Abstract
The RNA polymerase II C-terminal heptad repeat domain (CTD) is essential for normal transcription and co-transcriptional processing of mRNA precursors. The mammalian CTD comprises 52 heptads whose consensus, YSPTSPS, is conserved throughout eukaryotes, followed by a 10 amino acid C-terminal sequence that is conserved only among vertebrates. Here we show that surprisingly, the heptad repeats are not sufficient to support efficient transcription, pre-mRNA processing or full cell viability. In addition to the heptads, the 10 amino acid C-terminal motif is essential for high level transcription, splicing and poly(A) site cleavage. Efficient mRNA synthesis from a transiently transfected reporter gene required the C-terminal motif plus between 16 and 25 heptad repeats from either the N- or C-terminal half of the CTD. Twenty-seven consensus heptads plus the C-terminal motif also supported efficient mRNA synthesis but not cell viability.
Keywords: CTD/polyadenylation/RNA polymerase II/splicing/transcription
Introduction
The large subunit of RNA polymerase II (pol II) is distinguished by a conserved C-terminal heptad repeat domain (CTD) that is a unique feature of this enzyme. The vertebrate CTD comprises 52 heptad repeats, with the consensus YSPTSPS, followed by a unique 10 amino acid sequence. The CTD projects from the core of the enzyme at a point next to the RNA exit channel (Cramer et al., 2001). It is reversibly phosphorylated at Ser2 and Ser5 of the heptad repeats during transcription (Dahmus, 1996; Kobor and Greenblatt, 2002). This unusual domain functions in at least two aspects of RNA synthesis that are peculiar to mRNA production: (i) the response to sequence-specific transcriptional activators and repressors (Lee and Young, 2000); and (ii) co-transcriptional processing of the transcript (Hirose and Manley, 2000; Bentley, 2002). Little is known about how different segments of the CTD contribute to these functions.
The importance of the CTD in vivo is demonstrated by the fact that deletion of this domain inhibits enhancer-dependent transcription (Gerber et al., 1995), and capping, splicing and cleavage/polyadenylation of mRNA precursors in mammalian cells (McCracken et al., 1997a, b). The CTD makes direct protein–protein contacts with the mediator complex CRSP (Naar et al., 2002), the capping enzyme guanylyltransferase (McCracken et al., 1997a; Yue et al., 1997) and the poly(A) site cleavage stimulation factor, CstF (Fong and Bentley, 2001). Interactions also occur between pol II complexes and splicing factors, but the direct protein–protein contacts involved have not yet been defined (Kim et al., 1997; Misteli and Spector, 1999; Robert et al., 2002).
Co-transcriptional capping, splicing and 3′ end processing of mRNA precursors are interdependent; however, the CTD stimulates each of these steps independently in vivo (Fong and Bentley, 2001). The CTD also enhances capping, splicing and poly(A) site cleavage in reactions uncoupled from transcription in vitro (Yuryev et al., 1996; Hirose et al., 1999; Ho and Shuman, 1999; Zeng and Berget, 2000; Ryan et al., 2002). Deletion analysis of the CTD in Saccharomyces cerevisiae suggested that the functions of this domain are provided entirely by a minimum number of heptad repeats (Nonet et al., 1987; Scafe et al., 1990). C-terminal truncation from the full-length 26 heptads down to 11 heptads has no phenotype. Further deletion to nine or 10 heptads confers cold sensitivity and reduced response to activators, and <9 heptads is lethal (Nonet et al., 1987; Scafe et al., 1990). All the in vitro effects of the CTD on mammalian mRNA processing reactions are directly attributable to the heptad repeat sequences. The guanylyltransferase activity of mammalian capping enzyme is activated by binding to a peptide with four phosphorylated consensus heptad repeats (Ho and Shuman, 1999). The poly(A) site cleavage activity of the CTD is provided by synthetic polypeptides with 26 or more consensus heptad repeats (Ryan et al., 2002), and specific inhibition of in vitro splicing is caused by a peptide with four consensus heptad repeats (Yuryev et al., 1996). Unlike the yeast CTD, the mammalian CTD has many variant heptads with substitutions at position 7, particularly in its C-terminal half (heptads 27–52), and these variants could serve specific functions in vivo. Variant heptads enhance binding of CstF p50 (Fong and Bentley, 2001) and influence phosphorylation by CTD kinases (Rickert et al., 1999).
Using deletion mutants of the pol II large subunit expressed in transfected cells, we found that the N-terminal heptads 1–15 or 1–25 support capping but not splicing or 3′ end processing. In contrast, the C-terminal half of the CTD comprising heptads 27–52 plus the C-terminal 10 residues can support efficient splicing and 3′ processing as well as capping (Fong and Bentley, 2001). It is not clear from these experiments whether the functional difference between the N- and C-terminal halves of the CTD is due to the different heptad repeat sequences in these regions of the CTD or to the presence of the 10 residue motif C-terminal of heptad 52. The sequence of the C-terminal motif, ISPDDSDEEN, is strongly conserved among mammals, but not in Drosophila melanogaster, Caenorhabditis elegans or S.cerevisiae. This motif is phosphorylated in vitro by casein kinase II (CKII) (Payne et al., 1989), but its function in mRNA synthesis has not been studied.
Here we investigated the function of the unique 10 amino acid sequence that lies C-terminal of heptad 52. Our results lead to the unexpected conclusion that the heptad repeats of the CTD are not sufficient to support efficient transcription and pre-mRNA processing. We show that the C-terminal 10 residue motif of the CTD is essential for high level transcription, splicing and 3′ processing in mammalian cells. Moreover, this motif confers efficient transcription and processing capability to pol II with only heptads 1–25 or with 27 consensus heptads.
Results
Mutation of the CTD C-terminus inhibits splicing and 3′ end processing
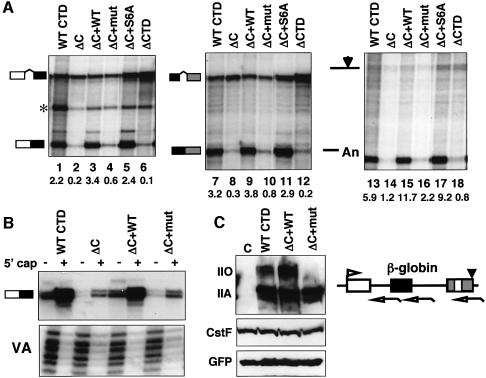
Why does the C-terminal half of the CTD support all three major mRNA processing steps whereas the N-terminal half supports only capping? To address this question, we asked whether the 10 residue C-terminal motif, ISPDDSDEEN, contributes to the functional difference between the two halves of the CTD. This motif, along with heptad 52, was deleted from the large pol II subunit (Rpb1) and the function of this mutant (Rpb1ΔC) was compared with intact Rpb1 (WT CTD) using the strategy of Gerber et al. (1995) (we were unable to make a C-terminal deletion that retained heptad 52). Cells were transiently transfected with expression vectors for α-amanitin-resistant Rpb1. After 12–16 h, α-amanitin was added to inhibit the host cell’s pol II. RNA was harvested 48 h after adding α-amanitin, at which time only polymerases that have incorporated the resistant large subunit are active. In Figure 1A, transcripts from a co-transfected β-globin reporter gene transcribed by WT or Rpb1ΔC pol II were analyzed by RNase protection to monitor splicing of introns 1 and 2 and cleavage at the poly(A) site. Surprisingly, Rpb1ΔC that retains 51 heptad repeats and lacks only 17 amino acids at the C-terminus produced transcripts that were poorly spliced and poorly cleaved at the poly(A) site relative to those made by WT pol II (Figure 1A, lanes 1, 7 and 13 versus 2, 8 and 14). The apparent splicing defect is not a result of transcript termination within introns as it was detected with probes spanning the 3′ end of intron 1 (Figure 1A) and intron 2 (see Supplementary data available at The EMBO Journal Online). The start site of transcription was unaffected by the mutants as determined by 5′ end mapping (Figure 1B). In contrast to splicing and cleavage, capping was not markedly inhibited by the C-terminal deletion (Figure 1B), consistent with previous results (Fong and Bentley, 2001). We conclude that the first 51 heptads of the CTD are not sufficient to support efficient splicing or 3′ processing in vivo.
Fig. 1. The CTD C-terminus is required for pre-mRNA processing. (A) Mutation of the C-terminus inhibits splicing and poly(A) site cleavage of β-globin transcripts. RNase protection of RNA from α-amanitin-treated 293 cells transfected with pSVβ128Rpbex5 and expression vectors for α-amanitin-resistant Rpb1 full-length (WT CTD), C-terminal deletion (ΔC), ΔC+WT Cter (ΔC+WT), ΔC+mut Cter (ΔC+mut), ΔC+S6ACter (ΔC+S6A), and CTD deletion (ΔCTD). The asterisk denotes an irrelevant undigested probe. Open arrows below the map of the globin reporter indicate RNase protection probes, and arrows above the map indicate the start site and poly(A) site. Ratios of spliced:unspliced and cleaved:uncleaved transcripts were calculated from phosphoimager data corrected for the [32P]U content of the protected fragments. (B) Capping is not significantly inhibited by mutation of the C-terminus. Total RNA was fractionated by GST–eIF4E pull-down prior to RNase protection with a 5′ end mapping probe. (C) Epitope-tagged Rpb1 with WT and mutant C-termini are expressed at approximately equal levels. Western blot of extracts from cells transfected with B10 epitope-tagged Rpb1 expression plasmid or no plasmid control (C) plus GFP control plasmid. Anti-B10 detects phosphorylated (II0) and unphosphorylated (IIA) Rpb1. Cellular CstF p77 is a loading control. Note the poor phosphorylation of Rpb1 with a mutant C-terminus (ΔC+mut).
To test whether the C-terminal 10 residue motif, ISPDDSDEEN, is necessary for mature mRNA production, we appended this sequence to the end of the Rpb1ΔC construct, making Rpb1ΔC+WT Cter. As a control for sequence specificity, a scrambled version of the same residues, NDSDSDPEIE, was also added to the C-terminus of Rpb1ΔC (ΔC+mut Cter). This experiment showed that splicing and poly(A) site cleavage of β-globin reporter transcripts were fully restored by adding back the wild-type C-terminus (Figure 1A, lanes 3, 9 and 15), whereas the scrambled C-terminus had no effect (Figure 1A, lanes 4, 10 and 16). The pol II large subunits with WT CTD, ΔC+WT Cter and ΔC+mut Cter were expressed at comparable levels as determined by western blotting with antibody against the N-terminal B10 epitope tag relative to green fluorescent protein (GFP) and CstF p77 controls (Figure 1C and Supplementary data). We conclude from the results in Figure 1 that the ISPDDSDEEN motif is the element at the C-terminus that is required for efficient splicing and 3′ processing of mRNA precursors.
CKII phosphorylation of the C-terminus is not required for mRNA processing
The C-terminal motif contains a consensus CKII site, DSDEE (Kuenzel et al., 1987), that is phosphorylated in vitro (Payne et al., 1989). We tested the importance of CKII phosphorylation at this site by mutating the Ser6 to alanine in the ISPDDSDEEN sequence and testing whether this mutant supported processing when it was fused to the C-terminus of Rpb1ΔC (ΔC+S6A). The S6A mutant was, in fact, capable of supporting wild-type levels of mRNA processing (Figure 1A, lanes 5, 11 and 17; Figure 2, lanes 3 and 7). We also tested the importance of Ser2, which is a weak potential CKII phosphorylation site. The single S2A mutant (data not shown) and the S2A,S6A double mutant both supported efficient splicing and 3′ processing (Figure 2, lanes 4 and 8). The scrambled C-terminus sequence present in Rpb1ΔC+mut Cter (NDSDSDPEIE) has no detectable ‘processing’ activity although it has potential CKII sites and is phosphorylated in HeLa nuclear extract almost as well as the wild-type C-terminal motif (data not shown). We conclude that phosphorylation of the C-terminal motif by CKII or other kinases is not essential for the mRNA processing functions we have assayed.
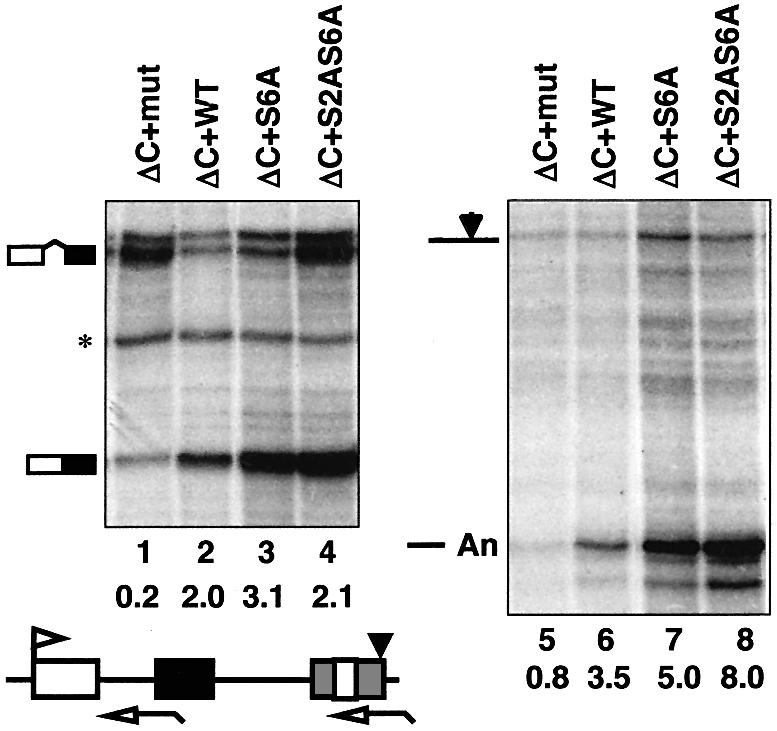
Fig. 2. Phosphorylation of the C-terminus is not essential for pre-mRNA processing. RNase protection of pSVβ128Rpbex5 transcripts from α-amanitin-treated cells expressing Rpb1 mutants ΔC+mut Cter, ΔC+WT Cter, ΔC+S6A Cter and ΔC+S2A,S6A Cter. The asterisk denotes an irrelevant undigested probe. S6A and S2A,S6A have alanine substituted for the serine residues in ISPDDSDEEN. Ratios of spliced:unspliced and cleaved:uncleaved transcripts corrected for [32P]U content are marked.
The function of the CTD C-terminal motif is conserved in Xenopus
The sequence of the CTD C-terminal motif is identical in human, mouse and Chinese hamster, but it is not present in invertebrate species. Analysis of two expressed sequence tags (ESTs; accession Nos CA971909.1 and AW635353.1) showed that the Xenopus pol II large subunit C-terminal motif, ISPDDSDEDN, differs from that in mammals by only one conservative glutamate to aspartate substitution at position 9. We reasoned that if its mechanism of action is also conserved, then the C-terminus of the CTD should also enhance RNA processing in amphibian cells. Therefore, we tested if the C-terminus of human pol II large subunit also stimulated mRNA processing when expressed in Xenopus oocytes. Expression vectors for Rpb1ΔC+WT Cter and Rpb1ΔC+mut Cter were injected into oocyte nuclei and several hours later cytomegalovirus (CMV) β-globin and VA reporter genes were injected along with α-amanitin to inhibit endogenous RNA pol II. The only functional pol II then is the hybrid that has incorporated human α-amanitin-resistant large subunit. Western blotting with anti-B10 showed equal expression of Rpb1ΔC+WT Cter and Rpb1ΔC+mut Cter (data not shown). The results in Figure 3A show that, as in human 293 cells, scrambling of the C-terminal motif inhibited splicing of β-globin intron 1 and cleavage at the poly(A) site. Similar results were obtained for intron 2 (data not shown). In summary, this experiment shows that, consistent with its sequence conservation, the RNA processing function of the CTD C-terminus is conserved between mammals and amphibians and is therefore likely to be of general significance among vertebrates.
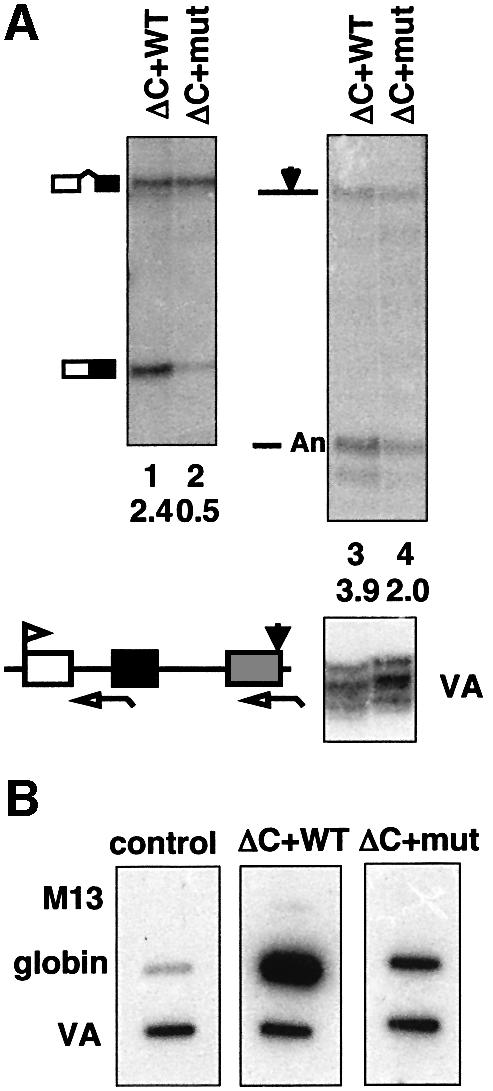
Fig. 3. (A) The mRNA processing function of the CTD C-terminus is conserved in Xenopus. RNase protection of pCMV β-globin transcripts made in α-amanitin-injected Xenopus oocytes expressing Rpb1ΔC+WT Cter or Rpb1ΔC+mut Cter. VA is a control for injection efficiency. Corrected ratios of spliced:unspliced and cleaved:uncleaved transcripts are marked below the lanes. (B) The CTD C-terminus is required for efficient transcription. Nuclear runons of 293 cells transfected with pCMV β-globin and pSPVA plus either no plasmid control, Rpb1ΔC+WT Cter or Rpb1ΔC+mut Cter. Runon transcripts made with α-amanitin were hybridized to M13 mp18 vector, M13 globin and M13 VA. Globin transcription by Rpb1ΔC+mut Cter was lower than WT but higher than the control.
The CTD C-terminus is required for efficient transcription in mammalian cells
The abundance of β-globin transcripts made by pol IIΔC or pol IIΔC+mut Cter is significantly reduced relative to WT pol II in 293 cells (Figure 1A, lanes 1, 2 and 4) due to reduced RNA stability and/or transcription rate. To determine if transcription is reduced by mutation of the C-terminal motif, runon assays were performed in nuclei from transfected cells expressing α-amanitin-resistant pol IIΔC+WT Cter or pol IIΔC+mut Cter plus CMV β-globin and VA reporter genes. Runon transcripts made in the presence of α-amanitin were hybridized to strand-specific M13 probes for β-globin and VA which is transcribed by pol III. This experiment showed that scrambling of the C-terminal sequence significantly reduced pol II transcription from the CMV promoter relative to the VA control (Figure 3B, compare panel ΔC+WT with ΔC+mut). We conclude that the C-terminal motif is required for wild-type levels of transcription from the CMV promoter in transiently transfected 293 cells as well as for efficient processing of the transcripts. Consistent with reduced transcription, the Rpb1ΔC+mut Cter polypeptide was less extensively phosphorylated on Ser5 residues in the CTD relative to Rpb1ΔC+WT Cter (Figure 1C and Supplementary data). In Xenopus oocytes, mutation of the C-terminus appears not to have a dramatic effect on transcription, at least 12–16 h after microinjection (Figure 3A, lanes 1 and 2). We do not fully understand this difference between the two cell types, but it could be related to differences in template chromatin structure. In the experiments described below, we focus on the role of the C-terminal motif in RNA processing.
The C-terminal motif is required for 3′ processing and splicing independently
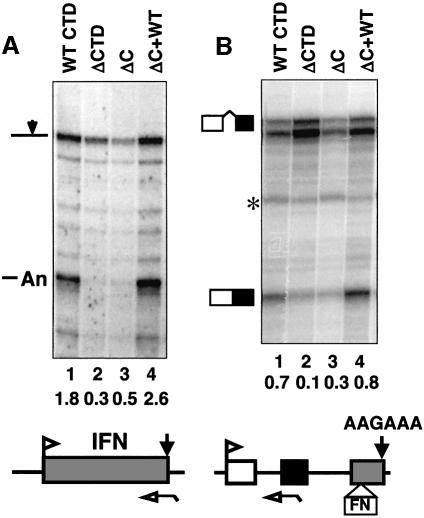
The results shown in Figures 1 and 2 demonstrate that the CTD C-terminus is required for splicing and 3′ processing, but they do not distinguish whether it is required for each of these events independently of the other. We therefore investigated the effects of the C-terminal motif on poly(A) site cleavage independent of splicing and on splicing independent of 3′ processing (Fong and Bentley, 2001). Cleavage at the poly(A) site of the intronless β-interferon (IFN) reporter gene driven by the HSV TK promoter, pIFpTK2, was assayed with full-length pol II (WT) and the C-terminal deletion (ΔC) as in Figure 1. The result in Figure 4A shows that the C-terminus is indeed essential for cleavage at this poly(A) site independently of splicing (compare Figure 4A, lanes 3 and 4). Deletion of the C-terminus reduced the ratio of cleaved:uncleaved transcripts (Figure 4A, lanes 1 and 3), and adding back the 10 residue C-terminal motif restored efficient cleavage (Figure 4A, lane 4). Adding back the scrambled C-terminus (Rpb1ΔC+mut Cter) did not restore poly(A) site cleavage (data not shown).
Fig. 4. The C-terminal motif stimulates poly(A) site cleavage and splicing independently. RNase protection of transcripts from α-amanitin-treated cells expressing Rpb1 mutants. Reporter genes: (A) intronless TK-βIFN, pIFpTK2 and (B) the poly(A) site mutant pSVβ128-AAGAAA-FN+. Corrected ratios of spliced:unspliced and cleaved: uncleaved transcripts are given below the lanes. The asterisk denotes an irrelevant undigested probe. Deletion of the CTD (ΔCTD, lanes 2) or the C-terminus (ΔC, lanes 3) inhibited poly(A) site cleavage without an intron (A) and splicing without a poly(A) site (B) relative to the control (ΔC+WT Cter, lanes 4).
The effect of the CTD C-terminus on splicing independent of 3′ processing was determined using a β-globin reporter with a mutant poly(A) site and a splicing enhancer derived from the fibronectin gene inserted into exon 3 (pSVβ128-AAGAAA-FN+). This reporter gives rise to predominantly poly(A)– transcripts in 293 cells (Fong and Bentley, 2001). The experiment in Figure 4B demonstrated that the C-terminus is required for efficient splicing of β-globin intron 1 independently of 3′ end processing. Similar results were obtained for intron 2 (data not shown). Deletion of the C-terminus (ΔC) reduced the ratio of spliced:unspliced transcripts by >2-fold relative to full-length CTD (WT), and adding back the wild-type 10 residue C-terminal motif (ΔC+WT) restored efficient splicing. Adding back the scrambled C-terminus (Rpb1ΔC+mut Cter) did not restore splicing (data not shown). In summary, the experiments in Figure 4 show that the CTD C-terminus is required for splicing and poly(A) site cleavage independently of one another.
The ISPDDSDEEN sequence confers mRNA processing function to pol II with 27 consensus heptads
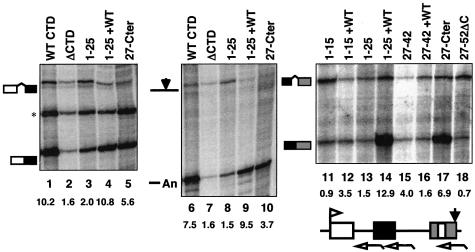
We asked whether the C-terminal 10 residue peptide is sufficient to restore processing of transcripts made by pol II that retains only the N-terminal half of the CTD (heptads 1–25). The ISPDDSDEEN sequence was fused to the C-terminus of heptads 1–25 in Rpb1 1–25+WT Cter and expressed in transiently transfected cells with a β-globin reporter. Remarkably, when the C-terminus was appended to heptads 1–25, splicing of globin introns 1 and 2 and poly(A) site cleavage increased by >5-fold (Figure 5, lanes 3, 4, 8 and 9, and 13 and 14). Similar results were obtained with Rpb1 1–25 and Rpb1 1–25+WT Cter in microinjected Xenopus oocytes (data not shown). These experiments showed that the N-terminal portion of the CTD comprising 18 consensus and seven variant heptads can support efficient mRNA processing provided it is supplemented with the C-terminal motif. To test if the C-terminal motif could enhance processing of transcripts made by pol II with fewer heptads, we fused it to the C-terminus of truncation mutants with heptads 1–15 or 27–42 (Fong and Bentley, 2001). In neither case was a large stimulation of processing efficiency observed (Figure 5, lanes 12 and 16). The stimulation of splicing in Rpb1 1–15+Cter relative to Rpb1 1–15 (Figure 5, lanes 11 and 12) was not consistently observed. Rpb1 40-Cter which has the last 12 heptads also did not support efficient processing (data not shown). The experiments in Figure 5 therefore show that for the C-terminal motif to provide maximal mRNA processing function, it must be fused to a minimum number of heptads >15 and ≤25.
Fig. 5. The C-terminal motif restores processing of transcripts made by pol II with heptads 1–25 but not 1–15 or 27–42. RNase protection of pSVβ128Rpbex5 transcripts from α-amanitin-treated cells expressing Rpb1 mutants. +WT indicates the presence of the wild-type C-terminus with the indicated hepad repeats. 27-Cter has heptads 27–52 plus the C-terminus, and 27–52ΔC has the C-terminus deleted. Corrected ratios of spliced:unspliced and cleaved:uncleaved transcripts are shown below the lanes. The asterisk marks an irrelevant undigested probe.
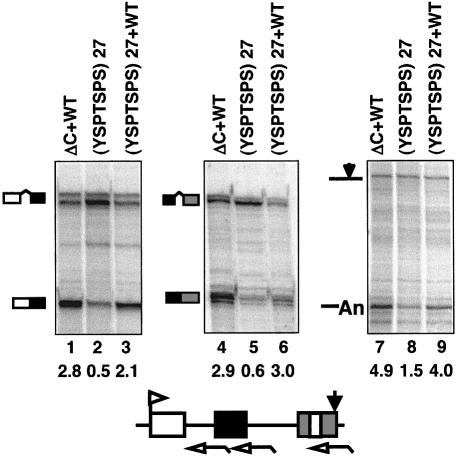
We asked if consensus heptad repeats alone are sufficient to support mRNA processing if they are supplemented with the C-terminal motif. α-amanitin-resistant Rpb1 constructs were made with 27 consensus heptads (YSPTSPS)27 plus and minus the C-terminal motif. These polymerases were tested for their ability to produce properly processed β-globin transcripts in 293 cells. As expected, Rpb1 (YSPTSPS)27 without the C-terminal motif did not produce efficiently processed transcripts (Figure 6, lanes 2, 5 and 8). Addition of the wild-type C-terminus, Rpb1 (YSPTSPS)27+WT Cter, however, restored splicing of introns 1 and 2 and poly(A) site cleavage to levels comparable with Rpb1ΔC+WT Cter which has 51 natural heptads (Figure 6, compare lanes 1, 3, 4, 6, 7 and 9). We conclude that 27 consensus heptads when supplemented with the C-terminal motif are sufficient to support efficient processing in vivo.
Fig. 6. The C-terminal motif restores splicing of transcripts made by pol II with 27 consensus heptads. RNase protection of pSVβ128Rpbex5 transcripts from α-amanitin-treated 293 cells expressing Rpb1ΔC+WT Cter, Rpb1 (YSPTSPS)27 or Rpb1 (YSPTSPS)27+WT Cter. Corrected ratios of spliced:unspliced and cleaved:uncleaved transcripts are shown below the lanes.
The C-terminal motif stimulates cell viability
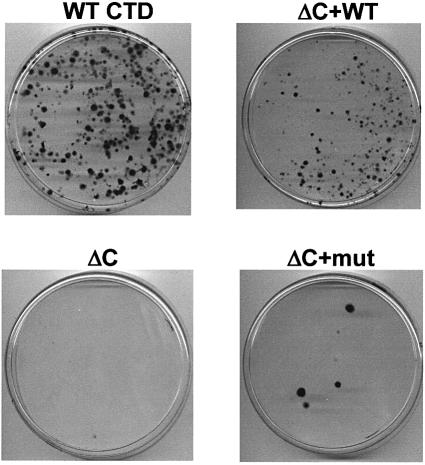
The importance of the Rpb1 C-terminal motif for cell viability was tested by asking whether cells stably expressing α-amanitin-resistant large subunits (N792D) with mutant C-termini can survive to make colonies in α-amanitin-containing medium (Bartolomei et al., 1988). 293 cells were co-transfected with Rpb1 expression vectors and a puromycin resistance plasmid. Duplicate plates were selected in puromycin or α-amanitin. α-amanitin-resistant colonies obtained with various C-terminal mutants are shown in Figure 7. As expected, numerous robust colonies were obtained with wild-type Rpb1 and Rpb1ΔC+WT Cter, but Rpb1ΔC and Rpb1ΔC+mut Cter gave far fewer colonies (Figure 7). The ratios of α-amanitin-resistant/puromycin-resistant colonies were: WT CTD, 0.24; Rpb1ΔC+WT Cter, 0.30; Rpb1ΔC, 0.0032; and Rpb1ΔC+mut Cter, 0.0005. Note that Rpb1ΔC+S6A Cter, Rpb1ΔC+S2A Cter, Rpb1 1–25+Cter, Rpb1 27-Cter and Rpb1 (YSPTSPS)27+WT Cter did not support efficient formation of α-amanitin-resistant colonies although they gave rise to properly processed β-globin mRNA in our assays (Figures 1, 2, 5 and 6).8
Fig. 7. Formation of stable α-amanitin-resistant colonies is inhibited by mutation of the C-terminal motif. 293 cells were transfected with α-amanitin-resistant Rpb1 expression plasmids plus a puromycin resistance plasmid (pBiC-S2N+puro), and duplicate plates were selected in α-amanitin or puromycin. Colonies were stained with crystal violet. Deletion (Rpb1ΔC) or scrambling of the C-terminus (Rpb1ΔC+mut Cter) reduced colony numbers relative to constructs with the wild-type C-terminus (Rpb1WT CTD and Rpb1ΔC+WT Cter). Ratios of α-amanitin-resistant/puromycin-resistant colonies determined in an independent experiment from that shown were WT CTD 0.24, Rpb1ΔC+WT Cter 0.30, Rpb1ΔC 0.0032 and Rpb1ΔC+mut Cter 0.0005.

Fig. 8. Summary of CTD mutants structure, and mRNA processing ability [splicing + poly(A) site cleavage]. Open rectangles indicate consensus heptads; green indicates variant heptads; red indicates WT C-terminus; black, blue and yellow indicate mutant C-termini.
Discussion
The CTD of the pol II large subunit is essential for coupling capping, splicing and 3′ processing of mRNA precursors to transcription. In this study, we report the unexpected finding that the conserved heptad repeats of the CTD are insufficient for high level transcription, splicing and 3′ processing of mRNA precursors. In addition to the heptad repeats, a 10 residue motif that lies C-terminal of heptad 52 is absolutely required for these key events in mRNA synthesis. Either deletion or scrambling of these 10 residues significantly reduced splicing and poly(A) site cleavage (Figure 1A) as well as pol II density on a transfected reporter gene in mammalian cells (Figure 3B). Although the ISPDDSDEEN sequence is absolutely required, it is not essential that it be located precisely at the C-terminus since constructs with an eight residue C-terminal extension ASHHHHHH (Rpb1 WT and Rpb1 27-Cter) produce well processed transcripts. The C-terminal motif is required for splicing and poly(A) site cleavage independently of one another (Figure 4), but it is not essential for capping (Figure 1B). The ISPDDSDEEN motif contains a consensus CKII phosphorylation site (Ser6); however, a S6A substitution did not inhibit processing nor did S2A (data not shown) or the S2A,S6A double mutant (Figure 2). Phosphorylation at these serines is therefore not essential for processing in our assays.
We previously observed that the N-terminal heptads 1–25 of the CTD do not support efficient splicing or poly(A) site cleavage, whereas heptads 27–52 plus the C-terminal motif do support these processing events (Fong and Bentley, 2001). This difference could be due to the predominance of variant heptads in the C-terminal half of the CTD in contrast to the predominance of consensus heptads in the N-terminal half. Our results show that, in fact, the functional distinction between N- and C-terminal portions of the CTD is explained by the 10 residue C-terminal motif. This sequence, ISPDDSDEEN, when added to heptads 1–25, restored splicing and poly(A) site cleavage to a level comparable with intact full-length CTD (Figure 5). The C-terminal peptide is therefore necessary and sufficient for pre-mRNA processing in vivo in the context of either the N- or C-terminal portions of the CTD. We conclude that mRNA processing does not have a strict requirement for a particular mix of heptad repeat sequences in the CTD. This conclusion is reinforced by the fact that a synthetic CTD comprising 27 consensus heptads plus ISPDDSDEEN at the C-terminus supported efficient splicing and 3′ end processing (Figure 6). We detected no requirement for particular heptad sequence variants for mRNA processing, but there is a requirement for a minimum number of heptads. CTD fragments with <25 heptads (heptads 40–52, 1–15 or 27–42) did not stimulate splicing or 3′ processing when supplied with the C-terminal motif. In summary, the minimal number of heptads required for efficient mRNA processing is between 15 and 25 in addition to the C-terminal motif. The minimal CTD required for mRNA processing is not sufficient for full cell viability under our conditions since Rpb1 1–25+Cter, Rpb1 27-Cter and Rpb1 (YSPTSPS)27+WT supported processing but not formation of stable α-amanitin-resistant transformants. This inviability suggests that a subset of essential genes is poorly processed or inadequately transcribed by these mutant polymerases. Our results are consistent with the fact that homozygous deletion of heptads 23–36 in mice is not lethal, although it causes growth retardation (Litingtung et al., 1999). They also agree with previous experiments showing that 36 or more heptads give wild-type viability in a myoblast cell line, 29–32 heptads give slow growth and 25 or fewer heptads is lethal (Bartolomei et al., 1988). Our results revealed a more important role for the C-terminal motif in supporting cell viability than was observed by Bartolomei et al. (1988). The reasons for this difference are not clear; however, different cell types and expression vectors were used, which could affect the amounts of Rpb1 expressed.
The C-terminal sequence, ISPDDSDEEN, is completely conserved between human, mouse and Chinese hamster pol II large subunits, but it is not conserved in C.elegans, Drosophila or unicellular eukaryotes, indicating that it may serve a function that is peculiar to vertebrates. Alternatively, it is possible that the same function is performed by the different sequences at the C-termini of Rpb1 in other species. In contrast to vertebrate cells, deletion of the yeast C-terminus including as many as 15 heptad repeats does not have an obvious deleterious phenotype (Nonet et al., 1987). In this regard, it is probably significant that in yeast, splicing and 3′ processing can occur independently of the CTD (Licatalosi et al., 2002).
How does the 10 amino acid C-terminal peptide work? It is possible that it affects transcription, splicing and 3′ end cleavage independently, but we think it more likely that this short motif has a single primary function which affects all three processes. One possibility is that the C-terminal motif is required for a concerted event involving transcription, splicing and 3′ end formation. An example of such a concerted event occurs on the Chironomus BR1 gene where splicing of the last intron and poly(A) site cleavage are tightly coupled with pol II release from the DNA template 600 bp downstream of the poly(A) site (Bauren et al., 1998). The C-terminal motif may work by binding to one or more common factors that enhance both transcription and processing. Interacting factors could recognize the C-terminal motif alone or in the context of adjacent heptad repeats. Recognition of the C-terminus by other proteins could serve to recruit transcription/processing factors to the elongating pol II transcription complex or to localize the polymerase in the correct nuclear compartment. So far, the essential function of the C-terminal motif has not been detected by in vitro assays for binding to processing factors (Fong and Bentley, 2001), enhancement of poly(A) site cleavage (Ryan et al., 2002) or assembly of splicing complexes (Hirose et al., 1999; Zeng and Berget, 2000). Therefore, the C-terminus may only function in the context of an intact elongating pol II ternary complex or another higher order nuclear structure.
Materials and methods
Transient transfections and RNA analysis
293 cells (150 mm plates) were transfected with 5–10 µg of reporter plasmid, 0.5 µg of pSPVA and 2.5 µg of pol II expression vectors by CaPO4 precipitation; however, identical results were obtained with Lipofectamine (Invitrogen)-transfected cells (see Supplementary data). α-amanitin (2.5 µg/ml) was added after 12–16 h and cells were harvested 48 h later. VA controls for transfection efficiency are not shown. Fractionation of capped and uncapped RNAs by binding to GST–eIF4E and RNase protection were as described (McCracken et al., 1997b). Fixed, dried gels were quantified by phosphoimager using Imagequant software. Ratios of spliced:unspliced and cleaved:uncleaved transcripts were corrected for the [32P]U content of the protected fragments.
Stable transfection
293 cells (100 mm plates) were co-transfected with α-amanitin-resistant Rpb1 expression plasmids (2.5 µg/plate) plus a puromycin resistance plasmid [0.5 µg of pBiC-S2N+puro (Yu et al., 2003)]. Duplicate plates were selected in puromycin (0.25 µg/ml for 3 days then 0.5 µg/ml) or α-amanitin (1 µg/ml for 3 days then 2.0 µg/ml for 14 days) (Bartolomei et al., 1988). Colonies were stained with crystal violet. Over 1200 puromycin-resistant colonies were obtained for each experiment.
Oocyte injection
Xenopus oocyte nuclei were injected with α-AmrRpb1 expression vectors (0.46 ng) in 23 nl. After overnight incubation, they were reinjected with the CMV β-globin (0.92 ng) and pSPVA (1.15 pg) reporter genes plus α-amanitin (25 µg/ml) in 23 nl. RNA was isolated 12–16 h later as described (Bentley and Groudine, 1988). No transcripts were made if the Rpb1 expression vector was omitted.
Nuclear runon analysis
Nuclear runon analysis was as described (Roberts and Bentley, 1992) except that nuclei were not pre-digested with RNase, and α-amanitin (2.0 µg/ml) was added 5 min before adding NTPs. Labeled RNA was hybridized to single-stranded M13 probes (1.0 µg) slot-blotted onto GeneScreen. Filters were digested with RNase A after washing.
Plasmids
pSVβ128-AAGAAA–FN+, pIFpTK2, pSPVA, pAT7Rpb1 with CTD deletions 1–15, 27–42, 1–25 and 27–52+Cter, and RNase protection probes were as described (Fong and Bentley, 2001). pSVβ128Rpbex5 (Fong and Bentley, 2001) contains the human β-globin gene with a 280 bp in-frame insertion from mouse pol II large subunit exon 5 in the BstXI site of exon 3. This insert modestly increased the efficiency of 3′ processing. pAT7Rpb1WTCTD is a CMV-driven expression vector identical to pAT7h1αAmr (Nguyen et al., 1996). Its C-terminus is ISPDDSDEENASHHHHHH-Ter. pAT7Rpb1Δ0 (Fong and Bentley, 2001) deletes the entire CTD. Versions with polylinkers in three frames were made.
pAT7Rpb1ΔC was made by inserting a 1.5 kb PCR fragment of human Rpb1 starting at amino acid 1366 into Eco52I–XbaI- cut pATRpb1Δ0. The C-terminus YSPTSPG YSPTSP AGV-Ter deletes 18 residues including heptad 52 and the last residue of heptad 51, and inserts unique SgrAI and XbaI sites upstream of the termination codon. pAT7Rpb1ΔC+WT Cter has an oligo nucleotide encoding ISPDDSDEEN-Ter in SgrAI–XbaI-cut pAT7Rpb1ΔC. pAT7Rpb1ΔC+mut Cter has an oligonucleotide en coding NDSDSDPEIE-Ter in SgrAI–XbaI-cut pAT7Rpb1ΔC. pAT7Rpb1ΔC+S6A has an oligonucleotide encoding ISPDDADEEN-Ter in SgrAI–XbaI-cut pAT7Rpb1ΔC. pAT7Rpb1ΔC+S2A has an oligonucleotide encoding IAPDDSDEEN-Ter in SgrA–XbaI-cut pAT7Rpb1ΔC. pAT7Rpb1ΔC+S2A,S6A has an oligonucleotide encoding IAPDDADEEN-Ter in SgrA–XbaI-cut pAT7Rpb1ΔC. pAT7Rpb1 1–25+WT Cter has an oligonucleotide encoding ISPDDSDEEN-Ter inserted at the SpeI site within heptad 26. pAT7Rpb1 1–15+WT Cter was made by cloning a PCR fragment encoding mouse heptads 1–15 fused to ISPDDSDEEN-Ter into PmeI–XbaI-cut pAT7Rpb1Δ0. Residues GLNAAALGSVGGAMSPS are inserted after residue 1492 at the junction with the CTD. pAT7Rpb1 27–42+WT Cter was made by cloning a PCR fragment encoding mouse heptads 27–42 fused to ISPDDSDEEN-Ter into PmeI–XbaI-cut pAT7Rpb1Δ0. Residues GLNAAVLGSS are inserted after residue 1492 at the junction with the CTD.
pAT7Rpb1 27-Cter was made by insertion of the SpeI–XbaI fragment of pAT7Rpb1WT CTD including the C-terminal His tag into pATRpb1Δ0. It has residues GLNAAALGSS inserted after residue 1492 at the junction with the CTD. pAT7Rpb1 27–52ΔC deletes the C-terminus of pAT7Rpb1 27-Cter and ends two residues after heptad 52, YSLTSPAGV-Ter. pAT7Rpb1 (YSPTSPS)27 was made by insertion of a PCR product with 27 consensus heptads into NotI–XbaI-cut pATRpb1Δ0. This polymer was made by several rounds of ligation and cloning of NheI–SpeI-cleaved products of the annealed oligonucleotides 5′-CTA GCCCGAGCTATTCTCCGA-3′ and 5′-CTAGTCGGAGAATAGC TCGGG-3′. Its C-terminus is YSPTSPSHHHHHQ-Ter. pAT7Rpb1 (YSPTSPS)27+WT Cter differs from pAT7Rpb1 (YSPTSPS)27 by addition of ISPDDSDEEN-Ter. M13 β-globin contains a 900 bp PCR product extending from the ATG to the BsrGI site in intron 2 into EcoRI–Acc65I-cut mp19. pCMV β-globin was made by three-way cloning of the human β-globin 2125 bp NcoI–PstI fragment starting at +48 with an oligonucleotide comprising bases +1+47 into pCI-neo (Promega) cut with SacI and BamHI. Globin base +1 is fused at +30 relative to the CMV TATA box.
Antibody
Anti-B10 antibody was purchased from Chemicon.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are particularly grateful to R.Harland for identifying Xenopus Rpb1 ESTs. We thank D.Zorio for help with the capping assay, D.Turner for plasmids, the UCHSC Cancer Center Sequencing Facility, A.M.Deshpande, D.Zorio, T.Blumenthal and S.Schroeder for valuable criticisms and suggestions, and T.Boudreau for secretarial help. This work was funded by grants from the Association pour la Recherche sur Ie Cancer (ARC 4521) to M.V., NIH T32GM08730 to G.B. and NIH GM58613 to D.B.
References
- Bartolomei M.S., Halden,N.F., Cullen,C.R. and Corden,J.L. (1988) Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol. Cell. Biol., 8, 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren G., Belikov,S. and Wieslander,L. (1998) Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev., 12, 2759–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. (2002) The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol., 14, 336–342. [DOI] [PubMed] [Google Scholar]
- Bentley D.L. and Groudine,M. (1988) Sequence requirements for premature termination of transcription in the human c-myc gene. Cell, 53, 245–246. [DOI] [PubMed] [Google Scholar]
- Cramer P., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science, 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- Dahmus M.E. (1996) Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem., 271, 19009–19012. [DOI] [PubMed] [Google Scholar]
- Fong N. and Bentley,D. (2001) Capping, splicing and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev., 15, 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber H.P., Hagmann,M., Seipel,K., Georgiev,O., West,M.A., Litingtung,Y., Schaffner,W. and Corden,J.L. (1995) RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature, 374, 660–662. [DOI] [PubMed] [Google Scholar]
- Hirose Y. and Manley,J.L. (2000) RNA polymerase II and the integration of nuclear events. Genes Dev., 14, 1415–1429. [PubMed] [Google Scholar]
- Hirose Y., Tacke,R. and Manley,J.L. (1999) Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev., 13, 1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. and Shuman,S. (1999) Distinct effector roles for Ser2 and Ser5 phosphorylation of the RNA polymerase II CTD in the recruitment and allosteric activation of mammalian capping enzyme. Mol. Cell, 3, 405–411. [DOI] [PubMed] [Google Scholar]
- Kim E., Du,L., Bregman,D.B. and Warren,S.L. (1997) Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-messenger RNA. J. Cell Biol., 136, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor M. and Greenblatt,J. (2002) Regulation of transcription elongation by phosphorylation. Biochim. Biophys Acta, 1577, 261–275. [DOI] [PubMed] [Google Scholar]
- Kuenzel E.A., Mulligan,J.A., Sommercorn,J. and Krebs,E.G. (1987) Substrate specificity determinants for casein kinase II as deduced from studies with synthetic peptides. J. Biol. Chem., 262, 9136–9140. [PubMed] [Google Scholar]
- Lee T.I. and Young,R.A. (2000) Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet., 34, 77–137. [DOI] [PubMed] [Google Scholar]
- Licatalosi D.D., Geiger,G., Minet,M., Schroeder,S., Cilli,K., McNeil,J.B. and Bentley,D.L. (2002) Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell, 9, 1101–1111. [DOI] [PubMed] [Google Scholar]
- Litingtung Y., Lawler,A.M., Sebald,S.M., Lee,E., Gearhart,J.D., Westphal,H. and Corden,J.L. (1999) Growth retardation and neonatal lethality in mice with a homozygous deletion in the C-terminal domain of RNA polymerase II. Mol. Gen. Genet., 261, 100–105. [DOI] [PubMed] [Google Scholar]
- McCracken S. et al. (1997a) 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev., 11, 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Fong,N., Yankulov,K., Ballantyne,S., Pan,G.H., Greenblatt,J., Patterson,S.D., Wickens,M. and Bentley,D.L. (1997b) The C-terminal domain of RNA polymerase II couples messenger RNA processing to transcription. Nature, 385, 357–361. [DOI] [PubMed] [Google Scholar]
- Misteli T. and Spector,D.L. (1999) RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell, 3, 697–705. [DOI] [PubMed] [Google Scholar]
- Naar A.M., Taatjes,D.J., Zhai,W., Nogales,E. and Tjian,R. (2002) Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev., 16, 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.T., Giannoni,F., Dubois,M.F., Seo,S.J., Vigneron,M., Kedinger,C. and Bensaude,O. (1996) In vivo degradation of RNA polymerase II largest subunit triggered by α-amanitin. Nucleic Acids Res., 24, 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Sweetser,D. and Young,R.A. (1987) Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell, 50, 909–915. [DOI] [PubMed] [Google Scholar]
- Payne J.M., Laybourn,P.J. and Dahmus,M.E. (1989) The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIA. J. Biol. Chem., 264, 19621–19629. [PubMed] [Google Scholar]
- Rickert P., Corden,J.L. and Lees,E. (1999) Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene, 18, 1093–1102. [DOI] [PubMed] [Google Scholar]
- Robert F., Blanchette,M., Maes,O., Chabot,B. and Coulombe,B. (2002) A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J. Biol. Chem., 277, 9302–9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S. and Bentley,D.L. (1992) Distinct modes of transcription read through or terminate at the c-myc attenuator. EMBO J., 11, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K., Murthy,K.G., Kaneko,S. and Manley,J.L. (2002) Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol. Cell. Biol., 22, 1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafe C., Chao,D., Lopes,J., Hirsch,J.P., Henry,S. and Young,R.A. (1990) RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature, 347, 491–494. [DOI] [PubMed] [Google Scholar]
- Yu J.Y., Taylor,J., DeRuiter,S.L., Vojtek,A.B. and Turner,D.L. (2003) Simultaneous inhibition of GSK3α and GSK3β using hairpin siRNA expression vectors. Mol. Ther., 7, 228–236. [DOI] [PubMed] [Google Scholar]
- Yue Z., Maldonado,E., Pilluta,R., Cho,H., Reinberg,D. and Shatkin,A. (1997) Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA pol II. Proc. Natl Acad. Sci. USA, 94, 12898–12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A., Patturajan,M., Litingtung,Y., Joshi,R., Gentile,C., Gebara,M. and Corden,J. (1996) The CTD of RNA polymerase II interacts with a novel set of SR-like proteins. Proc. Natl Acad. Sci. USA, 93, 6975–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C. and Berget,S.M. (2000) Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol., 20, 8290–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]