presented here and preliminary data on 50 additional cases). This methylation (type C) affects only a subset of tumors, which then evolve along a pathway of global hypermethylation. We propose then that CIMP leads to cancer development through the simultaneous inactivation of multiple tumor-suppressor genes such as p16, and induction of mismatch repair deficiency through inactivation of hMLH1. This model may be applicable to most human malignancies. In fact, we have observed evidence for type A and type C methylation in brain tumors (42), and we have preliminary evidence suggesting the presence of CIMP in multiple types of neoplasms, including stomach cancers, brain tumors, and hematopoietic malignancies. Deciphering the mechanisms underlying these phenomena should facilitate the early detection, prevention, and therapy of various neoplasms.
Figure 5.
A model integrating CGI methylation in colorectal carcinogenesis. See text for details.
Acknowledgments
We thank Dr. Stanley Hamilton for providing the colon tumor specimens and Dr. Bert Vogelstein for reviewing the manuscript. This work was supported by grants from the National Institutes of Health (National Cancer Institute Grants CA77045 and CA54396 and Colon Cancer Spore Grant CA62924). M.T. is a Postdoctoral Fellow of the Japan Society for the Promotion of Science. N.A. is supported by National Institutes of Health Training Grant 1-T32-DK07713. J.-P.J.I. is a Kimmel Foundation Scholar.
ABBREVIATIONS
- CRC
colorectal cancer
- MSI
microsatellite instability
- CGI
CpG island
- MCA
methylated CpG island amplification
- CIMP
CpG island methylator phenotype
References
- 1.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 2.White R L. Cell. 1998;92:591–592. doi: 10.1016/s0092-8674(00)81124-1. [DOI] [PubMed] [Google Scholar]
- 3.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1994;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 4.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed J C, Perucho M. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 5.Peltomaki P, de la Chapelle A. Adv Cancer Res. 1997;71:93–119. doi: 10.1016/s0065-230x(08)60097-4. [DOI] [PubMed] [Google Scholar]
- 6.Bird A P. Nature (London) 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 7.Latham K E. Trends Genet. 1996;112:134–138. doi: 10.1016/0168-9525(96)10017-2. [DOI] [PubMed] [Google Scholar]
- 8.Hansen R S, Gartler S M, Scott C R, Chen S H, Laird C D. Hum Mol Genet. 1992;8:571–578. doi: 10.1093/hmg/1.8.571. [DOI] [PubMed] [Google Scholar]
- 9.Issa J-P, Ottaviano Y L, Celano P, Hamilton S R, Davidson N E, Baylin S B. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 10.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J-P. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 11.Jones P A. Cancer Res. 1996;56:263–267. [PubMed] [Google Scholar]
- 12.Ahuja N, Mohan A L, Li Q, Stolker J M, Herman J G, Hamilton S R, Baylin S B, Issa J-P. Cancer Res. 1997;57:3370–3374. [PubMed] [Google Scholar]
- 13.Issa J-P, Baylin S B, Belinsky S A. Cancer Res. 1996;56:3655–3658. [PubMed] [Google Scholar]
- 14.Toyota M, Ho C, Ahuja N, Jair K-W, Li Q, Ohe-Toyota M, Baylin S B, Issa J-P J. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- 15.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J-P, Davidson N E, Sidransky D, Baylin S B. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 16.Sadri R, Hornsby P J. Nucleic Acids Res. 1996;24:558–559. doi: 10.1093/nar/24.24.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong Z, Laird P W. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardiner-Garden M, Frommer M. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 19.Ahuja N, Li Q, Mohan A L, Baylin S B, Issa J-P. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 20.Kamb A. Trends Genet. 1995;11:136–140. doi: 10.1016/s0168-9525(00)89027-7. [DOI] [PubMed] [Google Scholar]
- 21.Dameron K M, Volpert O V, Tainsky M A, Bouck N. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 22.Sheibani N, Frazier W A. Proc Natl Acad Sci USA. 1995;92:688–692. doi: 10.1073/pnas.92.15.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kane M F, Loda M, Gaida G M, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Cancer Res. 1996;57:808–811. [PubMed] [Google Scholar]
- 24.Herman J G, Umar A, Polyak K, Graff J R, Ahuja N, Issa J-P J, Markowitz S, Willson J K, Hamilton S R, Kinzler K W, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham J M, Christensen E R, Tester D J, Kim C Y, Roche P C, Burgart L J, Thibodeau S N. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 26.Samowitz W S, Slattery M L. Gastroenterology. 1997;112:1516–1519. doi: 10.1016/s0016-5085(97)70032-5. [DOI] [PubMed] [Google Scholar]
- 27.Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Onda A, Okumura Y, Kishi N, Iwama T, Mori T, Koike M, et al. Gastroenterology. 1997;111:307–317. doi: 10.1053/gast.1996.v111.pm8690195. [DOI] [PubMed] [Google Scholar]
- 28.Antequera F, Boyes J, Bird A. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 29.Issa J-P, Vertino P M, Boehm C D, Newsham I F, Baylin S B. Proc Natl Acad Sci USA. 1996;93:1757–1762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mummaneni P, Bishop P L, Turker M S. J Biol Chem. 1993;268:552–558. [PubMed] [Google Scholar]
- 31.Mummaneni P, Walker K A, Bishop P L, Turker M S. J Biol Chem. 1995;270:788–792. doi: 10.1074/jbc.270.2.788. [DOI] [PubMed] [Google Scholar]
- 32.Magewu A N, Jones P A. Mol Cell Biol. 1994;14:4225–4332. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vertino P M, Yen R W, Gao J, Baylin S B. Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macleod D, Charlton J, Mullins J, Bird A P. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 35.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Nature (London) 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 36.Turker M S, Bestor T H. Mutat Res. 1997;386:119–130. doi: 10.1016/s1383-5742(96)00048-8. [DOI] [PubMed] [Google Scholar]
- 37.Chen F Y, Harris L C, Remack J S, Brent T P. Proc Natl Acad Sci USA. 1997;94:4348–4353. doi: 10.1073/pnas.94.9.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipkin M. Cancer Res. 1988;48:235–245. [PubMed] [Google Scholar]
- 39.Holt P R, Yeh K Y. Gastroenterology. 1988;95:1556–1563. doi: 10.1016/s0016-5085(88)80077-5. [DOI] [PubMed] [Google Scholar]
- 40.Roncucci L, Ponz de Leon M, Scalmati A, Malagoli G, Pratissoli S, Perini M, Chahin N J. Cancer. 1988;62:2373–2377. doi: 10.1002/1097-0142(19881201)62:11<2373::aid-cncr2820621120>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 41.Laird P W, Jackson-Grusby L, Fazeli A, Dickinson S L, Jung W E, Li E, Weinberg R A, Jaenisch R. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 42.Li Q, Jedlicka A, Ahuja N, Gibbons M C, Baylin S B, Burger P C, Issa J-P J. Oncogene. 1998;16:3197–3202. doi: 10.1038/sj.onc.1201831. [DOI] [PubMed] [Google Scholar]