Abstract
Post-transcriptional synthesis of 2′-O-methylated nucleotides and pseudouridines in Sm spliceosomal small nuclear RNAs takes place in the nucleoplasmic Cajal bodies and it is directed by guide RNAs (scaRNAs) that are structurally and functionally indistinguishable from small nucleolar RNAs (snoRNAs) directing rRNA modification in the nucleolus. The scaRNAs are synthesized in the nucleoplasm and specifically targeted to Cajal bodies. Here, mutational analysis of the human U85 box C/D-H/ACA scaRNA, followed by in situ localization, demonstrates that box H/ACA scaRNAs share a common Cajal body-specific localization signal, the CAB box. Two copies of the evolutionarily conserved CAB consensus (UGAG) are located in the terminal loops of the 5′ and 3′ hairpins of the box H/ACA domains of mammalian, Drosophila and plant scaRNAs. Upon alteration of the CAB boxes, mutant scaRNAs accumulate in the nucleolus. In turn, authentic snoRNAs can be targeted into Cajal bodies by addition of exogenous CAB box motifs. Our results indicate that scaRNAs represent an ancient group of small nuclear RNAs which are localized to Cajal bodies by an evolutionarily conserved mechanism.
Keywords: box C/D RNA/box H/ACA RNA/Cajal body/RNA localization/scaRNA
Introduction
In eukaryotes, most aspects of the nuclear biogenesis of cellular RNAs can be linked to distinct subnuclear domains, also called nuclear organelles (Lamond and Earnshaw, 1998; Misteli and Spector, 1998; Lewis and Tollervey, 2000). The structural and functional compartmentalization of the nucleus requires specific targeting of many RNAs to their correct destination. Therefore, intranuclear RNA transport plays a fundamental role in eukaryotic gene expression. However, in contrast to the extensively studied nucleo-cytoplasmic RNA transport, little is known about the principles governing RNA localization within the nucleus. In this study, we investigate the mechanism directing specific localization of small Cajal body (CB)-specific RNAs (scaRNAs) to the nucleoplasmic CBs.
CBs are evolutionarily conserved nucleoplasmic organelles that contain many protein and ribonucleoprotein (RNP) factors involved in mRNA and rRNA biogenesis (Bohmann et al., 1995; Matera, 1999; Gall, 2000; Dundr and Misteli, 2001; Ogg and Lamond, 2002). The list of proteins and RNPs detected in CBs includes RNA polymerase I, II and III subunits, basal transcription factors, small nuclear RNPs (snRNPs) mediating mRNA splicing and small nucleolar RNPs (snoRNPs) required for rRNA modification and nucleolytic processing. CBs, however, do not contain nascent mRNAs and rRNAs, indicating that neither synthesis nor processing of these RNAs occur in this organelle. Instead, the available data are most consistent with the idea that the major function of CBs is in maturation, assembly and/or trafficking of snRNPs, snoRNPs and transcription complexes which later function in mRNA and rRNA biogenesis in other nuclear compartments (Gall et al., 1999; Sleeman and Lamond, 1999; Gall, 2001; Carmo-Fonseca, 2002; Ogg and Lamond, 2002; Verheggen et al., 2002; Jády et al., 2003; Stanek et al., 2003).
We recently have demonstrated that an important step of maturation of spliceosomal snRNPs, namely post-transcriptional modification of small nuclear RNAs (snRNAs), takes place in CBs (Jády et al., 2003). We found that site-specific synthesis of 2′-O-methylated nucleotides and pseudouridines in the RNA polymerase II-synthesized U1, U2, U4 and U5 Sm snRNAs is directed by guide RNAs that specifically localize to CBs (Kiss, 2001, 2002; Darzacq et al., 2002; Kiss et al., 2002). Intriguingly, the scaRNAs are structurally and functionally indistinguishable from small nucleolar RNAs (snoRNAs) that direct 2′-O-methylation and pseudouridylation of rRNAs within the nucleolus (Tollervey and Kiss, 1997; Weinstein and Steitz, 1999; Bachellerie et al., 2000, 2002; Kiss, 2001; Filipowicz and Pogacic, 2002; Kiss et al., 2002; Terns and Terns, 2002). The 2′-O-methylation guide scaRNAs and snoRNAs feature the conserved box C (RUGAUGA) and D (CUGA) elements located close to their 5′ and 3′ ends, respectively, and also internal copies of these elements, called C′ and D′ boxes. The 5′ and 3′ termini of 2′-O-methylation guide RNAs form a common terminal core motif that encompasses the C and D boxes. The pseudouridylation guide snoRNAs and scaRNAs fold into a common ‘hairpin–hinge–hairpin–tail’ secondary structure. The single-stranded hinge and tail regions carry the conserved box H (AnAnnA) and ACA elements which, together with the basal stem of the 3′ hairpin, form the minimal core structure of box H/ACA RNAs. Both 2′-O-methylation and psedouridylation guide RNAs select the substrate nucleotides via formation of transient base pairing interactions with their target RNAs (Kiss, 2001; Bachellerie et al., 2002; Filipowicz and Pogacic, 2002; Terns and Terns, 2002; Decatur and Fournier, 2003).
The box C/D and H/ACA core motifs, through directing binding of two distinct sets of RNP proteins, play a crucial role in both accumulation and function of snoRNAs and scaRNAs. The box C/D snoRNAs are associated with fibrillarin, Nop58p, Nop56p and 15.5 kDa snoRNP proteins, while the box H/ACA snoRNAs are complexed with dyskerin, Nhp2p, Gar1p and Nop10p (reviewed in Filipowicz and Pogacic, 2002; Terns and Terns, 2002; Decatur and Fournier, 2003). Amongst the snoRNP proteins, fibrillarin and dyskerin are the catalysts of the 2′-O-methylation and pseudouridylation reactions, respectively. The box C/D and H/ACA scaRNPs are believed to contain all of the RNP proteins known to be associated with box C/D and H/ACA snoRNAs, although this assumption has been confirmed experimentally only for fibrillarin and Gar1p (Darzacq et al., 2002).
Previous works proved that snoRNAs represent an excellent model system to obtain insights into the molecular mechanism underlying intranuclear RNA targeting. It has been demonstrated that the common box C/D and H/ACA core motifs, and probably the associated snoRNP proteins, are the key determinants of the nucleolar localization of snoRNAs (Lange et al., 1998, 1999; Samarsky et al., 1998; Narayanan et al., 1999a,b). Since scaRNAs also contain the box C/D or H/ACA core motif or, most frequently, both of them (Darzacq et al., 2002), they would be predicted to accumulate in the nucleolus. In this study, we demonstrate that box H/ACA scaRNAs share a common, evolutionarily conserved sequence motif that is responsible for targeting these RNAs into CBs. We show that addition of an exogenous CB localization signal is sufficient to target snoRNAs specifically into CBs and that authentic scaRNAs can be directed into the nucleolus by alteration of their CB localization elements.
Results
ScaRNAs are evolutionarily conserved and specific components of CBs
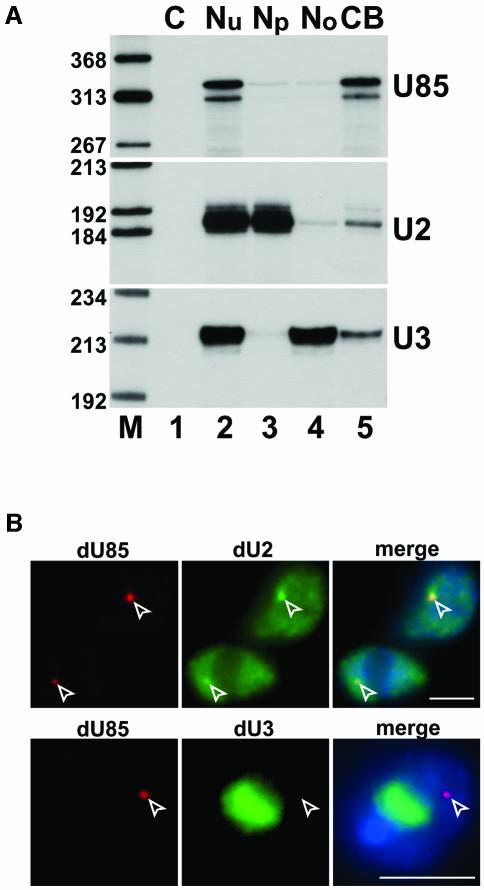
Although previous fluorescent in situ hybridization (FISH) experiments failed to detect scaRNAs outside CBs (Darzacq et al., 2002; Kiss et al., 2002; Jády et al., 2003), we cannot exclude the possibility that a portion, or even the majority of scaRNAs are dispersed in the nucleoplasm, as has been demonstrated for p80-coilin, the commonly used marker protein of CBs (Andrade et al., 1991). To test this possibility, we performed cell fractionation experiments, taking advantage of the recently reported CB purification protocol of the Lamond laboratory (Lam et al., 2002). Nuclei of human HeLa cells were disrupted and fractionated into nucleolar, nucleoplasmic and CB fractions by differential centrifugation (see Materials and methods). RNA was extracted from each fraction and the distribution of the U85 scaRNA and, as a control, the nucleoplasmic U2 spliceosomal snRNA and the nucleolar U3 snoRNA was determined by RNase A/T1 protection analysis (Figure 1A). We found that the U85 scaRNA co-purified with the CB fraction (lane 5), and it was hardly detectable in the nucleoplasmic (lane 3) and nucleolar (lane 4) fractions. As expected, the U2 snRNA and U3 snoRNA accumulated predominantly in the nucleoplasm and nucleolus, respectively. These results indicate that U85 and perhaps all scaRNAs are tightly associated with CBs and, in contrast to the other known components of CBs, are not present in other nuclear compartments.
Fig. 1. U85 scaRNA is a specific molecular marker for CBs in both human and Drosophila cells. (A) Human U85 scaRNA co-purifies with CBs. Nuclei (Nu) of human HeLa cells were subfractionated into nucleoplasmic (Np), nucleolar (No) and CB fractions (Lam et al., 2002). RNA extracted from each fraction corresponding to 5 × 106 cells was mapped by RNase A/T1 protection by using an excess of sequence-specific antisense RNA probes as indicated on the right. Protected RNAs were separated on a 6% sequencing gel. Lane C, control mapping with E.coli tRNA. Lane M, size markers in nucleotides (HaeIII- and TaqI-digested pBR322). (B) In situ localization of Drosophila U85 scaRNA. Drosophila Schneider II cells were hybridized with fluorescent oligonucleotide probes specific for the U85 scaRNA (dU85), U2 snRNA (dU2) and U3 snoRNA (dU3). CB-like structures are indicated by open arrowheads. Nuclei were visualized by DAPI staining. Scale bar, 5 µm.
Thus far, scaRNAs have been demonstrated to exist only in mammalian cells (Darzacq et al., 2002; Kiss et al., 2002). Since CBs or CB-like structures have been found in many species, including amphibians, insects and plants (Gall, 2000), scaRNAs may not be confined to mammalian cells. We previously have identified a functional homologue of the human U85 scaRNA in Drosophila melanogaster (Jády and Kiss, 2001). Here, to determine the intracellular localization of Drosophila U85 (dU85), we used in situ fluorescent microscopy (Figure 1B). Hybridization of Drosophila Schneider II cells with a dU85-specific fluorescent oligonucleotide probe showed that dU85 concentrates within one or, less frequently, two discrete dot-like nucleoplasmic domains. Probing the same cells with a fluorescent oligonucleotide complementary to the Drosophila U3 snoRNA demonstrated that the U85-containing domain is located outside the nucleolus of Schneider cells. In contrast, in situ localization of the U2 spliceosomal snRNA (dU2) showed that regions accumulating dU85 also concentrate U2. Since spliceosomal snRNAs are known to be enriched in CBs (Bohmann et al., 1995; Matera, 1999), we conclude that the nucleoplasmic loci accumulating U85 represent Drosophila CBs (Yannoni and White, 1997). Based upon the above results, we propose that scaRNAs represent an evolutionarily conserved group of snRNAs and they are the most specific molecular markers for CBs identified so far.
The box H/ACA domain is required for localization of the human U85 scaRNA to CBs
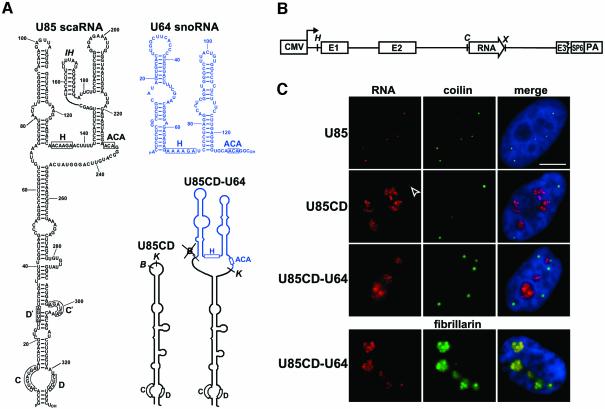
The box C/D and H/ACA core motifs have been demonstrated to be essential and sufficient to direct nucleolar accumulation of snoRNAs (Lange et al., 1998, 1999; Samarsky et al., 1998; Narayanan et al., 1999a,b). Thus, we assumed that scaRNAs might possess additional localization element(s) necessary to override the nucleolar localization function of the box C/D and H/ACA motifs. To define this putative cis-acting CB localization signal, we performed a mutational analysis of the human U85 scaRNA that is composed of box C/D and H/ACA domains (Figure 2A). Altered versions of the intron-encoded U85 RNA were transiently expressed in human HeLa cells by using the pCMV/globin expression vector (Figure 2B) (Jády and Kiss, 2001; Darzacq et al., 2002). Accumulation and correct processing of each RNA was confirmed by RNase A/T1 mapping (data not shown), and intracellular localization of the accumulating RNA was determined by in situ fluorescent microscopy. CBs were stained with an antibody directed against p80-coilin, and green fluorescent protein (GFP)-tagged fibrillarin was expressed to mark both nucleoli and CBs (Lapeyre et al., 1990; Aris and Blobel, 1991; Jansen et al., 1991).
Fig. 2. The box H/ACA domain is essential for CB-specific localization of human U85 C/D-H/ACA scaRNA. (A) Computer-predicted two- dimensional structures of the human U85 scaRNA (black) and U64 box H/ACA snoRNA (blue) and schematic structures of the box C/D domain of U85 (U85CD) and the U85CD–U64 chimeric RNAs. Positions of the conserved C, D, H and ACA box elements and the relevant restriction sites are indicated (B, BglII; K, KpnI). (B) Schematic structure of the pCMV/globin expression cassette. The test RNA genes (open arrow) were inserted into the second intron of the human β-globin gene that had been placed under the control of the cytomegalovirus promoter (CMV). The exons (E1, E2 and E3) and the polyadenylation site (PA) of the globin gene as well as the SP6 promoter used to transcribe antisense RNA probes are indicated. Relevant restriction sites are shown (H, HindIII; C, ClaI; X, XhoI). (C) In situ localization of transiently expressed human U85 scaRNA, the box C/D domain of U85 (U85CD) and the U85CD–U64 composite RNA in HeLa cells. The RNAs were detected by sequence-specific fluorescent oligonucleotide probes (see Materials and methods). CBs were detected by an anti-coilin antibody and nucleoli were visualized by co-expression of GFP-tagged fibrillarin (fibrillarin). Nuclei were visualized by DAPI staining. A CB accumulating U85CD is indicated by an open arrow. Scale bar, 10 µm.
Because box C/D snoRNAs transit through CBs before accumulating in the nucleolus (Narayanan et al., 1999b; Verheggen et al., 2002), we first tested whether the box C/D domain of U85 alone could localize to CBs. Upon removal of the box H/ACA domain of U85, the remaining U85CD RNA (Figure 2A) was expressed efficiently and correctly in transfected HeLa cells (data not shown). However, in contrast to the wild-type U85 scaRNA that accumulated exclusively in CBs, the truncated U85CD RNA localized mainly to nucleoli, although it was also detectable in CBs (Figure 2C). Since authentic box C/D snoRNAs show a similar intranuclear distribution (Samarsky et al., 1998; Narayanan et al., 1999b; Verheggen et al., 2002), we conclude that U85CD can be considered as a canonical box C/D snoRNA. Unfortunately, the H/ACA domain of U85 alone lacks metabolic stability (Jády and Kiss, 2001). Therefore, we could not assess whether this domain of U85 could localize to CBs. However, when the H/ACA domain of U85 was replaced with the U64 box H/ACA snoRNA, the resulting U85CD–U64 composite RNA (Figure 2A) co-accumulated with GFP–fibrillarin in the nucleoli (Figure 2C). These results demonstrate that the box H/ACA domain plays an essential role in the CB-specific localization of the human U85 scaRNA.
Identification of a CB localization element in the box H/ACA domain of U85
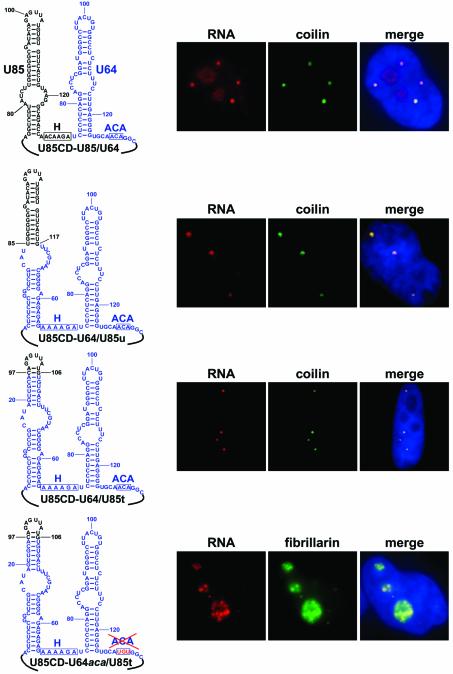
To define the putative CB localization element(s) in the box H/ACA domain of U85, we tried to restore the CB-specific accumulation of the U85CD–U64 artificial composite snoRNA by adding back various segments of the box H/ACA domain of the U85 scaRNA (Figure 3). First, the 5′ hairpin and the H box of U85 were fused to the 3′-terminal half of U64 to produce U85CD–U85/U64. Upon transient expression in HeLa cells, the U85CD–U85/U64 chimeric RNA accumulated mainly in CBs, although it was also detected in nucleoli (see below). Replacement of the upper part (U85CD–U64/U85u) or the terminal stem–loop (U85CD–U64/U85t) of the 5′ hairpin of U64 with the corresponding regions of U85 resulted in chimeric RNAs that accumulated exclusively in CBs. This demonstrates that the terminal stem–loop region (C97–G106) of the 5′ hairpin of U85 contains signal element(s) that are sufficient to support CB-specific accumulation of U85CD–U64.
Fig. 3. Intranuclear localization of transiently expressed composite U85–U64 RNAs in HeLa cells. Structures of the artificial box H/ACA domains composed of U85- (black) and U64-specific (blue) sequences are shown. The sequence and structure of the common box C/D domain (U85CD) of the expressed chimeric RNAs is shown in Figure 2A. Localization of RNAs was determined by in situ hybridization with sequence-specific fluorescent oligonucleotide probes complementary to the 5′ (U85CD–U64/U85u, U85CD–U64/U85t, U85CD–U64aca/U85t) and 3′ (U85CD–U85/U64) junction sequences of U85 and U64. For other details, see the legend to Figure 2C.
Appropriate structural arrangement of the box H/ACA core motif is critical for efficient assembly of box H/ACA snoRNPs (Ganot et al., 1997; Bortolin et al., 1999). The observation that the U85CD–U85/U64 chimeric RNA, in contrast to U85CD–U64/U85u and U85CD–U64/U85t, failed to localize fully to CBs suggested that assembly of box H/ACA RNP might be important for the function of the CB-specific localization signal of U85. We hypothesized that the suboptimal structure of the chimeric H/ACA core of U85CD–U85/U64 might not support efficient binding of box H/ACA proteins, and molecules not associated with H/ACA proteins are probably targeted into the nucleolus by the box C/D domain of the RNA. To test this hypothesis, we altered the ACA motif of the U85CD–U64/U85t RNA (Figure 3). Consistent with expectations (Balakin et al., 1996; Ganot et al., 1997; Jády and Kiss, 2001), immunoprecipitation with an antibody directed against the Gar1 snoRNP protein demonstrated that the expressed U85CD–U64aca/U85t RNA failed to associate with box H/ACA RNP proteins (data not shown). FISH showed that U85CD–U64aca/U85t, contrary to the presence of the CB-specific localization signal of U85, behaved as a canonical box C/D snoRNA: it co-localized with GFP-tagged fibrillarin to the nucleolus and CBs. Similar results were obtained when the H box of U85CD–U64/U85t was altered (data not shown), demonstrating that assembly of box H/ACA RNP is required for the efficient function of the CB-specific localization signal of U85.
Targeting of U64 box H/ACA snoRNA to CBs
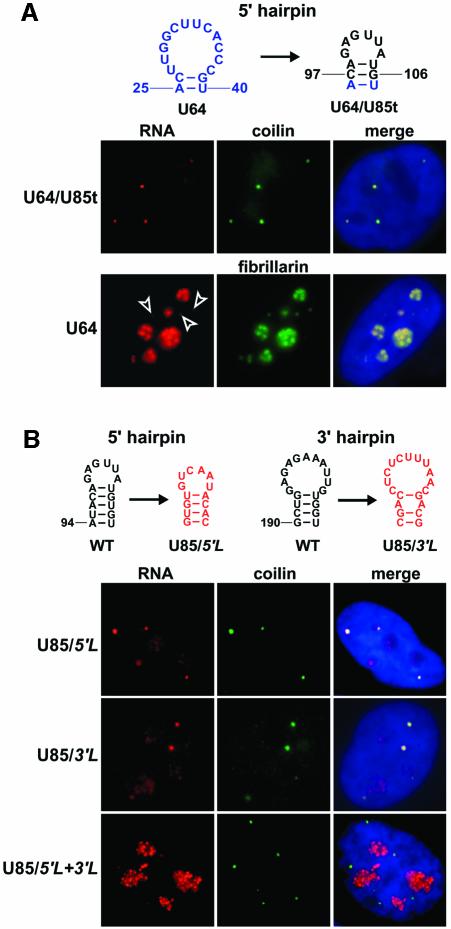
The experiments described above demonstrate that the C97–G106 terminal stem–loop region of U85, in concert with the box H/ACA core motif of U64, can localize the U85CD–U64/U85t chimeric RNA to CBs (Figure 3). Since the U85CD–U64/U85t RNA contains the box C/D domain of U85, we cannot exclude the possibility that the box C/D core motif or other element(s) located in this region of U85 may also contribute to the CB-specific localization of U85CD–U64/U85t. This possibility is especially appealing when one considers that box C/D snoRNAs pass through CBs before reaching nucleoli (Samarsky et al., 1998; Narayanan et al., 1999b; Verheggen et al., 2002). Therefore, we tested whether fusion of the C97–G106 terminal stem–loop of U85 to the U64 box H/ACA snoRNA can alter the intracellular localization of the resulting U64/U85t RNA (Figure 4A). In situ hybridization demonstrated that the transiently expressed U64/U85t RNA co-localized with p80-coilin-containing CBs. This result shows that no box C/D domain is required for the correct function of the CB localization signal of U85 and that addition of an external CB localization element can direct box H/ACA snoRNAs into CBs. It is also noteworthy that in a control experiment, overexpressed wild-type U64 snoRNA was also detected within CBs, raising the intriguing possibility that box H/ACA snoRNAs may also transit through CBs (see Discussion).
Fig. 4. The terminal stem–loop structures of the H/ACA domain of U85 carry elements essential for localization to CBs. (A) Targeting of human U64 snoRNA into CBs by addition of the terminal stem–loop of the 5′ hairpin of U85. Transiently expressed chimeric U64/U85t and wild-type U64 RNAs were localized by in situ hybridization. (B) Alteration of the terminal stem–loop sequences in the 5′ and/or 3′ hairpin of the box H/ACA domain of human U85. The intranuclear distribution of mutant U85 RNAs overexpressed in HeLa cells was determined by in situ hybridization. For other details, see the legend to Figure 2C.
U85 carries two CB-specific localization elements
Next, we wanted to confirm that the C97–G106 stem–loop region indeed functions in the CB-specific localization of the wild-type U85 RNA. Nucleotides A94–U109 encompassing the alleged CB localization element of U85 were altered, and localization of the transiently expressed U85/5′L RNA was determined by in situ hybridization (Figure 4B). To our surprise, the mutant U85/5′L RNA, apart from a small fraction that localized to the nucleoli, showed a predominant CB-specific accumulation. We speculated that U85 might carry another CB localization signal that could compensate abolition of the localization element in the 5′ hairpin of the RNA. In fact, the appearance of U85/5′L in the nucleolus indicated that the C97–G106 terminal stem–loop contributes to the CB-specific localization of U85. The terminal stem–loop region of the 3′ hairpin seemed to be the most likely region to accommodate a second CB localization signal in the box H/ACA domain of U85. Therefore, we changed the sequence of this stem–loop region (G190–U208) of U85 (Figure 4B). In situ hybridization revealed that the expressed mutant U85/3′L RNA, like U85/5′L, accumulated in both CBs and nucleoli, indicating that the terminal stem–loop of the 3′ hairpin also contributes to localization of U85 to CBs. Finally, when the 5′ and 3′ hairpin mutations were combined, the double mutant U85/5′L+3′L RNA showed a predominant nucleolar localization. We conclude that the human U85 scaRNA carries two CB localization elements located in the terminal regions of the 5′ and 3′ hairpins of its box H/ACA domain.
CAB box, a common sequence motif for box H/ACA scaRNAs
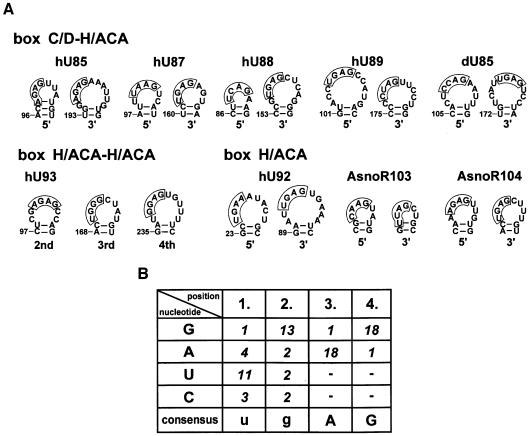
In the hope of identifying a common sequence or structural motif responsible for targeting scaRNAs into CBs, we have collected the sequences of the 5′- and 3′-terminal stem–loop regions of the available box H/ACA scaRNAs (Figure 5A). Thus far, nine authentic or potential box H/ACA scaRNAs have been reported from human, Drosophila and Arabidopsis. Accumulation of the human (h) U85, U87, U88, U89, U92 and U93 (Darzacq et al., 2002; Kiss et al., 2002) and Drosophila (d) U85 (this study) RNAs in CBs has been demonstrated already. The Arabidopsis (A) snoR103 and snoR104 RNAs, since they are predicted to direct pseudouridylation of the U5 and U2 spliceosomal snRNAs (Marker et al., 2002), may represent the first scaRNAs identified in plants. Inspection of the compiled terminal stem–loop sequences revealed the presence of a common sequence motif that showed significant evolutionary conservation. This putative CB localization element, called the CAB (Cajal body) box, can be described by the ugAG consensus in human, Drosophila and Arabidopsis scaRNAs (Figure 5A and B). Deviations from the ugAG consensus sequence are found frequently in the first and second positions of the CAB boxes. In contrast, the A and G residues in the third and fourth positions show a striking conservation. So, we propose that the terminal loops of the 5′ and 3′ hairpins share a common sequence element, the CAB box, that probably determines the CB-specific accumulation of box H/ACA scaRNAs.
Fig. 5. Identification of a putative CB localization signal for box H/ACA scaRNAs. (A) Compilation of the terminal stem–loop regions of the 5′ and 3′ hairpins of box H/ACA scaRNAs. Sequences that show significant conservation are boxed. (B) Nucleotide conservation in the proposed cis-acting CB localization elements of box H/ACA scaRNAs.
The CAB box is essential for localization of scaRNAs to CBs
We have demonstrated above that the C97–G106 terminal stem–loop of U85, when fused to the U64 (Figure 4A) or the U85CD–U64 (Figure 3) snoRNA, is capable of directing these, otherwise nucleolar RNAs to CBs. To test the functional importance of the 98-AGAG-101 putative CAB box motif, we performed a systematic mutational analysis of the C97–G106 stem–loop of U85 in the U85CD–U64/U85t composite RNA (see Figure 3). Selection of U85CD–U64/U85t, rather than U64/U85t, as a test RNA was justified by the fact that all mutant versions of U85CD–U64/U85t could be readily distinguished from the endogenous U85 and U64 RNAs by utilization of a single fluorescent oligonucleotide probe complementary to the U85–U64 junction sequences.
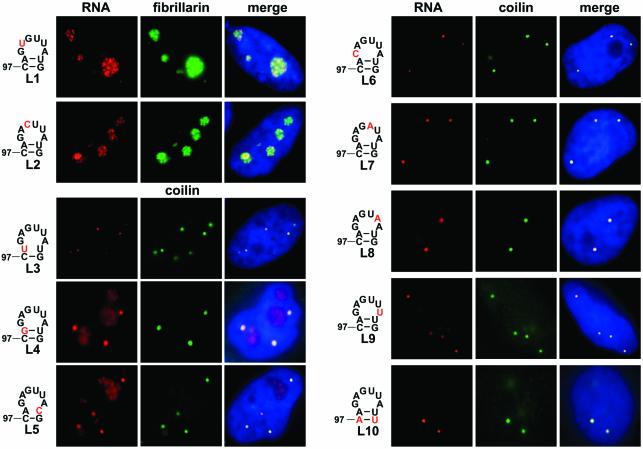
First, we tested the functional importance of the strongly conserved A100 and G101 residues in the last two positions of the putative CAB box of U85CD–U64/U85t (Figure 6). Upon replacement of these nucleotides with a U (A100) or C (G101) residue, the resulting L1 and L2 derivatives of U85CD–U64/U85t, as demonstrated by co-expression of GFP–fibrillarin, accumulated in the nucleolus and were hardly detectable in CBs. These results support the notion that the conserved A3 and G4 residues in the CAB box motif are fundamental to the localization of box H/ACA scaRNAs to CBs.
Fig. 6. Mutational analysis of the minimal stem–loop region of U85 directing localization of the U85CD–U64/U85t chimeric RNA to CBs. Altered nucleotides in the terminal stem–loop region of the U85CD–U64/U85t chimeric RNA are indicated. Subnuclear localization of transiently expressed mutant RNAs was determined by in situ hybridization with a fluorescent oligonucleotide probe complementary to the 5′ junction of U85 and U64 sequences. For other details, see the legend to Figure 2C.
As expected, an A to U transversion in the first position of the CAB box of U85CD–U64/U85t, that actually restored the ugAG consensus sequence, did not influence the CB-specific localization of L3 RNA. However, the L4 derivative of U85CD–U64/U85t that carried a G residue in the first position of its CAB box motif, although accumulated mainly in CBs, was also detectable in the nucleolus. Interestingly, disruption of the A98–U105 base pair provoked a partial nucleolar accumulation of the mutant L5 RNA, contrary to the fact that the CAB box motif (AGAG) of this RNA remained unchanged. This observation suggests that the structural organization of the terminal loop carrying the CAB box motif is also important for the efficient localization of scaRNAs to CBs. Substitution of G99 for a C (mutant L6) in the second position of the CAB box had no detectable effect on the intranuclear distribution of U85CD–U64/U85t. Likewise, alteration of the non-conserved U102 (L7), A103 (L8) or A104 (L9) residues in the loop failed to change the CB-specific accumulation of U85CD–U64/U85t. Finally, replacement of the C97–G106 base pair by an A–U pair in the L10 RNA did not alter the CB-specific accumulation of U85CD–U64/U85t, indicating that the base composition of the terminal helix has no influence on the function of the CB localization signal of scaRNAs. Taken together, our results demonstrate that the conserved CAB box element plays a fundamental role in the CB-specific accumulation of scaRNAs. In the ugAG consensus, the conserved A3 and G4 residues are indispensable, while the U1 and G2 residues in the first two positions seem to be less important for targeting scaRNAs to CBs.
Discussion
In eukaryotes, post-transcriptional modification of rRNAs and snRNAs is directed by a vast array of box C/D 2′-O-methylation and box H/ACA pseudouridylation guide RNAs that specifically accumulate and function in either the nucleolus (snoRNAs) or the nucleoplasmic CBs (scaRNAs). Since both snoRNAs and scaRNAs are synthesized in the nucleoplasm, specific transport mechanisms are required to target these RNAs to the nucleolus or CBs. Previous studies by several laboratories demonstrated that the common box C/D or box H/ACA core motifs are responsible for the nucleolar accumulation of snoRNAs (Lange et al., 1998, 1999; Samarsky et al., 1998; Narayanan et al., 1999a,b). In this work, we have investigated the determinants of the CB-specific accumulation of human scaRNAs.
We have demonstrated that the CB-specific localization of box H/ACA scaRNAs, which represent the majority of scaRNAs identified so far (Darzacq et al., 2002), is directed by a newly discovered cis-acting sequence element, called the CAB box. Each box H/ACA scaRNA carries two copies of the CAB box consensus (ugAG) that are located in the terminal loops of their 5′ and 3′ hairpins (Figure 5A). Mutational analysis of the human U85 scaRNA has revealed that both CAB boxes contribute to the efficient CB-specific localization of scaRNAs. Mutant U85 RNAs lacking either the 5′ or 3′ CAB box motif can only partially localize to CBs (Figure 4B). The same results were obtained upon mutational analysis of the 5′ and 3′ CAB box elements of the human U87 scaRNA (our unpublished data), leading to the conclusion that the 5′ and 3′ CAB boxes of scaRNAs function in an additive manner. In contrast to the C, D, H and ACA boxes, the CAB boxes are not required for RNA expression: scaRNAs with diminished CAB boxes are metabolically stable, but they accumulate in the nucleolus, as do canonical snoRNAs. The CAB boxes therefore function exclusively as CB localization elements. A previous localization study demonstrated that the CB-specific accumulation of U7 snRNA is directed by its unusual Sm core motif that is also essential for the accumulation and function of U7 (Wu et al., 1996).
The finding that CAB boxes support CB-specific accumulation only in concert with the box H/ACA core motif of scaRNAs (Figure 3) suggests that the putative protein factor that probably binds to the CAB box (tentatively called CAB protein) may also interact with box H/ACA core proteins. Earlier, electron microscopy revealed a symmetric bipartite structure for box H/ACA snoRNPs and, based on the predicted molecular mass of the snoRNP particles, suggested the presence of two copies of each of the Gar1, Nop10, Nhp2 and dyskerin (Cbf5) core proteins (Watkins et al., 1998). Our finding that box H/ACA scaRNAs possess two CB localization signals that probably bind two copies of the putative CAB protein further emphasizes the bipartite structural organization of box H/ACA RNPs. In this context, it was somewhat unexpected that the 5′ CAB box of the U85 scaRNA, when fused to the U64 authentic and the U85CD–U64 artificial snoRNAs, could alone support efficient accumulation of these RNAs in CBs (Figures 3 and 4A). However, closer inspection of the terminal loop of the 3′ hairpin of the U64 snoRNA revealed the presence of a putative cryptic CAB box motif (101-UGUG-104) (Figure 2A). It seems conceivable that in the U64/U85t RNP, binding of one copy of the CAB protein to the 5′ CAB box originating from the U85 scaRNA might facilitate recruitment of another copy of the CAB protein by the cryptic 3′ CAB box of U64, for example, by protein–protein interaction.
It has been observed that box C/D snoRNAs transiently localize to CBs before being transported to the nucleolus (Samarsky et al., 1998; Narayanan et al., 1999b). Most probably, nucleolytic processing of mature termini and assembly into snoRNP particles take place in CBs (Verheggen et al., 2002). Notably, in this study, we found that the overexpressed U64 box H/ACA snoRNA also appears in CBs of transiently transfected HeLa cells (Figure 4A). Consistent with our data, earlier microinjection experiments showed that the human telomerase RNA that has a box H/ACA snoRNA domain (Mitchell et al., 1999) travels through CBs before accumulating in the nucleoli of Xenopus oocytes (Lukowiak et al., 2001). Hence, both classes of snoRNAs might pass through CBs to undergo maturation. This notion raises the intriguing possibility that the CAB box motif of scaRNAs in fact could be considered as a CB retention element rather than a CB targeting signal. According to this scenario, the putative CAB box protein would be likely to bind to scaRNAs only within CBs and would tightly anchor the mature scaRNPs to CB structures.
Nucleotides in the ugAG consensus CAB box sequence are unequally conserved (Figure 5). Mutational analysis demonstrated that the A3 and G4 residues in the last two positions of the CAB box consensus, consistent with their strong conservation, are indispensable for CB localization (Figure 6). In contrast, the loosely conserved U1 and G2 residues seem to be less important. Some of our observations suggest that not only the nucleotide composition, but also the three-dimensional structure of the CAB box motif and the entire loop region contribute to the efficient CB-specific localization of scaRNAs. Disruption of the base pairing ability of the first nucleotide in the CAB box of U85 by changing its complementary base altered the CB localization capacity of the mutant U85CD–U64/U85L5 RNA (Figure 6). Similar results were obtained when more than one nucleotide was altered simultaneously in the non-conserved U102–A104 loop region of U85 (our unpublished data). Understanding the optimal structural conformation of the ‘CAB box loop’ of scaRNAs that support efficient CAB protein binding requires further mutational and structural analysis of scaRNAs.
Sequence motifs identical to or highly reminiscence of the CAB box consensus of human scaRNAs are also present in the terminal loops of the 5′ and 3′ hairpins of the Drosophila U85 and Arabidopsis AsnoR103 and AsnoR104 box H/ACA RNAs (Figure 5A). Indeed, in situ hybridization demonstrated that the Drosophila U85 RNA specifically accumulates in CB-like structures in the nucleoplasm of Schneider cells (Figure 1B). Thus, we propose that scaRNAs are not confined to mammals, but represent an evolutionarily conserved group of snRNAs. Although the functional importance of the alleged CAB boxes of the Drosophila U85 and Arabidopsis RNAs has not yet been confirmed, the strong sequence conservation of these elements suggests that the putative CAB box-binding protein(s), similar to the box C/D and H/ACA core proteins, might also be conserved during evolution. In the future, identification of scaRNAs in phylogenetically distant species might help us to understand the evolutionary origin of scaRNAs. According to the most obvious scenario, scaRNAs probably evolved from snoRNAs by acquiring CB-specific localization signals.
Most known components of the CB localize only transiently or partially to this dynamic organelle (Matera, 1998). This applies even to the autoantigen p80-coilin that has long been used as a marker protein for CBs, contrary to the fact that most of the cellular coilin is dispersed in the nucleoplasm (Andrade et al., 1991; Raska et al., 1991). Our results demonstrate that scaRNAs represent highly specific molecular markers for CBs, that can unambiguously indicate CB structures even in those organisms where a homologue of p80-coilin has not yet been identified. Therefore, future identification of scaRNAs from lower eukaryotes may also lead to identification of CBs or CB-related structures which, due to a shortage of appropriate molecular markers, remain hidden thus far.
Finally, we have to mention that although the great majority of scaRNAs feature a box H/ACA domain and possess putative CAB boxes, two human scaRNAs, the U90 and U91 box C/D RNAs, lack H and ACA boxes and are not associated with box H/ACA snoRNPs (Darzacq et al., 2002). The elements responsible for the CB-specific accumulation of these scaRNAs remain totally unknown. This raises the intriguing possibility that beside the CAB box-supported localization mechanism of box H/ACA scaRNAs, there might exist other molecular devices to sequester specifically modification guide RNAs into CBs.
Materials and methods
Plasmid constructs
Unless otherwise stated, standard manipulations of Escherichia coli, nucleic acids and oligodeoxynucleotides were performed as described (Sambrook et al., 1989). The identity of all constructs was verified by sequence analysis. Construction of the pCMV/globin expression vector has been described (Darzacq et al., 2002). To obtain pCMV/globin/U85, the coding region of human U85 scaRNA together with upstream and downstream flanking sequences of 64 and 52 nucleotides, repectively, was PCR amplified with oligonucleotide primers 1 (ATAATCGATGGAAGGTGTTTGTTATC) and 2 (ATACTCGAGTTTCACTCACTTCTTTC) by using human genomic DNA as a template. The amplified fragment was digested with ClaI and XhoI and inserted into the same sites of pCMV/globin. The same approach was used to generate pCMV/globin/U64, except that the coding region of human U64 was amplified with oligonucleotides 3 (TATATCGATACTCTCTCGGCTCTGCATAG) and 4 (TTACTCGAGGCCTGTTGCACCCCTCAAGG). To generate pCMV/globin/U85CD, the 5′- and 3′-terminal portions of U85 were amplified with oligonucleotides 1 and 5 (ATAAGATCTTTAACAGGCCAAAGGTCCCT) or 2 and 6 (TAAAGATCTATAGGTACCGGGCATGTTCAGGGTATC), respectively. After digestion with BglII, the two fragments were ligated together, digested with ClaI and XhoI and inserted into the same sites of pCMV/globin. To obtain pCMV/globin/U85CD–U64, the coding region of U64 was amplified with oligonucleotides 7 (ATAGGATCCACTCTCTCGGCTCTGCAT) and 8 (ATAGGTACCGCCTGTTGCACCCCTCAAGG), digested with BamHI and KpnI and inserted into the BglII and KpnI sites of pCMV/globin/U85CD. To construct pCMV/globin/U85CD–U85/U64, the 5′ half of U85 was amplified with oligonucleotides 1 and 9 (ATAAGATCTTGTTGGTCTGCCCTTAC). The amplified fragment was digested with ClaI and BglII and used for replacement of the ClaI–BglII fragment of pCMV/globin/U85CD–U64.
Construction of recombinant pCMV/globin vectors expressing U85CD–U64/U85u, U85CD–U64/U85t, U85/5′L, U85/3′L, U85/5′L+3′L and U85CD–U64/U85L1 to U85CD–U64/U85L10 RNAs was performed by the megaprimer amplification approach (Datta, 1995). In the first amplification reaction, oligonucleotide 1 or 2 was used together with an appropriately designed mutagenic internal oligonucleotide (available upon request) to amplify the 5′ (oligonucleotide 1) or 3′ (oligonucleotide 2) part of the desired mutant RNA gene. The resulting products were used as megaprimers in the second amplification step in combination with oligonucleotide 1 or 2. Finally, the amplified products were cut with ClaI and XhoI and inserted into the same sites of pCMV/globin. To generate pCMV/globin/U64/U85t, the coding region of U64/U85t was amplified with oligonucleotides 3 and 4 using pCMV/globin/U85CD–U64/U85t as a template. The amplified DNA was digested with ClaI and XhoI and cloned into the same sites of pCMV/globin. To construct pCMV/globin/U85CD–U64aca/U85t, the coding region of U64/U85t was amplified with primer 7 and a mutagenic 3′ end-specific oligonucleotide (ATAGGTACCGCCACATGCACCCCTCAAGG). The resulting DNA was digested with BamHI and KpnI and inserted into the BglII and KpnI sites of pCMV/globin/U85CD. Construction of pFibrillarin–GFP has been reported (Dundr et al., 2000). Transfection of human HeLa cells was performed with Fugene 6 (Roche) transfection reagent as recommended by the manufacturer.
Cell fractionation and RNA analysis
Nuclei of human HeLa cells were isolated as described by Tyc and Steitz (1989). Subfractionation of nuclei into nucleoplasmic, nucleolar and CB fractions was performed according to the protocol of the Lamond laboratory (Lam et al., 2002). Briefly, nuclei were suspended in 0.35 M sucrose and 0.5 mM MgCl2 and disrupted by ultrasonication. After addition of sucrose to 1 M final concentration, nucleoli were pelleted by centrifugation for 10 min at 3000 g. The supernatant was supplemented with 15.4% Percoll and 1% Triton X-100 and centrifuged in an SW41 rotor at 170 000 g for 2 h at 4°C. The upper part of the Percoll gradient was removed and used as the nucleoplasmic fraction; the last 1 ml at the bottom of the tube, containing a loose pellet, was considered as a crude CB fraction. RNA isolation from the nuclear and nucleolar fractions of HeLa cells was performed by the guanidine thiocyanate–phenol–chloroform extraction procedure (Goodall et al., 1990). RNA from the nucleoplasmic and CB fractions of HeLa cells was obtained by phenol–chloroform extraction after a 2-fold dilution with TE pH 8.0. After adding carrier glycogen at 50 µg/ml final concentration, RNA was collected by ethanol precipitation. RNase A/T1 protection analysis was performed according to Goodall et al. (1990). Synthesis of internally labelled antisense RNA probes complementary to the human U85, U2 and U3 RNAs has been described (Jády and Kiss, 2001). Before utilization, each probe was purified on a 6% sequencing gel.
In situ hybridization
Synthesis and chemical conjugation of amino-modified oligonucleotides with FluoroLink Cy3 monofunctional dye (Amersham), FISH of human HeLa cells and image acquisition and processing were performed according to the protocols of the laboratory of Dr R.Singer (http://singerlab.aecom.yu.edu; Darzacq et al., 2002). The following oligonucleotide probes were used to detect endogenous or transiently expressed RNAs: dU85, AT*GACGTCGTCACCAT*GACAAACAGCTTAGACCTAACT*A; dU2, CT*ACACTTTGATCTTAGCCAT*AAGGCCGAGAAGCT*A; dU3, AT*AGAACGATTCAACT*CATC AACGCGGGT*GCACTTGGCTTTCT*A; hU85, CT*TAGCCAAACCAACT*GAATCACAACAGCCT*TGATATCATCATGT*G; U85CD, CT*GAACATGCCCGGTACCT*ATAGATCTTT*AACAGGCCAAAT*G; U85CD–U64, U85CD–U64/U85u, U85CD–U64/U85t, U85CD–U64aca/U85t and U85CD–U64/U85L1 to U85CD–U64/U85L10, AT*GCAGAGCCGAGAGAGT*GGATCTTT*AACAGGCCAAAT*G; U85CD–U85/U64, AT*CCTGAACAT*GCCCGGTACCGCCT*GTTGCACCCCT*C; U85/5′L and U85/5′L+3′L, TT*ACAGTGAACAGTGT*ATTGACACACT*CGCCCACCAAGAT*T; U85/3′L, CT*CACTC CAGTTCGT*CGTTAAAGAGAGGT*CGAACTGGAAGAAT*C; U64, TT*ACGAAAGTCACACGGGT*GAAGCCAAGTGCAACT*A; and U64/U85t, TT*ACGAAAGTCACACAT*AACTCTGTGCAACTAT*G. Amino-allyl-modified T residues are indicated by asterisks. p80-coilin was detected with polyclonal rabbit anti-coilin antibody (kindly provided by Dr A.Lamond) as described earlier (Darzacq et al., 2002). Nuclear DNA was stained with 0.1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI).
Acknowledgments
Acknowledgements
We are grateful to Y.de Préval for synthesis of oligonucleotides. X.D. was funded by la Fondation pour la Recherche Médicale. B.E.J. is supported by a long-term EMBO fellowship. Our work was supported by grants from la Ligue Nationale contre le Cancer, Association pour la Recherche contre le Cancer and the French MNRT (ACI).
References
- Andrade L.E., Chan,E.K., Raska,I., Peebles,C.L., Roos,G. and Tan,E.M. (1991) Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med., 173, 1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris J.P. and Blobel,G. (1991) cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl Acad. Sci. USA, 88, 931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J.P., Cavaille,J. and Qu,L.H. (2000) Nucleotide modification of eukaryotic rRNAs: the world of small nucleolar RNAs revisited. In Garrett,R.A., Douthwaite,S.R., Matheson,A.T., Moure,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 191–203. [Google Scholar]
- Bachellerie J.P., Cavaille,J. and Huttenhofer,A. (2002) The expanding snoRNA world. Biochimie, 84, 775–790. [DOI] [PubMed] [Google Scholar]
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- Bohmann K., Ferreira,J., Santama,N., Weis,K. and Lamond,A.I. (1995) Molecular analysis of the coiled body. J. Cell Sci. Suppl., 19, 107–113. [DOI] [PubMed] [Google Scholar]
- Bortolin M.L., Ganot,P. and Kiss,T. (1999) Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J., 18, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M. (2002) New clues to the function of the Cajal body. EMBO Rep., 3, 726–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Jády,B.E., Verheggen,C., Kiss,A.M., Bertrand,E. and Kiss,T. (2002) Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J., 21, 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A.K. (1995) Efficient amplification using ‘megaprimer’ by asymmetric polymerase chain reaction. Nucleic Acids Res., 23, 4530–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur W.A. and Fournier,M.J. (2003) RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem., 278, 695–698. [DOI] [PubMed] [Google Scholar]
- Dundr M. and Misteli,T. (2001) Functional architecture in the cell nucleus. Biochem. J., 356, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Misteli,T. and Olson,M.O. (2000) The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol., 150, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W. and Pogacic,V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol., 14, 319–327. [DOI] [PubMed] [Google Scholar]
- Gall J.G. (2000) Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol., 16, 273–300. [DOI] [PubMed] [Google Scholar]
- Gall J.G. (2001) A role for Cajal bodies in assembly of the nuclear transcription machinery. FEBS Lett., 498, 164–167. [DOI] [PubMed] [Google Scholar]
- Gall J.G., Bellini,M., Wu,Z. and Murphy,C. (1999) Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell, 10, 4385–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- Goodall G.J., Wiebauer,K. and Filipowicz,W. (1990) Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol., 181, 148–161. [DOI] [PubMed] [Google Scholar]
- Jády B.E. and Kiss,T. (2001) A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády B.E., Darzacq,X., Tucker,K.E., Matera,A.G., Bertrand,E. and Kiss,T. (2003) Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal bodies following import from the cytoplasm. EMBO J., 22, 1878–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.P., Hurt,E.C., Kern,H., Lehtonen,H., Carmo-Fonseca,M., Lapeyre,B. and Tollervey,D. (1991) Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol., 113, 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell, 109, 145–148. [DOI] [PubMed] [Google Scholar]
- Kiss A.M., Jády,B.E., Darzacq,X., Verheggen,C., Bertrand,E. and Kiss,T. (2002) A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res., 30, 4643–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Y.W., Lyon,C.E. and Lamond,A.I. (2002) Large-scale isolation of Cajal bodies from HeLa cells. Mol. Biol. Cell, 13, 2461–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A.I. and Earnshaw,W.C. (1998) Structure and function in the nucleus. Science, 280, 547–553. [DOI] [PubMed] [Google Scholar]
- Lange T.S., Borovjagin,A., Maxwell,E.S. and Gerbi,S.A. (1998) Conserved boxes C and D are essential nucleolar localisation elements of U14 and U8 snoRNAs. EMBO J., 17, 3176–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T.S., Ezrokhi,M., Amaldi,F. and Gerbi,S.A. (1999) Box H and box ACA are nucleolar localisation elements of U17 small nucleolar RNA. Mol. Biol. Cell, 10, 3877–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B., Mariottini,P., Mathieu,C., Ferrer,P., Amaldi,F., Amalric,F. and Caizergues-Ferrer,M. (1990) Molecular cloning of Xenopus fibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol. Cell. Biol., 10, 430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D. and Tollervey,D. (2000) Like attracts like: getting RNA processing together in the nucleus. Science, 288, 1385–1389. [DOI] [PubMed] [Google Scholar]
- Lukowiak A.A., Narayanan,A., Li,Z.H., Terns,R.M. and Terns,M.P. (2001) The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. RNA, 7, 1833–1844. [PMC free article] [PubMed] [Google Scholar]
- Marker C., Zemann,A., Terhorst,T., Kiefmann,M., Kastenmayer,J.P., Green,P., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2002) Experimental RNomics. identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol., 12, 2002–2013. [DOI] [PubMed] [Google Scholar]
- Matera A.G. (1998) Of coiled bodies, gems and salmon. J. Cell. Biochem., 70, 181–192. [PubMed] [Google Scholar]
- Matera A.G. (1999) Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol., 9, 302–309. [DOI] [PubMed] [Google Scholar]
- Misteli T. and Spector,D.L. (1998) The cellular organization of gene expression. Curr. Opin. Cell Biol., 10, 323–331. [DOI] [PubMed] [Google Scholar]
- Mitchell J.R., Cheng,J. and Collins,K. (1999) A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol., 19, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A., Lukowiak,A., Jády,B.E., Dragon,F., Kiss,T., Terns,R.M. and Terns,M.P. (1999a) Nucleolar localisation signals of box H/ACA small nucleolar RNAs. EMBO J., 18, 5120–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A., Speckmann,W., Terns,R. and Terns,M.P. (1999b) Role of the box C/D motif in localisation of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell, 10, 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S.C. and Lamond,A.I. (2002) Cajal bodies and coilin—moving towards function. J. Cell Biol., 159, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I., Aandrade,L.E., Ochs,R.L., Chan,E.K., Chang,C.M., Roos,G. and Tan,E.M. (1991) Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res., 195, 27–37. [DOI] [PubMed] [Google Scholar]
- Samarsky D.A., Fournier,M.J., Singer,R.H. and Bertrand,E. (1998) The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localisation. EMBO J., 17, 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sleeman J.E. and Lamond,A.I. (1999) Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol., 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Stanek D., Rader,S.D., Klingauf,M. and Neugebauer,K.M. (2003) Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol., 160, 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M.P. and Terns,R.M. (2002) Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr., 10, 17–39. [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- Tyc K. and Steitz,J.A. (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C., Lafontaine,D.L., Samarsky,D., Mouaikel,J., Blanchard,J.M., Bordonne,R. and Bertrand,E. (2002) Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J., 21, 2736–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J., Gottschalk,A., Neubauer,G., Kastner,B., Fabrizio,P., Mann,M. and Luhrmann,R. (1998) Cbf5p, a potential pseudouridine synthase and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA, 4, 1549–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein L.B. and Steitz,J.A. (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Murphy,C. and Gall,J.G. (1996) The Sm binding site targets U7 snRNA to coiled bodies (spheres) of amphibian oocytes. RNA, 2, 811–823. [PMC free article] [PubMed] [Google Scholar]
- Yannoni Y.M. and White,K. (1997) Association of the neuron-specific RNA binding domain-containing protein ELAV with the coiled body in Drosophila neurons. Chromosoma, 105, 332–341. [DOI] [PubMed] [Google Scholar]