Abstract
Hypoxia-inducible factor (HIF), a transcriptional complex conserved from Caenorhabditis elegans to vertebrates, plays a pivotal role in cellular adaptation to low oxygen availability. In normoxia, the HIF-α subunits are targeted for destruction by prolyl hydroxylation, a specific modification that provides recognition for the E3 ubiquitin ligase complex containing the von Hippel–Lindau tumour suppressor protein (pVHL). Three HIF prolyl-hydroxylases (PHD1, 2 and 3) were identified recently in mammals and shown to hydroxylate HIF-α subunits. Here we show that specific ‘silencing’ of PHD2 with short interfering RNAs is sufficient to stabilize and activate HIF-1α in normoxia in all the human cells investigated. ‘Silencing’ of PHD1 and PHD3 has no effect on the stability of HIF-1α either in normoxia or upon re-oxygenation of cells briefly exposed to hypoxia. We therefore conclude that, in vivo, PHDs have distinct assigned functions, PHD2 being the critical oxygen sensor setting the low steady-state levels of HIF-1α in normoxia. Interestingly, PHD2 is upregulated by hypoxia, providing an HIF-1-dependent auto-regulatory mechanism driven by the oxygen tension.
Keywords: angiogenesis/HIF prolyl-hydroxylases/hypoxia signalling/oxygen sensor/small interfering RNA
Introduction
All organisms possess mechanisms to maintain oxygen homeostasis, which are essential for survival. The hypoxia-inducible factor-1 (HIF-1), conserved during evolution from worms to flies to vertebrates, is central to adaptation to low oxygen availability. HIF-1 in turn regulates transcription of many genes involved in cellular and systemic responses to hypoxia, including breathing, vasodilation, anaerobic metabolism, erythropoiesis and angiogenesis. Therefore, hif represents a ‘master’ gene in oxygen homeostasis during embryonic development and postnatal life in both physiological and pathophysiological processes such as tumour growth and metastasis (for a review, see Semenza, 1998).
HIF-1 is a heterodimer consisting of one of three α-subunits (HIF-1α, HIF-2α or HIF-3α) and the β-subunit (HIF-1β, also called aryl hydrocarbon nuclear translocator, or ARNT) (Wang et al., 1995; Ema et al., 1997; Tian et al., 1997; Gu et al., 1998). HIF-1β is a constitutive nuclear protein, which also participates in the cellular response to environmental toxins such as aryl hydrocarbons, whereas HIF-α is specific to the response to hypoxia (Hoffman et al., 1991). Although oxygen availability regulates multiple steps on HIF-1 transcriptional activation, the dominant control mechanism occurs through oxygen-dependent proteolysis of HIF-α (Huang et al., 1996). The most extensively studied isoform of the α-subunits is the ubiquitous HIF-1α.
In normoxia, HIF-1α is constitutively synthesized and sent to destruction by the ubiquitin–proteasome pathway (half-life <5 min) (Salceda and Caro, 1997; Huang et al., 1998; Kallio et al., 1999). This process is mediated by the specific binding of pVHL, the product of the von Hippel–Lindau tumour suppressor gene, which is mutated in most sporadic clear cell carcinomas and in VHL disease (Kaelin and Maher, 1998; Maxwell et al., 1999; Cockman et al., 2000; Kamura et al., 2000; Ohh et al., 2000). pVHL is part of a multiprotein complex that includes elongin B, elongin C, Rbx1 and Cul2 (Kamura et al., 1999; Lisztwan et al., 1999; Stebbins et al., 1999). This complex functions as an E3 ubiquitin ligase which, only in the presence of oxygen, binds directly to and targets HIF-1α for polyubiquitylation and proteasome-dependent degradation. Decreased oxygen levels result in the stabilization of HIF-1α and the activation of the transcriptional complex leading to the expression of target genes such as vegf, epo and glut-1 (Semenza, 1998).
Recent major advances have shown that prolyl hydroxylation and acetylation, by controlling HIF-1α–pVHL physical interaction, are critical in the regulation of HIF-1α steady-state levels (Ivan et al., 2001; Jaakkola et al., 2001; Jeong et al., 2002). The proline residues subjected to hydroxylation reside in the HIF-1α oxygen-dependent degradation domain (ODDD) within an LXXLAP sequence motif, which is strongly conserved between the HIF-α isoforms. In the same degradation domain, Lys532, when acetylated by ARD1, cooperates with the hydroxyl group in the recruitment of pVHL and subsequent HIF-1α degradation (Jeong et al., 2002).
In mammalian cells, three isoforms, PHD1, PHD2 and PHD3, have been identified and shown to hydroxylate in vitro the key proline residues (Pro402 and Pro564) of HIF-1α (Epstein et al., 2001). These three orthologues of the Caenorhabditis elegans Egl-9 have also been called EGLN2, EGLN1, EGLN3 and HPH3, HPH2, and HPH1, respectively (Bruick and McKnight, 2001; Ivan et al., 2002). Hereafter, they will be referred to using the PHD nomenclature. PHDs are dioxygenases that utilize oxygen as co-substrate providing the molecular basis for the oxygen-sensing function of these enzymes. Indeed, the activity of the purified PHDs has been reported to be strikingly sensitive to graded levels of hypoxia in vitro, mirroring the progressive increases in HIF-1α protein and DNA binding activity that are observed when cells are exposed to gradual hypoxia in culture (Epstein et al., 2001). In addition, the prolyl hydroxylation reaction requires 2-oxoglutarate and iron as cofactors, thereby accounting for the well known ‘hypoxia-mimic’ effects of iron chelators (such as desferrioxamine) and transition metals (such as Co2+, Mn2+ and Ni2+) on HIF-1α induction. Each PHD isoform differs in the relative abundance of their mRNA, but all three show a ubiquitous pattern of expression (Lieb et al., 2002; Cioffi et al., 2003).
The co-expression of the three highly conserved PHDs raises the following intriguing questions: why three isoforms? Do they have overlapping or unique activities? Do they respond to different pO2 values? To evaluate the role of the three mammalian Egl-9 orthologues with respect to HIF-1α hydroxylation and expression in vivo, we specifically ablated each isoform by exploiting the small interfering RNA (siRNA) approach, developed with great success by Tuschl and co-workers (Elbashir et al., 2001). Here we show that specific silencing of PHD2 is sufficient to: (i) stabilize HIF-1α steady-state levels in normoxia in all the human cells analysed so far; (ii) fully protect HIF-1α degradation upon re-oxygenation of hypoxia-stressed cells; and (iii) trigger HIF-1α nuclear accumulation and HIF-dependent transcriptional activation in normoxia. Finally, we present evidence that PHD2 is upregulated by hypoxia, supporting the auto-regulatory mechanism we have previously proposed for oxygen-driven HIF-1α regulation (Berra et al., 2001a).
Results
Specific silencing of PHD isoforms using a siRNA approach
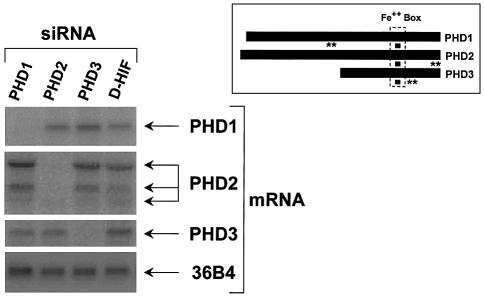
To evaluate the role of the three mammalian PHDs in the stability of HIF-1α in vivo, we proceeded to ablate each isoform by transfecting HeLa cells with siRNAs. Two independent sets of 21 bp siRNA duplexes were chosen. We first targeted the sequence coding for the iron-binding site within PHD1, PHD2 and PHD3 (inset Figure 1). This region is 100% conserved at the amino acid level between the three isoforms. However, degeneration of the codons within this region allowed design of three siRNAs differing by five nucleotides; this variation has been described to be more than sufficient to target specifically the different isoforms (Elbashir et al., 2001). As shown in Figure 1, HeLa cells transiently transfected with the siRNA duplex corresponding to PHD1 displayed a remarkably specific and a virtually complete loss of the PHD1 mRNA, whereas the same siRNA had no effect on PHD2, PHD3 or control 36B4 mRNA levels. Equivalent results were obtained following transfection with the siRNAs targeting PHD2 or PHD3. As a control, we used an irrelevant siRNA (D-HIF) which, as expected, had no effect on any mRNA. The same results were also obtained with transfection of a second set of siRNA duplexes targeting an independent and non-conserved region within PHD1, PHD2 and PHD3 (inset of Figure 1). We have also examined the siRNA action at the protein level. In Supplementary figure 1S (see Supplementary data available at The EMBO Journal Online), we show the remarkable efficiency of PHDs siRNAs, used under the same conditions, in lowering protein levels of the corresponding recombinant expressed PHDs. In fact, presumably due to differences in protein turnover, PHD1 and PHD3 are much more reduced than PHD2. Therefore, as previously published (Elbashir et al., 2001), our results confirm the efficiency and specificity of the siRNA silencing strategy.
Fig. 1. Specific silencing of PHD isoforms using siRNAs. Northern blot showing total RNA isolated from HeLa cells after 48 h of transfection with the indicated siRNAs (20 nM). Blots were hybridized with 32P- labelled probes specific for PHD1, PHD2, PHD3 or 36B4 (loading control). Inset shows localization of the PHD region targeted by the two independent sets of siRNAs (filled squares and double asterisks).
Silencing of PHD2 upregulates HIF-1α steady-state levels in normoxia
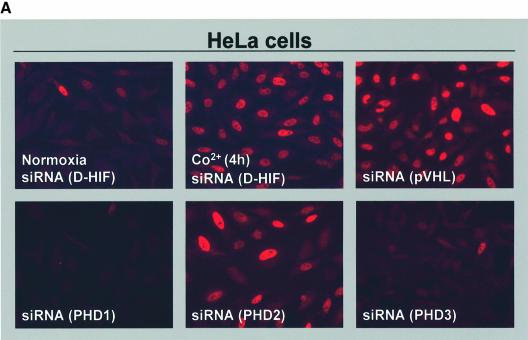
We next evaluated the impact of the specific silencing of each PHD isoform on the steady-state levels of the HIF-1α subunit, monitored by immunofluorescence microscopy or western blotting. Under the same conditions shown in Figure 1, we transiently transfected HeLa cells with siRNAs corresponding to PHD1, PHD2 or PHD3. As a control, HeLa cells transfected with the irrelevant D-HIF siRNA were incubated either in normoxia, in the presence of Co2+ (200 µM) or in hypoxia for 4 h (data not shown). Immunofluorescence depicted in Figure 2A revealed that extinction of either PHD1 or PHD3 had no impact on HIF-1α expression in normoxia. However, silencing of PHD2 upregulated HIF-1α similarly to hypoxia or Co2+. In addition, as expected, transfection of a siRNA targeting pVHL mimicked PHD2 silencing (Figure 2A).
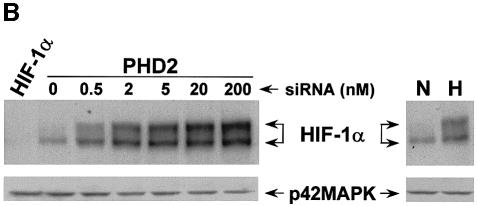
Fig. 2. Role of PHD isoforms in HIF-1α regulation. (A) Silencing of PHD2 upregulates HIF-1α expression in normoxia. HeLa cells were transfected with siRNAs (20 nM) and, 48 h later, were analysed by immunofluorescence microscopy using the anti-HIF-1α antibody. As a control for HIF-1α induction, cells were incubated in the presence of Co2+ (200 µM) for 4 h. (B) PHD2 silencing upregulates HIF-1α in a dose-dependent manner. HeLa cells were transfected with either human HIF-1α siRNA (200 nM) or increasing doses of PHD2 siRNA, and HIF-1α expression was analysed by western blotting after 48 h of transfection (left panel). Extracts of cells incubated in normoxia (N; 20% O2) or hypoxia (H; 1–2% O2) for 4 h were resolved as a control (right panel). Western blots were re-probed using the anti-p42MAPK antibody to check for total protein loading.
Moreover, HIF-1α steady-state upregulation was dependent upon the amount of PHD2 siRNA transfected (Figure 2B). Interestingly, we detected HIF-1α induction at a concentration as low as 0.5 nM siRNA; at 2 nM, the level of HIF-1α surpassed that achieved after 4 h of hypoxic stress, and slightly increased up to 200 nM. In addition, transfection of an siRNA targeting the human HIF-1α isoform completely abolished the signal we detected by immunoblotting using our anti-HIF-1α antibody in normoxia as well as in hypoxia (left lane of Figure 2B; data not shown). This result also validates that the two immunoreactive species shown in the SDS–gel correspond to the human HIF-1α isoform. In addition, it is important to note that none of the transfected siRNA had an impact on the expression of p42MAPK.
PHD2 silencing upregulates HIF-1α in all the human cells investigated
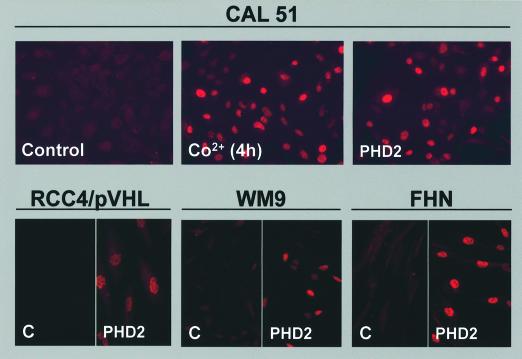
We analysed whether specific silencing of PHD2-induced HIF-1α upregulation was a common mechanism. Thus, we investigated a battery of human cells of different origin. First we tested immortalized cell lines such as CAL27 (derived from a squamous cell carcinoma of the tongue), CAL51 (derived from a breast cancer), HaCAT (keratinocyte cell line), HT29 (derived from a colon carcinoma), RCC4/pVHL (derived from a clear cell renal carcinoma in which we have re-introduced wild-type pVHL) and WM9 (melanoma cell line) (Gioanni et al., 1990). Secondly, we assessed non-immortalized cultures of fibroblasts (FHN), keratinocytes and vascular endothelial cells from the umbilical vein (HUVECs). As for HeLa cells, only PHD2 siRNA was able to upregulate HIF-1α in all the cells so far evaluated (Figure 3; Supplementary figures 3S1, 3S2, 3S3 and 3S4). Although the efficiency of transfection varied greatly among the cell types, neither PHD1 nor PHD3 siRNAs had, under these conditions, an impact on HIF-1α. We therefore suggest that PHD2 controls steady-state levels of HIF-1α in all cell types examined.
Fig. 3. PHD2 silencing upregulates HIF-1α in all of the human cell lines so far analysed. Immunofluorescence staining showing cells transfected with 20 nM of an irrelevant siRNA (Control or C) or the PHD2 siRNA (PHD2) using the anti-HIF-1α antibody. As a positive control, cells were incubated in the presence of Co2+ (200 µM) for 4 h. The following cells are shown: CAL51 (breast cancer cell line), RCC4/pVHL (RCC4 stably transfected with pCDNA3pVHL), WM9 (melanoma cell line) and FHN (primary fibroblasts).
HIF-1α induced in normoxia by PHD2 silencing is functionally active
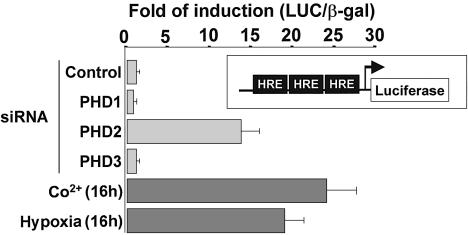
It has been reported that oxygen deprivation affects the subcellular localization, DNA-binding capacity and transcriptional activation function of HIF-1α in addition to regulating its stability (Kallio et al., 1999). However, results shown in Figures 2A and 3 demonstrate that silencing of PHD2 also triggers HIF-1α nuclear accumulation. To characterize the functionality of HIF-1α induced in normoxia by PHD2 silencing, we performed luciferase assays by using a hypoxia-sensitive reporter gene vector (pRE-Δtk-LUC) coding for the LUC gene under the control of a minimal promoter containing three copies of the hypoxia response element (HRE) from the erythropoietin gene. Thus, HeLa cells were transfected with the reporter construct in the presence of either the irrelevant D-HIF siRNA, as a control, or siRNAs targeting each of the PHD isoforms, and an expression vector coding for β-galactosidase for normalization. Luciferase activity was measured after 48 h of transfection. As a positive control, cells transfected with D-HIF siRNA were incubated for 16 h either in hypoxia or in the presence of Co2+ (200 µM). As shown in Figure 4, LUC reporter gene expression was induced in both conditions by ∼20-fold over the basal luciferase activity detected in extracts from non-stimulated cells. In agreement with the above reported findings, PHD1 or PHD3 silencing did no affect the luciferase activity of the reporter vector. However, the specific silencing of PHD2 markedly activated the HREs (14-fold stimulation), showing that upregulated HIF-1α is functionally active in normoxia. Moreover, in parallel experiments, we showed that luciferase activity correlated with the levels of HIF-1α achieved by transfection of PHD2 siRNA at different concentrations (data not shown).
Fig. 4. HIF-1α induced by PHD2 silencing is functionally active. The indicated siRNAs (20 nM) were transfected into HeLa cells with 50 ng of a reporter vector (pRE-Δtk-LUC) containing three copies of the HRE from the erythropoietin gene. In all cases, 100 ng of a plasmid for β-galactosidase were co-transfected to normalize for transfection efficiency. After 48 h of transfection, luciferase activity was measured. As a control for HIF-1 activation, cells were incubated in the presence of Co2+ (200 µM) or in hypoxia (1–2% O2) for 16 h. Results are representative of three independent experiments performed in triplicate.
PHD2 is essential for HIF-1α degradation
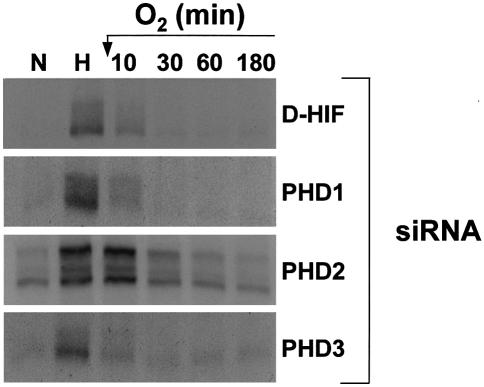
The finding that PHD1 and PHD3 fail to regulate HIF-1α levels, whereas both isoforms share with PHD2 the ability to hydroxylate in vitro the critical Pro564 within the HIF-1α ODDD, is rather intriguing. So far, all experiments were performed in normoxia. We therefore wanted to explore whether low oxygen availability might not be required to ‘activate’ PHD1 and/or PHD3 in vivo. To investigate this possibility, we stressed cells for 4 h in hypoxia followed by re-oxygenation. We next measured the HIF-1α half-life in total cell extracts from cells transfected with the siRNA control or the specific siRNAs invalidating each PHD isoform. As previously reported, re-oxygenation triggers immediate and dramatic destruction of HIF-1α in control cells (Figure 5). Interestingly, cells silenced for either PHD1 or PHD3 displayed identical kinetics for HIF-1α degradation compared with control cells. Once again, only the silencing of PHD2 had a significant effect on oxygen-dependent HIF-1α degradation. As expected, cells transfected with PHD2 siRNA showed upregulated levels of HIF-1α in normoxia. This level was increased further with 4 h of hypoxic stress. Re-oxygenation for 10 min did not modify HIF-1α expression, whereas the protein level progressively decreased to reach that found in normoxia after 60 min. These results clearly excluded the contribution of PHD1 and PHD3 to HIF-1α regulation even under these stress conditions. We therefore conclude that PHD2 is the critical oxygen sensor regulating oxygen-dependent HIF-1α degradation either in normoxia or following a short exposure to hypoxia.
Fig. 5. PHD2 controls HIF-1α stability upon re-oxygenation of hypoxic cells. HeLa cells transfected with siRNAs (200 nM) were incubated in normoxia (N; 20% O2) or hypoxia (H; 1–2% O2) for 4 h, after which hypoxic cells were returned to 20% O2 for 0–180 min. Total cellular extracts were immunoblotted with the anti-HIF-1α antibody.
PHD2 is a hypoxia-inducible gene product
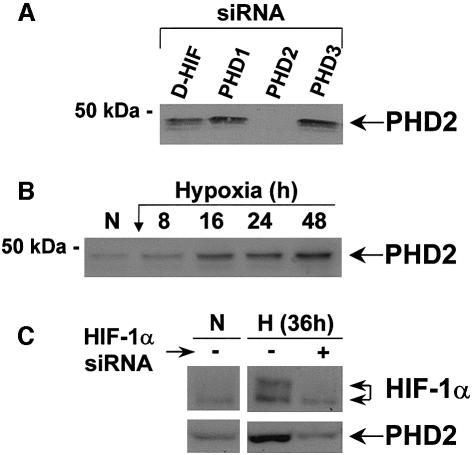
We have shown previously that HIF-1α is subjected to an autoregulatory mechanism, and we have postulated that HIF-1α is targeted for degradation by a factor we called HIF-1α proteasome targeting factor (HPTF) (Berra et al., 2001a). This factor accumulates in an inactive form during long periods of hypoxia, whereas, upon re-oxygenation, its reactivation triggers HIF-1α degradation at a rate that is inversely proportional to the length of hypoxia. Therefore, we propose that HPTF is PHD2. As previously reported (Epstein et al., 2001), we have demonstrated that PHD2 mRNA is upregulated by hypoxia (data not shown). To evaluate this hypothesis further, we raised antibodies against the last 16 amino acids of the human form of PHD2. The antibodies recognized faint bands at ∼50 kDa, which all disappeared following transfection of the siRNA targeting PHD2 (Figure 6A). In contrast, a siRNA control or those silencing PHD1 and PHD3 had no effect on PHD2 protein levels, showing the specificity of the antibody we have generated. HeLa cells were subjected to different periods of hypoxia and total cellular extracts analysed by western blotting. As shown in Figure 6B, PHD2 protein expression increased progressively with the duration of the hypoxic stress. Upregulation detected after 8 h of hypoxia reached a maximal level after 48 h of incubation at low pO2. Identical results were obtained using the CAL51 tumour cell line (data not shown). Interestingly, this hypoxic induction of PHD2 is mediated via HIF-1 transcriptional activity since depletion of HIF-1α with the appropriate siRNA abolishes hypoxia-dependent PHD2 induction (Figure 6C).
Fig. 6. phd2 is a hypoxia-inducible gene. (A) Specificity of the anti-PHD2 antibody. Western blot of total cell extracts of HeLa cells after 48 h of transfection with the indicated siRNAs (20 nM) using an anti-PHD2 antibody. (B) PHD2 is upregulated by hypoxia. Western blot showing the hypoxic induction of PHD2. Immunoblotting was performed using total cellular extracts of HeLa cells incubated in normoxia (N; 20% O2) or hypoxia (1–2% O2) for different periods of time. (C) Hypoxic induction of PHD2 is HIF-1 dependent. Western blot of total cell extracts of HeLa cells transfected in either the absence or presence of a human HIF-1α siRNA. Immunoblotting was performed with antibodies against HIF-1α and PHD2.
Discussion
In recent years, a number of remarkable advances have shed intense light on the stagnating field of hypoxia signalling, leading to the emergence of a universal model for oxygen sensing. Post-translational modification by prolyl hydroxylation was found to target HIF-α subunits to the pVHL ubiquitylation complex, leading to rapid proteasomal degradation (Ivan et al., 2001; Jaakkola et al., 2001). The relevant enzyme(s), predicted to belong to the 2-oxoglutarate-dependent dioxygenase superfamily, with dioxygene as a co-substrate, were expected to be the ‘oxygen sensor(s)’. This prediction was elegantly validated soon after (Epstein et al., 2001). Ratcliffe and co-workers first identified in C.elegans a single dioxygenase, Egl-9, that regulates HIF-1α in vivo. Invalidation of the lone egl-9 gene in C.elegans and Drosophila melanogaster led to HIF-α stabilization in vivo (Bruick and McKnight, 2001; Epstein et al., 2001). In mammals, however, three orthologues of the unique Egl-9 gene product, PHD1, PHD2 and PHD3, were characterized and found to hydroxylate the key proline residues of the HIF-α subunit in vitro (Bruick and McKnight, 2001; Epstein et al., 2001). Here, we addressed the intriguing question of the multiplicity of these isoforms relative to HIF-1α instability in vivo. We showed by silencing specifically each PHD isoform that only PHD2 (EGLN1) controls the steady-state levels of HIF-1α in HeLa cells and in all human cells examined. Therefore, we conclude that, under these normoxic conditions, PHD1 and PHD3 do not contribute to hydroxylation of HIF-1α in vivo. Re-oxygenation of hypoxia-stressed cells was unable to reveal any implication of PHD1 or PHD3 in the degradation of HIF-1α.
This is an important point which underscores a profound physiological difference between the three PHD isoforms. However, since RNA interference (RNAi), in contrast to the traditional knockout approach, does not completely eliminate the protein of interest, it was critical to evaluate the efficiency of siRNAs at the protein level. Figure 1S (Supplementary data) has nicely revealed that both PHD1 and PHD3 siRNA were fully competent to knock-down the transfected recombinant protein. In fact, PHD2, was the most ‘resistant’ PHD protein to siRNA-induced ablation. The critical role of PHD2 in controlling HIF-1α levels was demonstrated further by examination of the dose response of PHD2 siRNA on the levels of endogenous PHD2 protein (Supplementary figure 6S). PHD2 protein levels inversely correlate with the levels of HIF-1α (compare Figure 2B with Supplementary figure 6S). Interestingly, a slight reduction of the PHD2 protein, attained by 0.2 nM PHD2 siRNA, is sufficient to impact on HIF-1α stability. We therefore conclude that in contrast to PHD1 and PHD3, PHD2 (EGLN1) is a rate-limiting enzyme setting the low normoxic levels of HIF-1α. Three recent studies provide some strength to this conclusion. First, Kaelin and co-workers, identified PHD2 (EGLN1) as the mammalian PHD by biochemical purification of the PHD activity present in rabbit reticulocyte lysate (Ivan et al., 2002). Secondly, Lee and co-workers, comparing the three human PHD isoforms, reported that in vitro, PHD2 has the highest specific activity toward the primary hydroxylation site of HIF-1α (Huang et al., 2002). Thirdly, and perhaps most importantly, Taylor showed, by phylogenetic analysis and domain organization, that PHD2 (EGLN1) represents the ancestral form of the phd (egln) gene family (Taylor, 2001). Indeed the MYND-type zinc finger found in the N-terminal domain of Egl-9 is only conserved in PHD2 (EGLN1). This result indicates that PHD2/Egl-9, the ancestral form of the enzyme, ensures HIF-1α instability in C.elegans, D.melanogaster and mammals (this study). We could speculate that by gene duplication and divergence, phd2/egl-9 has generated the isoforms phd1 and phd3 to fulfil close but distinct physiological functions in vertebrates.
So which could be the role and the substrates of these close PHD isoforms? The LXXLAP motif targeted by these PHDs is not unique to HIF-1α. It is also found in the two other members of the HIF-α family (HIF-2α and HIF-3α), as well as in other unrelated proteins such as the large subunit of RNA polymerase II (Kuznetsova et al., 2003). However, we cannot exclude the possibility that PHD1 and PHD3 could, in some tissues or under appropriate physiological conditions, target and regulate HIF-1α levels as well. Indeed, during the investigation of long-term maintenance of siRNA action (5–6 days), we uncovered an unexpected role for PHD1 (Supplementary figure 7S). In cells permanently exposed to PHD2 siRNA, leading to constitutive expression of HIF-1α in normoxia, HIF-1α levels progressively declined to low levels in spite of a total depletion of PHD2 protein (compare lanes 2 and 5, Supplementary figure 7S). Under these conditions only, co-addition of PHD1 siRNA re-established the high HIF-1α levels (compare lanes 5 and 8, Supplementary figure 7S). Our interpretation is that PHD1 has been ‘activated’ during this long-term PHD2 depletion, presumably by an HIF-1-dependent mechanism. This preliminary new finding suggests an interplay between PHD2 and PHD1 and announces a more complex and fine tuning mechanism in HIF-1α regulation.
The in vitro studies into the sequence determinants for hydroxylation of HIF-1α by the three enzymes PHD1, PHD2 and PHD3 have revealed a high degree of tolerance within the LXXLAP motif (Huang et al., 2002). As has been shown for protein kinases and their substrates, we agree with Lee and co-workers that docking and/or recognition sites distant from the hydroxyl acceptor residue and from the catalytic site of the hydroxylase are likely to be found to govern in vivo substrate specificity within the PHD family members (Huang et al., 2002). In this regard, we anticipate that forced expression of each PHD in cells, a reverse approach to that of RNAi-mediated knockdown, will reveal that every PHD has the potential to downregulate HIF-1α in vivo. These findings have already started to appear in the literature as for PHD3 expressed in 293 cells (Bruick and McKnight, 2001), in HeLa cells (Cioffi et al., 2003) or even more dramatic, overexpression of PH4, a more distant dioxygenase (Oehme et al., 2002). Results from forced expression of enzymes that often ‘violate’ in vivo rules that govern specificity (alterations in cellular compartmentation, in stoïchiometry of the complexes) have to be interpreted with extreme caution.
Two additional findings were obtained from the present study. First, we showed that PHD2 is an HIF-1 regulated gene product that fulfils the criteria of the putative HPTF, the existence of which we previously postulated (Berra et al., 2001a). In a similar manner to p53, which controls its own protein level via the expression of the E3-ligase MDM2, HIF-1α governs its own stability by controlling the expression of PHD2. Besides the auto-control by the pO2, PHD2 could also be a target for growth factors and/or oncogenic transformation. In this context, it is interesting that insulin, insulin-like growth factor 1 (IGF-1), platelet-derived growth factor (PDGF) or vasoactive hormones such as angiotensin II and thrombin, in vascular smooth muscle cells, or Ras V12, v-Src or PTEN, have been reported to increase HIF-1α in normoxia (Jiang et al., 1997; Zelzer et al., 1998; Richard et al., 2000; Zundel et al., 2000). In this context, it has been reported that normoxic induction of HIF-1α by oncogenes RasV12 and v-Src appears to be mediated by inhibition of prolyl hydroxylation on residue Pro564, while Akt-induced stabilization appears to be independent of the prolyl hydroxylation event (Chan et al., 2002). It would be of interest to investigate whether these physiological settings change the expression and/or the intrinsic activity of PHD2.
The other finding that emerged from silencing PHD2 in normoxic cells is that not only was HIF-1α stabilized but it was also translocated into the nucleus and was found to be transcriptionally active (as measured by an HRE- dependent luciferase reporter gene). HIF-1α contains two transactivation domains called N-TAD (N-terminal transactivation domain) and C-TAD (C-terminal transactivation domain). A recent study showed that the C-TAD is inactive in normoxia due to hydroxylation of a specific asparagine residue via the dioxygenase FIH-1 (factor inhibiting HIF) (Hewitson et al., 2002; Lando et al., 2002a,b; McNeill et al., 2002). We thus conclude that the N-TAD is active in normoxia. This result is consistent with a previous report from our group in which we showed that a HIF-1α splice variant that retains the N-TAD, but lacks the C-TAD, is still able to activate transcription (Gothié et al., 2000). Transcriptional activation via the N-TAD has also been invoked to account for the finding that inactivation of pVHL leads to robust activation of HIF-1 target genes despite the presence of oxygen. Increased luciferase activity by PHD2 silencing (14-fold over the basal level) was however lower than that attained in hypoxia or following incubation with Co2+ (∼20-fold induction). We think that this difference in transcriptional efficiency reflects the contribution of the C-TAD. Using the same reporter system used in this study, we are currently testing whether co-silencing of PHD2 and FIH-1 will recapitulate, in normoxia, the full transcriptional activity revealed by hypoxic stress.
Finally, another point worth discussion is the apparent additive effect of hypoxia on PHD2-silenced cells in HIF-1α expression (Figure 5). We have clearly excluded the contribution of PHD1 and PHD3 in this process; however, three other possibilities can be envisaged: (i) although PHD2 mRNA and the 50 kDa protein appear to be lacking, we cannot exclude a residual low amount of PHD2; (ii) the contribution of the acetyl transferase ARD1, involved in HIF-1α stability, whose expression is downregulated by hypoxia (Jeong et al., 2002); or (iii) the rate of HIF-1α synthesis is increased in hypoxia-stressed cells. The third possibility is favoured by the presence of the IRES motif within the HIF-1α 5′-untranslated region (Lang et al., 2002). Whatever the issue, it is remarkable that in cells lacking PHD2 only, HIF-1α remains fully ‘immune’ to destruction for the first 10 min following re-oxygenation.
This study has highlighted a specific role for PHD2, apparently not shared by the two other isoforms, PHD1 and PHD3. Considering that the three PHDs appear to be expressed in all tissues, although at different levels (Lieb et al., 2002; Cioffi et al., 2003), one of the next challenges will be to identify the in vivo substrates, the function(s), fine tuning and interplay of these three PHD isoforms in various physiological settings. As far as new PHD substrates are concerned, it is interesting to see that rat SM-20, the homologue of PHD3, is targeted via a specialized N-terminal motif to the mitochondria (Lipscomb et al., 2001). Knowing the central role played by mitochondria in oxygen metabolism and cellular induced apoptosis, novel key substrates in oxygen signalling are likely to be discovered.
Materials and methods
siRNA preparation
The 21-nucleotide RNAs were chemically synthesized by Eurogentec. The two sets of siRNAs targeting PHD1 (GenBank accession No. XP_040482) correspond to the coding region 538–558 and 835–855 relative to the start codon. siRNA sequences targeting PHD2 (accession No. AAG33965) correspond to regions 885–905 and 1250–1270. PHD3 (accession No. NP_071356) siRNAs were from positions 351–371 and 389–409. The siRNA sequences targeting human HIF-1α (accession No. U22431) and SIMA (accession No. U43090), used as an irrelevant control (D-HIF), correspond to the coding regions 1691–1711 and 2558–2578, respectively, relative to the start codon. pVHL (accession No. NM 000551) siRNA corresponds to positions 609–629 of the coding region. Annealing of siRNAs was performed as previously described by Tuschl and co-workers (Elbashir et al., 2001). For sequences of the corresponding siRNAs, see Supplementary data.
Cell culture
HeLa, CAL27, CAL51, HaCAT and HT29 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 7.5% fetal bovine serum (FBS), and antibiotics (penicillin G, 50 U/ml; and streptomycin, 50 µg/ml) (Gibco-BRL). RCC4/pVHL cells were grown in DMEM/Ham’s F10 (1:1) medium supplemented with 10% FBS and antibiotics. HUVECs were grown in human endothelial SFM medium supplemented with fibroblast growth factor-2 (FGF-2; 10 ng/ml), epidermal growth factor (EGF; 10 ng/ml), heparin (1 µg/ml), 20% FBS and antibiotics.
Hypoxic conditions were produced by incubation of cells in a sealed ‘Bug-Box’ anaerobic workstation (Ruskin Technologies, Jouan). The oxygen in this workstation was maintained at 1–2%, with the residual gas mixture being 93–94%, and 5% carbon dioxide.
Transient transfection and luciferase assay
Transfections were carried out using the calcium phosphate precipitation method (Chen and Okayama, 1987). Cells were transfected with siRNAs twice at 24 h intervals (except for the experiment described in Supplementary figure 7S) and were analysed on the day following the last transfection.
For luciferase assays, HeLa cells were transfected with siRNAs (20 nM), 50 ng of the reporter vector (pRE-Δtk-LUC) containing three copies of the HRE from the erythropoietin gene, and 100 ng of a plasmid for β-galactosidase. pRE-Δtk-LUC has been generated by deletion of the –105/–80 region within the tk promoter of the original reporter vector pRE-tk-LUC, generously provided by Dr S.L.McKnight (Tian et al., 1997). As a control for HIF-1 activation, cells were incubated in the presence of Co2+ (200 µM) or in hypoxia (1–2% O2) for 16 h. At 48 h after transfection, cells were lysed, and luciferase and β-galactosidase activity was measured as previously described (Richard et al., 1999). Results were quantified with a MicroBeta TRILUX luminescence counter (Wallac) and normalized values were expressed as the fold induction over control cells.
Northern blotting
Subconfluent cells were lysed, and RNA was purified using the RNeasy Mini Kit (Qiagen). A 20 µg aliquot of total mRNA was resolved on agarose/formaldehyde gel, and transferred to Hybond N+ nylon membrane (Pharmacia Biosciences). Membranes were hybridized with the indicated 32P-labelled cDNA probe, and results were visualized using a phosphorimager (Storm 840; Amersham Biosciences). cDNA probes were obtained by RT–PCR using the primers allowing the amplification of nucleotides 298–563, 3800–4438 and 327–1048, corresponding to PHD1 (accession No. XP_040482), PHD2 (accession No. AAG33965) and PHD3 (accession No. NP_071356), respectively. 36B4 (accession No. M178885) was used to normalize for mRNA loading.
Antibodies
Anti-HIF-1α (antiserum 2087) and p42MAPK (antiserum E1B4) were produced and characterized in our laboratory (Brondello et al., 1997; Richard et al., 1999). Anti-PHD2 (antiserum 804) was raised in rabbits immunized against the last 16 amino acids of the C-terminal end of human PHD2 (accession No. AAG33965). The anti-haemagglutinin (HA) epitope monoclonal antibody HA.11 (clone 16B12) was from BAbCO.
Immunofluorescence microscopy
Cells grown on glass coverslips were fixed with paraformaldehyde and processed using the anti-HIF-1α antibody as previously described (Berra et al., 2001b). Immunofluorescence was then analysed with a Leica DM-R microscope equipped with a DC-100 digital camera.
Western blotting
Subconfluent cells were lysed in Laemmli buffer. The protein concentration was determined using the Lowry assay and 50 µg of whole-cell extracts were resolved by SDS–PAGE (7.5% for HIF-1α detection, and 12% in the case of PHD2 and HA-epitope tagged PHDs) and electrophoretically transferred onto a PVDF membrane (Millipore). Immunoreactive bands were visualized with the ECL system (Amersham Biosciences).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Professor W.Kaelin for the generous gift of the HA-tagged PHD expression plasmids, and Drs R.Balloti, L.Gagnoux and G.Ponzio for kindly providing WM9, keratinocytes and FHN, respectively. CAL27 and CAL51 cell lines were a gift from C.Mazeau. We thank Dr C.Brahimi-Horn for carefully reading the manuscript. Financial support was from the Centre National de la Recherche Scientifique (CNRS), Ministère de l’Education, de la Recherche et de la Technologie, Ligue Nationale Contre le Cancer (Equipe labellisée) and the GIP HMR (contract No. 1/9743B-A3). E.B. is a recipient of a fellowship from ‘Centre Hospitalier Universitaire de Nice’.
References
- Berra E., Richard,D.E., Gothié,E. and Pouysségur,J. (2001a) HIF-1-dependent transcriptional activity is required for oxygen-mediated HIF-1α degradation. FEBS Lett., 491, 85–90. [DOI] [PubMed] [Google Scholar]
- Berra E., Roux,D., Richard,D.E. and Pouysségur,J. (2001b) Hypoxia-inducible factor-1α (HIF-1α) escapes O2-driven proteasomal degradation irrespective of its subcellular localization: nucleus or cytoplasm. EMBO Rep., 2, 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondello J.M., Brunet,A., Pouysségur,J. and McKenzie,F.R. (1997) The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J. Biol. Chem., 272, 1368–1376. [DOI] [PubMed] [Google Scholar]
- Bruick R.K. and McKnight,S.L. (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science, 294, 1337–1340. [DOI] [PubMed] [Google Scholar]
- Chan D.A., Sutphin,P.D., Denko,N.C. and Giaccia,A.J. (2002) Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1α. J. Biol. Chem., 277, 40112–40117. [DOI] [PubMed] [Google Scholar]
- Chen C. and Okayama,H. (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol., 7, 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi C.L., Qin Liu,X., Kosinski,P.A., Garay,M. and Bowen,B.R. (2003) Differential regulation of HIF-1α prolyl-4-hydroxylase genes by hypoxia in human cardiovascular cells. Biochem. Biophys. Res. Commun., 303, 947–953. [DOI] [PubMed] [Google Scholar]
- Cockman M.E. et al. (2000) Hypoxia inducible factor-α binding and ubiquitylation by the von Hippel–Lindau tumor suppressor protein. J. Biol. Chem., 275, 25733–25741. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Ema M., Taya,S., Yokotani,N., Sogawa,K., Matsuda,Y. and Fujii-Kuriyama,Y. (1997) A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl Acad. Sci. USA, 94, 4273–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein A.C. et al. (2001) C.elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell, 107, 43–54. [DOI] [PubMed] [Google Scholar]
- Gioanni J., Le Francois,D., Zanghellini,E., Mazeau,C., Ettore,F., Lambert,J.C., Schneider,M. and Dutrillaux,B. (1990) Establishment and characterisation of a new tumorigenic cell line with a normal karyotype derived from a human breast adenocarcinoma. Br. J. Cancer, 62, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothié E., Richard,D.E., Berra,E., Pagès,G. and Pouysségur,J. (2000) Identification of alternative spliced variants of human hypoxia-inducible factor-1α. J. Biol. Chem., 275, 6922–6927. [DOI] [PubMed] [Google Scholar]
- Gu Y.Z., Moran,S.M., Hogenesch,J.B., Wartman,L. and Bradfield,C.A. (1998) Molecular characterization and chromosomal localization of a third α-class hypoxia inducible factor subunit, HIF3α. Gene Expr., 7, 205–213. [PMC free article] [PubMed] [Google Scholar]
- Hewitson K.S. et al (2002) Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem., 277, 26351–267355. [DOI] [PubMed] [Google Scholar]
- Hoffman E.C., Reyes,H., Chu,F.F., Sander,F., Conley,L.H., Brooks,B.A. and Hankinson,O. (1991) Cloning of a factor required for activity of the Ah (dioxin) receptor. Science, 252, 954–958. [DOI] [PubMed] [Google Scholar]
- Huang J., Zhao,Q., Mooney,S.M. and Lee,F.S. (2002) Sequence determinants in hypoxia inducible factor-1α for hydroxylation by the prolyl hydroxylases PHD1, PHD2 and PHD3. J. Biol. Chem., 277, 39792–39800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.E., Arany,Z., Livingston,D.M. and Bunn,H.F. (1996) Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its α subunit. J. Biol. Chem., 271, 32253–32259. [DOI] [PubMed] [Google Scholar]
- Huang L.E., Gu,J., Schau,M. and Bunn,H.F. (1998) Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin–proteasome pathway. Proc. Natl Acad. Sci. USA, 95, 7987–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M. et al. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science, 292, 464–468. [DOI] [PubMed] [Google Scholar]
- Ivan M. et al. (2002) Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl Acad. Sci. USA, 99, 13459–13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P. et al. (2001) Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science, 292, 468–472. [DOI] [PubMed] [Google Scholar]
- Jeong J.W. et al. (2002) Regulation and destabilization of HIF-1α by ARD1-mediated acetylation. Cell, 111, 709–720. [DOI] [PubMed] [Google Scholar]
- Jiang B.H., Agani,F., Passaniti,A. and Semenza,G.L. (1997) V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumor progression. Cancer Res., 57, 5328–5335. [PubMed] [Google Scholar]
- Kaelin W.G. Jr and Maher,E.R. (1998) The VHL tumour-suppressor gene paradigm. Trends Genet., 14, 423–426. [DOI] [PubMed] [Google Scholar]
- Kallio P.J., Wilson,W.J., O’Brien,S., Makino,Y. and Poellinger,L. (1999) Regulation of the hypoxia-inducible transcription factor 1α by the ubiquitin–proteasome pathway. J. Biol. Chem., 274, 6519–6525. [DOI] [PubMed] [Google Scholar]
- Kamura T. et al. (1999) Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science, 284, 657–661. [DOI] [PubMed] [Google Scholar]
- Kamura T., Sato,S., Iwai,K., Czyzyk-Krzeska,M., Conaway,R.C. and Conaway,J.W. (2000) Activation of HIF1α ubiquitination by a reconstituted von Hippel–Lindau (VHL) tumor suppressor complex. Proc. Natl Acad. Sci. USA, 97, 10430–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A.V., Meller,J., Schnell,P.O., Nash,J.A., Ignacak,M.L., Sanchez,Y., Conaway,J.W., Conaway,R.C. and Czyzyk-Krzeska,M.F. (2003) von Hippel–Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc. Natl Acad. Sci. USA, 100, 2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D., Peet,D.J., Gorman,J.J., Whelan,D.A., Whitelaw,M.L. and Bruick,R.K. (2002a) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev., 16, 1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D., Peet,D.J., Whelan,D.A., Gorman,J.J. and Whitelaw,M.L. (2002b) Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science, 295, 858–861. [DOI] [PubMed] [Google Scholar]
- Lang K.J., Kappel,A. and Goodall,G.J. (2002) Hypoxia-inducible factor-1α mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell, 13, 1792–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb M.E., Menzies,K., Moschella,M.C., Ni,R. and Taubman,M.B. (2002) Mammalian EGLN genes have distinct patterns of mRNA expression and regulation. Biochem. Cell Biol., 80, 421–426. [DOI] [PubMed] [Google Scholar]
- Lipscomb E.A., Sarmiere,P.D. and Freeman,R.S. (2001) SM-20 is a novel mitochondrial protein that causes caspase-dependent cell death in nerve growth factor-dependent neurons. J. Biol. Chem., 276, 5085–5092. [DOI] [PubMed] [Google Scholar]
- Lisztwan J., Imbert,G., Wirbelauer,C., Gstaiger,M. and Krek,W. (1999) The von Hippel–Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev., 13, 1822–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P.H. et al. (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature, 399, 271–275. [DOI] [PubMed] [Google Scholar]
- McNeill L.A., Hewitson,K.S., Claridge,T.D., Seibel,J.F., Horsfall,L.E. and Schofield,C.J. (2002) Hypoxia-inducible factor asparaginyl hydroxylase (FIH-1) catalyses hydroxylation at the β-carbon of asparagine-803. Biochem. J., 367, 571–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehme F., Ellinghaus,P., Kolkhof,P., Smith,T.J., Ramakrishnan,S., Hutter,J., Schramm,M. and Flamme,I. (2002) Overexpression of PH-4, a novel putative proline 4-hydroxylase, modulates activity of hypoxia-inducible transcription factors. Biochem. Biophys. Res. Commun., 296, 343–349. [DOI] [PubMed] [Google Scholar]
- Ohh M., Park,C.W., Ivan,M., Hoffman,M.A., Kim,T.Y., Huang,L.E., Pavletich,N., Chau,V. and Kaelin,W.G. (2000) Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel–Lindau protein. Nat. Cell Biol., 2, 423–427. [DOI] [PubMed] [Google Scholar]
- Richard D.E., Berra,E., Gothié,E., Roux,D. and Pouysségur,J. (1999) p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1α (HIF-1α) and enhance the transcriptional activity of HIF-1. J. Biol. Chem., 274, 32631–32637. [DOI] [PubMed] [Google Scholar]
- Richard D.E., Berra,E. and Pouysségur,J. (2000) Nonhypoxic pathway mediates the induction of hypoxia-inducible factor 1α in vascular smooth muscle cells. J. Biol. Chem., 275, 26765–71. [DOI] [PubMed] [Google Scholar]
- Salceda S. and Caro,J. (1997) Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin–proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem., 272, 22642–22647. [DOI] [PubMed] [Google Scholar]
- Semenza G.L. (1998) Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr. Opin. Genet. Dev., 8, 588–594. [DOI] [PubMed] [Google Scholar]
- Stebbins C.E., Kaelin,W.G.,Jr and Pavletich,N.P. (1999) Structure of the VHL–elonginC–elonginB complex: implications for VHL tumor suppressor function. Science, 284, 455–461. [DOI] [PubMed] [Google Scholar]
- Taylor M.S. (2001) Characterization and comparative analysis of the EGLN gene family. Gene, 275, 125–132. [DOI] [PubMed] [Google Scholar]
- Tian H., McKnight,S.L. and Russell,D.W. (1997) Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev., 11, 72–82. [DOI] [PubMed] [Google Scholar]
- Wang G.L., Jiang,B.H., Rue,E.A. and Semenza,G.L. (1995) Hypoxia-inducible factor 1 is a basic-helix–loop–helix–PAS heterodimer regulated by cellular O2 tension. Proc. Natl Acad. Sci. USA, 92, 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer E., Levy,Y., Kahana,C., Shilo,B.Z., Rubinstein,M. and Cohen,B. (1998) Insulin induces transcription of target genes through the hypoxia-inducible factor HIF-1α/ARNT. EMBO J., 17, 5085–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zundel W. et al. (2000) Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev., 14, 391–396. [PMC free article] [PubMed] [Google Scholar]