Abstract
Glutamate decarboxylase is a vitamin B6-dependent enzyme, which catalyses the decarboxylation of glutamate to γ-aminobutyrate. In Escherichia coli, expression of glutamate decarboxylase (GadB), a 330 kDa hexamer, is induced to maintain the physiological pH under acidic conditions, like those of the passage through the stomach en route to the intestine. GadB, together with the antiporter GadC, constitutes the gad acid resistance system, which confers the ability for bacterial survival for at least 2 h in a strongly acidic environment. GadB undergoes a pH-dependent conformational change and exhibits an activity optimum at low pH. We determined the crystal structures of GadB at acidic and neutral pH. They reveal the molecular details of the conformational change and the structural basis for the acidic pH optimum. We demonstrate that the enzyme is localized exclusively in the cytoplasm at neutral pH, but is recruited to the membrane when the pH falls. We show by structure-based site-directed mutagenesis that the triple helix bundle formed by the N-termini of the protein at acidic pH is the major determinant for this behaviour.
Keywords: bacterial acid resistance/cellular localization/Escherichia coli/glutamate decarboxylase/pyridoxal 5′-phosphate
Introduction
Glutamate decarboxylase (Gad; EC 4.1.1.15) is a pyridoxal 5′-phosphate (PLP)-dependent enzyme, which catalyses the irreversible α-decarboxylation of L-glutamate to γ-aminobutyrate (GABA). This enzyme is widely distributed amongst eukaryotes and prokaryotes, but its function varies in different organisms.
Gad has a crucial role in the vertebrate central nervous system where it is responsible for the synthesis of GABA, the major inhibitory neurotransmitter. In the majority of vertebrates Gad occurs in two isoforms, GAD65 and GAD67, both active at neutral pH (Soghomonian and Martin, 1998).
A unique feature of plant and yeast Gad is the presence of a calmodulin (CaM)-binding domain in the C-terminal region. In Saccharomyces cerevisiae, Gad expression is required for normal oxidative stress tolerance (Coleman et al., 2001). In plants, Gad is thought to be a stress-adapter chaperonin sensing Ca2+ signals. Removal of the CaM-binding domain causes a deregulation of the activity, leading to severe developmental problems (Baum et al., 1996). The plant enzyme has a pH optimum of 5.8, but its activity is also significant at pH 7.3 in the presence of the Ca2+/CaM complex (Zik et al., 1998), which is ∼500 kDa in size (Baum et al., 1996).
Bacterial Gad has some features similar to those of the plant enzyme: it exhibits an acidic pH optimum (3.8–4.6), forms a hexamer (Sukhareva et al., 1994) and is expressed in response to environmental stresses (Blankenhorn et al., 1999; De Biase et al., 1999). Gad isoforms (GadA and GadB) have also been reported in some bacterial species, including the Gram-negative bacterium Escherichia coli (Smith et al., 1992) and the Gram-positive bacterium Listeria monocytogenes (Cotter et al., 2001). Separate expression of the two isoforms demonstrated that they are biochemically indistinguishable (De Biase et al., 1996). In E.coli and in other enteric bacteria—both commensal and pathogenic—GadA and GadB have been identified as structural constituents of the gad system (De Biase et al., 1999; Cotter et al., 2001). Of the three acid resistance systems known in E.coli (Lin et al., 1995, 1996), the gad system is by far the most potent and is involved in conferring acid resistance to the bacteria in stationary phase, giving them survival capacity for at least 2 h in a strongly acidic environment (pH < 2.5), such as that of the stomach. GadB and GadA belong to the degradative amino acid decarboxylases, acid-induced in E.coli, which include arginine (AdiA), lysine (CadA) and ornithine (SpeF) decarboxylases (Slonczewski and Foster, 1996). However, only the gad and the adi systems provide acid protection at an external pH of 2.5 (Lin et al., 1995), when the internal pH is estimated to be near 5. This fits with the hypothesis that the relevant systems for acid protection must be those with the optimum pH of the corresponding decarboxylases closer to pH 5, i.e. gad and adi (Foster, 2000).
The genes encoding the two Gad isoforms, gadA and gadB, are 2100 kb apart in the E.coli chromosome (Blattner et al., 1997). Additionally, downstream of gadB, another gene called gadC is found, which is co-transcribed with gadB and codes for GadC, a putative glutamate/GABA antiporter. In different micro-organisms, the integrity of the system is essential for coping with acidic stress (Hersh et al., 1996; Waterman and Small, 1996; De Biase et al., 1999; Cotter et al., 2001). The Gad system is assumed to control the acidification of the cytosolic environment by decarboxylating an acidic substrate (glutamate) into a neutral compound (GABA) via incorporation of H+. GABA would then be exported into the extracellular medium through the GadC protein, thereby contributing to local alkalinization of the extracellular environment. A similar mechanism was also proposed for the lysine decarboxylase system (CadB, transporter; and CadA, decarboxylase), the first decarboxylase operon to be elucidated (Meng and Bennett, 1992).
The expression of the gad structural genes in E.coli involves a number of protein factors that act as repressors and activators (De Biase et al., 1999; Tramonti et al., 2002b). The molecular cascade leading to the activation of the gad system is rather complex and possibly involves proteins acting as molecular switches for a number of stress response systems, including the one for acid resistance.
Herein, the crystal structure of E.coli GadB, the Gad isoform co-expressed with GadC, is described and provides evidence for the well-known pH-dependent conformational change involved in GadB activation (O’Leary and Brummund, 1974). Several active site determinants involved in substrate binding and coenzyme positioning, some of which have an unexpected chemical nature, were also identified. Moreover, evidence is provided that the enzyme is localized exclusively in the cytoplasm at neutral pH, but is recruited to the membrane when the pH falls.
Results
Overall structure of GadB and pH-dependent conformational change
The structure of recombinant E.coli GadB was determined by a combination of the multiple isomorphous replacement with anomalous scattering (MIRAS) and multiple anomalous dispersion (MAD) methods. Two different crystal forms of GadB were refined (Table I), one at low pH (form A, at 2.0 Å resolution, pH 4.6) and one at neutral pH (form C, at 2.3 Å resolution, pH 7.6), both exhibiting excellent geometry and stereochemistry.
Table I. Data collection and phasing statistics.
| Crystal form | A (nativelow pH) | B (nativereducedneutral pH) | B (Ta6Br12) I | B (Ta6Br12) II | B (HgAc2remote) | B (HgAc2peak) | B (HgAc2inflection) | C (nativereducedneutral pH) |
|---|---|---|---|---|---|---|---|---|
| Space group | P1 | P3 | P3 | P3 | P3 | P3 | P3 | P32 |
| Cell constants(a, b, c, Å; α, β, γ, °) | 90.8, 91.4,93.7, 76.8,77.1, 78.2 | 193.1, 193.1,66.1, 90,90, 120 | 193.6, 193.6,66.1, 90, 90,120 | 192.8, 192.8,66.0, 90, 90,120 | 192.7, 192.7,66.0, 90, 90,120 | Same asremote | Same asremote | 116.0, 116.0,207.4, 90, 90,120 |
| Source | PX-SLS | X31 DESY | Rotating anode | X31 DESY | BW7A DESY | BW7A DESY | BW7A DESY | ID14–1 ESRF |
| λ (Å) | 0.918 | 1.200 | 1.5418 | 1.200 | 0.9190 | 1.0052 | 1.0074 | 0.934 |
| Soaking conc.and time | Saturated, 3 h | Saturated,4 h 20 min | 2 mM, 2 h | 2 mM, 2 h | 2 mM, 2 h | |||
| Resolution range (Å),(last shell) | 20–2.0(2.07–2.0) | 18–2.45 | 20–3.0 | 20–2.5 | 25.0–3.0 | 25.0–3.25 | 25–3.25 | 20–2.3(2.35–2.3) |
| Unique reflections | 178 481 | 48 795 | 26 794 | 87 293a | 26 828 | 41 713a | 42 338a | 136 345 |
| Completeness (%) | 95.8 | 93.7 | 97.1 | 91.6 | 94.2 | 96.4 | 97.9 | 98.9 |
| Multiplicity | 2.6 | 3.8 | 5.4 | 3.6 | 2.9 | 3.0 | 3.0 | 2.2 |
| Rmerge (%)b | 8.7 (16.2) | 11.1 | 14.4 | 12.7 | 8.5 | 7.5 | 9.5 | 9.7 (42.2) |
| I/σ(I) | 9.3 (6.2) | 8.1 | 9.8 | 10.6 | 11.3 | 14.4 | 12.6 | 8.1 (2.1) |
| Phasing power iso/ano | 0.14/0.10 | 0.39/0.29 | 0.00/1.47 | 1.63/1.87 | 1.08/1.02 |
aIn these data sets, Bijvoet pairs were kept separated in scaling and in the output.
bRmerge = ΣhklΣj|Ij,hkl – <Ihkl>|/ΣhklΣjIj,hkl where <Ihkl> is the average of the intensity Ij,hkl over j = 1, …, N observations of symmetry equivalent reflections hkl.
The 330 kDa GadB hexamer (466 residues/subunit) has an approximate 32 symmetry and is composed of three dimers, the symmetry axes of which coincide with the 2-fold axes of the point group (Figure 1A). The hexamer is arranged in two layers of three subunits each, with the dimers contributing one subunit to each layer. A search for structurally related proteins in the Protein Data Bank (Berman et al., 2000), performed with DALI (Holm and Sander, 1993), indicated that the subunits of pig DOPA decarboxylase (DDC) (Burkhard et al., 2001), pig cytosolic aspartate aminotransferase (AAT) (Rhee et al., 1997) and Thermotoga maritima NifS-like protein (Kaiser et al., 2000) are most similar to E.coli GadB, allowing three-dimensional alignments of 387, 335 and 288 Cα atoms, respectively, out of 453, with r.m.s. deviations of 2.9, 4.8 and 3.4 Å, respectively.
Fig. 1. Surface (top) and cartoon (bottom) representation of the GadB hexamer at neutral pH (A) and low pH (B). Figures were prepared with MSMS (Sanner et al., 1996), DINO (www.dino3d.org), MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Murphy, 1997).
Comparison between the low-pH (form A) and neutral-pH (form C) structures of GadB shows that the overall structure of the hexamer remains the same, although very significant changes occur at the N- and C-termini and in a β-hairpin region spanning residues 300–313. Met1 and Asp2 are not visible in either of the structures. Residues 3–15 do not form any regular secondary structure in the neutral-pH crystal form and take up different conformations for each of the six subunits (Figures 1A and 2). Instead, in the low-pH form residues 3–15 of all subunits assume an α-helical conformation, forming two short three-helical bundles parallel to the hexamer 3-fold axis (Figures 1B and 2). The average B-factor for these residues (22.2 Å2) is higher than that for all protein atoms (14.9 Å2). At the C-terminus, residues 452–466 are well structured in the neutral-pH crystal form and point inside the active site funnel, thus reaching the substrate binding site (Figure 3). In form A, electron density corresponding to those residues is not visible. Correspondingly, the 300–313 β-hairpin, which narrows the active site funnel in form C, is shifted towards the centre of the funnel in form A.
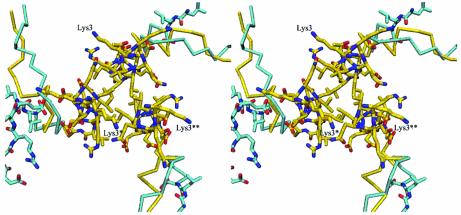
Fig. 2. Superposition of the three N-termini from one layer of the GadB hexamer at neutral pH (cyan and atom colors) and at low pH (yellow and atom colors). The N-termini for the low-pH form are labelled.
Fig. 3. Superposition of the GadB (cyan, neutral-pH form) and DDC (red) Cα traces. N- and C- termini are labelled, as are the DDC residues (327 and 340) flanking the region for which no electron density is visible.
Subunit structure
A superposition of the GadB monomer on DDC, the closest structural homologue, reveals that they indeed share a similar fold (Figure 3). However, structural differences are found in three places: at the N-terminal domain, towards the end of the large domain, and at the C-terminal end. GadB and DDC are both composed of three domains: N-terminal, large and small. In DDC, the N-terminal domain (residues 1–85), which is composed of two parallel helices linked by an extended strand, contacts the top of the opposite subunit thereby providing an additional dimer interface (Burkhard et al., 2001). In GadB, the N-terminal domain (residues 1–57) is shorter and is clearly also functional to hexamer formation. It is shaped like a hook and contains little secondary structure. This ‘hook’, instead of simply extending the interface between two monomers, as in DDC (Burkhard et al., 2001), departs nearly perpendicularly from the subunit surface and allows each GadB subunit of the hexamer to contact both its counterpart in the dimer as well as a subunit placed on a different layer and belonging to another dimer (Figure 3). Thus, this portion of GadB contributes residues involved in both dimerization and hexamerization. Residues from Lys3 to Lys30 display different conformations in the low- and neutral-pH forms (Figures 1 and 2).
The large domains of GadB (residues 58–346) and DDC are more alike: both contain the cofactor binding site and exhibit an α/β fold consisting of a central seven-stranded mixed β-sheet surrounded by eight α-helices, typical of PLP-dependent enzymes belonging to the type I fold (Jansonius, 1998). Moderate differences in both length and orientation of these secondary structural elements are observed between the two structures. A significant difference is found in the region encompassing residues 300–313 in GadB and residues 327–340 in DDC. While in DDC this region is mostly disordered and lacks electron density in the stretch 328–339 (Burkhard et al., 2001), in GadB it forms a β-hairpin, which protrudes towards the small domain of the other subunit of the dimer and narrows the active site funnel (Figure 3). This structural element is also involved in the above-described pH-dependent conformational change.
The GadB small domain (residues 347–466) is also similar to the DDC small domain: both consist of a four-stranded antiparallel β-sheet with three helices packed against the face opposite to the large domain. Again, differences exist in both the length and the orientation of the secondary structural elements. A significant structural difference is seen at the C-terminal end. Unlike DDC, GadB does not end with the last helix of the small domain protruding into the solvent, but contains a 16-amino acid stretch, which, within the same subunit, loops around the small domain, enters the active site funnel, contacts the β-hairpin 300–313, and ends within the active site region, approximately at the level of the substrate binding site (Figure 3). The C-terminal extension encompassing residues 452–466 is also involved in the pH-dependent conformational change.
The dimer interface of the neutral-pH structure, calculated following Janin (Janin, 1997), is rather large (5580 Å2) if compared with that of AAT (3250 Å2), representing 25% of the subunit total accessible surface area. Also, the interface area between dimers is fairly extended and amounts to 2880 Å2, ∼8% of the dimer total accessible surface area. The values calculated for the low-pH structure are 4850 and 3040 Å2 for the monomer–monomer and the dimer–dimer interfaces, respectively.
The pH-dependent membrane association of GadB and role of the N-terminal region
Escherichia coli Gad has an acidic pH optimum for activity, which dramatically falls above pH 5.0 (Shukuya and Schwert, 1960a). The enzyme’s pH-dependent conformational change involves the reversible uptake of 4–6 protons upon acidification (O’Leary and Brummund, 1974; Tramonti et al., 2002a). The conformational change is detectable in both isoforms by the blue-shifted cofactor absorbance from 420 to 340 nm upon changing the pH from 5 to 6 (Shukuya and Schwert, 1960b; Tramonti et al., 2002a). These features are remarkable given that the enzyme catalyses its reaction intracellularly, i.e. in an environment with an overall neutral pH (Booth, 1999). Thus, the possibility exists that upon exposure to strong acids E.coli Gad becomes active in the cell only where the internal pH falls below a threshold level. The cytoplasmic side of the inner membrane is the part of the cell where incoming protons should be significantly more abundant and in which both the proton-consuming activity and the proton-uptake ability of GadB would be beneficial to the cell. To test the possibility that GadB has different pH-dependent cellular localizations, a culture of the E.coli strain JM109 overexpressing GadB was grown and treated as described in Materials and methods to produce two cell extracts of identical origin, but with a different final pH. These extracts were used for electrophoretic and immunoblot analyses, as well as for activity measurements. The results shown in Figure 4 (left panels) clearly indicate that 92% of GadB is localized in the cytoplasmic fraction at neutral pH. However, at mildly acidic pH, only 36% of GadB is present in the cytoplasm; the remainder (at least 55%) is both active and detectable in the detergent-solubilized membrane fraction (Figure 4, left panels, lanes M1 + M2). The pH-dependent conformational change observed in the crystal structure suggests that the triple helix bundle observed at acidic pH might be involved, directly or indirectly, in the observed membrane association of GadB. A deletion mutant of GadB lacking the first 14 residues (GadBΔ1–14) was constructed, and overexpressed in E.coli JM109 as described in Materials and methods. Using the same approach as for wild-type GadB, two aliquots of the cell extract containing mutant GadBΔ1–14 at different pHs were generated and analysed (Figure 4, right panels). Although the GadBΔ1–14 mutant is two times less abundant in the total cell extract, it is clear that its localization is cytoplasmic (95%) at neutral pH, as observed for GadB wild type. However, at mildly acidic pH, the majority of GadBΔ1–14 mutant (76%) is both active and detectable in the cytoplasmic fraction. An amount accounting for <25% is still detectable in the detergent-solubilized membrane fraction. Thus, when compared with the wild type, the GadBΔ1–14 mutant has an almost reversed partition ratio between the cytoplasmic and the membrane fractions.
Fig. 4. Effect of pH on the cellular localization of GadB wt (left panels) and GadBΔ1–14 (right panels). Upper panels: 10% SDS–PAGE of cell supernatants (S, 20 µg), obtained after cell lysis and centrifugation to remove cell debris; the cytoplasmic (C, 20 µg) and membrane (M1, 20 µg) fractions were obtained as described in Materials and methods. For GadB wt, an additional lane (M2) represents the 1% lauroyl sarcosine-solubilized membrane fraction after the last ultracentrifugation, still containing some GadB wt. Molecular weight (kDa) standards are shown in the middle of the right panel and on the left hand side of the left panel. The vertical bar between panels refers to the anti-GadB probed region. Lower panels: immunoblot analysis of samples as in upper panel, probed with anti-GadB polyclonal antibodies. Due to the different amount of GadB (wt or mutant) present in the cell supernatant (S) the loadings were 2.5–2.8 µg for GadB wt and 8.4–8.9 µg for GadBΔ1–14. The C, M1 and M2 fractions were loaded according to the total protein partition between these fractions, with respect to S. The decarboxylase activity is in total units (U × 103, upper line) and as a percentage (%, lower line) with respect to the cell supernatant (S = 100%). The values with an asterisk refer to the decarboxylase activity present in the resuspended membrane fraction before separation from the insoluble material, thus allowing measurement of the enzyme still present in the M2 fraction. The reported activity values represent the means of three independent experiments, with a variation that does not exceed 10% of the stated value.
The striking acidic pH-driven conformational change observed in wild-type GadB (Figure 1) involves the uptake of 4–6 protons (Tramonti et al., 2002a). This suggests that Asp and Glu residues located within the regions undergoing conformational changes might become protonated upon exposure. The N-terminal region of GadB (residues 1–15) contains four acidic residues (Asp2, Asp8, Glu12 and Asp15); these are all likely candidates for the reversible proton uptake occurring in GadB upon acidification. To test this possibility, the cofactor absorbance change of GadBΔ1–14 (missing all the acidic residues listed above) was analysed in the pH range 3–7.2 (Figure 5). The absorbance change with decreasing pH in this mutant fits adequately to a model that requires the uptake of no more than three protons (Figure 5, inset). Thus, the deleted region contributes at least two of the residues, the protonation of which is associated with the conformational change.
Fig. 5. Effect of pH on the absorbance spectra of GadBΔ1–14. Spectra of GadB (6.9 µM) were determined in 0.1 M Na acetate in the pH range 3–7.15. Spectra at pH 3.20, 5.05, 5.25, 5.34, 5.47, 5.55, 5.88 and 7.15 are shown. In the inset, the pH variation at 418 nm is represented. The solid line is that of best fit to equation 1 (Materials and methods), with pK = 5.46 ± 0.01, AbsHnE = 0.0604 ± 0.0005, AbsE = 0.0095 ± 0.0007 and n = 3.0 ± 0.2. Curve fitting obtained by using the 338-nm absorbance readings gave similar pK and n values (pK = 5.45 ± 0.017; n = 2.4 ± 0.2).
The active site
The active site of GadB (Figure 6A, neutral pH; Figure 6B, low pH) is similar to that of DDC, with several conserved amino acids [Phe63(80), Ser128(149), Thr212(246), Asp243(271), Ala245(273), His275(302), Lys276(303) and Arg422(447)] occupying structurally equivalent positions in both enzymes. Figure 6A shows the two active sites superimposed. A number of differences between GadB and DDC are immediately apparent. (i) While in DDC the pyridine ring of PLP is sandwiched between Ala273 and His192, in the GadB active site Gln163 takes the place of DDC His192. (ii) Three cysteine residues are in the GadB active site: Cys64, replacing Ala83 in DDC and Cys130 and 165, corresponding to Thr152 and Ser194 in DDC. The latter two are at the back of the active site, not far from the N1 and C6 atoms of the cofactor; their electron density indicates that they are reduced, although the distance between the two -SH groups (3.8 Å) suggests that upon a slight conformational change they could form a disulfide bridge. (iii) As already mentioned, in the neutral-pH GadB structure, a 16-residue-long C-terminal extension after the last helix ends within the active site and the C-terminus (Thr466) occupies approximately the same position where the natural substrate is expected to bind. (iv) An important residue for substrate binding in GadB is contributed by the neighbouring subunit (Asp86*, corresponding spatially to Ser104* in DDC) and belongs to a loop taking up rather different conformations in the two enzymes.
Fig. 6. Active site views. (A) Active sites of GadB (neutral pH, grey and atom colours) and DDC (yellow and atom colours) superimposed. For clarity, no water molecules are shown. Asterisks indicate the two Cys residues of GadB (Cys130 and Cys165), which may form a disulfide bridge. (B) Active site of GadB at low pH. The final 2mFo – DFc electron density for PLP, Lys276 and the bound acetate is contoured in green at 1σ level. (C) Model for the external aldimine intermediate with glutamate. Relevant hydrogen bonds appear as green dotted lines, bound water molecules as red spheres.
Binding of acetate and modelling of the PLP–glutamate complex (external aldimine)
In the low-pH structure of GadB, buffered with Na acetate at pH 4.6, an acetate molecule is clearly bound in the active site of all six subunits (Figure 6B). Acetate is an inhibitor of GadB (Shukuya and Schwert, 1960a). The acetate electron density is very well defined and indicates the binding site and mode of the distal carboxylate of glutamate, the natural substrate. The carboxylate group of acetate, which is probably protonated in the experimental conditions, is firmly kept in place by a cluster of interactions with the enzyme: it receives hydrogen bonds from the side chain OH of Thr62 and from the amide nitrogen of Phe63. It is also hydrogen-bonded to the side-chain carboxylate of Asp86* (belonging to the neighbouring subunit). The methyl group of acetate does not exhibit relevant interactions with the protein.
Based on the acetate-bound structure, it was possible to model the PLP–glutamate external aldimine into the GadB active site, under the following assumptions. (i) The main-chain conformation of the enzyme does not undergo significant changes with respect to the conformation observed in the GadB–acetate complex. Strong harmonic restraints were imposed on the Cα atoms in order to satisfy this condition. (ii) The pyridine ring of the cofactor in the external aldimine structure was rotated by ∼20° with respect to the experimental internal aldimine model, with the phosphate moiety acting as a nearly immobile anchor. This cofactor movement was observed in many PLP-dependent enzymes (see for instance Jansonius and Vincent (1987). (ii) A second anchoring point is provided by the side chain carboxylate of glutamate, the position of which is known from the GadB–acetate complex.
The final energy-minimized external aldimine with glutamate satisfies these conditions and is shown in Figure 6C. The α-carboxylate group of the external aldimine is perpendicular to the cofactor π system and receives a hydrogen bond from the carboxamide side chain of Gln163. The side-chain carboxylate of the adduct retains the interactions with Thr62, Phe63 and Asp86*, which were observed in the acetate complex (Figure 6B). An additional hydrogen bonding interaction is predicted to occur between Cys64 and the γ-carboxylate of glutamate.
Discussion
Enteric bacteria, both commensal and pathogenic, are periodically exposed to environmental stress (nutrient limitation, temperature and pH extremes, osmolarity fluctuations, high levels of metal ions or oxidative compounds) which threaten their survival. As a consequence, both survival in the natural ‘non-host’ environment and colonization of the host gut strongly rely on the ability to promptly perceive harmful stimuli and to activate an efficient cellular response. This ability is particularly relevant for human health when enteric pathogens, i.e. E.coli, Shigella flexneri and Salmonella typhimurium, with an oral–faecal route of transmission, are considered. Their ability to colonize the intestine largely relies on their capacity to survive the extremely acidic secretions (pH < 3) of the stomach, the primary bactericidal barrier in the gastrointestinal tract (Giannella et al., 1972). Moreover, enteric bacteria must endure the deleterious effects of short-chain fatty acids (butyrate, propionate, acetate), the fermentation products of the intestinal microflora. In fact, short-chain fatty acids in their protonated form are able to cross the cytoplasmic membrane and then dissociate in the cytoplasm; in doing so they decrease the intracellular pH, even when the external pH is neutral (Cherrington et al., 1991). Thus, a correlation exists between the response to acid stress (acid resistance) and the onset of the pathogenic process. In E.coli, the glutamate-based system is by far the most efficient: it guarantees survival against gastric secretions (HCl, pH 2) as well as against the short-chain fatty acids (Lin et al., 1996). For the system to function, the coordinated action of Gad and of the putative antiporter GadC is required. The present work was undertaken with the aim of gaining further insight into the mechanism of action of E.coli GadB, the Gad isoform co-expressed with GadC. The two GadB crystal structures have been instrumental in identifying important conformational changes and functional features, which would have gone unnoticed had only one structure been available.
GadB is a compact hexamer of approximate 32 symmetry, in agreement with the original hypothesis based on electron microscopy studies (To, 1971). The hexamer is composed of three dimers, which are not very dissimilar from those observed in many constitutively dimeric PLP-dependent enzymes of type I fold (Jansonius, 1998). The distinctive features of the GadB dimers from those of DDC, the closest structural homologue, reside in the N- and C-termini of their subunits. While the N-termini of the DDC subunits serve to enlarge the dimer interface, the N-termini of GadB have evolved two different functions. First, they act as hexamerization arms (residues 13–57) and connect the two layers (three subunits each) of the GadB quaternary assembly. Second, residues 1–15 play a significant role in the acid pH-driven association of GadB to the membrane, as shown by our site-directed mutagenesis studies. From a structural point of view, residues 3–15 assume different conformations in the neutral- and low-pH forms. In the former, they possess conformational flexibility (different conformations for each of the six chains, no secondary structure, electron density missing for some residues); in the latter, they are folded into two short three-helical bundles perpendicular to the plane of the hexamer. This phenomenon is accompanied by a slight conformational change in residues 16–30. Packing analysis of the neutral-pH crystal form shows that lattice effects may cause the flexible, unbundled state of the N-terminus. Mutagenesis studies, however, clearly demonstrate the importance of this region not only for membrane association but also for the pH-dependent conformational change. Since this reversible event involves an average of five protons/subunit in wild-type GadB, it is clearly not due to the protonation of the chromophore alone, but is also due to a change in the overall protein conformation. Comparative spectroscopic analysis of GadB wild type (Tramonti et al., 2002a) and GadBΔ1–14 shows that two of the protons involved in the conformational change must come from residues 1–14.
The mechanism through which the N-terminus of GadB influences the partition of the enzyme between membrane and cytosolic fraction is unclear. The triple helix bundle has a hydrophobic core, formed through one and a half heptad repeats (Val-Thr-Asp-Leu-Arg-Ser-Glu-Leu), and is polar/charged on the outside. One intriguing possibility is that the triple helix bundle is recognized by a so-far unidentified membrane-associated protein, which would serve as an anchor; this protein might also be the antiporter GadC (De Biase et al., 1999).
The response to low pH involves also the last 16 C-terminal residues (C-terminal tail) and the β-hairpin 300–313. At neutral pH, the C-terminal tail is well defined and is inserted in the active site so that the last residue occupies the putative substrate-binding site. In the low-pH crystal form, the absence of electron density corresponding to residues 452–466 raises the question of whether the tail is still inserted in the active site funnel and is just disordered or if it is really outside. Two lines of evidence support the second hypothesis. First, the β-hairpin 300–313, which at neutral pH flanks the active site funnel and contacts residues 460*–463* of the tail from a neighbouring subunit, is shifted towards the centre of the funnel in the low-pH form. It is improbable that such a shift could occur if residues 452–466 were still in the funnel, albeit in a disordered state. Secondly, the acetate molecule bound at low pH defines the position of the substrate-binding site, which corresponds approximately to the volume occupied by Thr466 in the neutral-pH crystal form. Thus, the acidic pH is the driving force for the exit of the C-terminus, which makes the active site accessible to the substrate. In other words, the C-terminal region sterically modulates the enzyme activity.
The active site of GadB exhibits an unprecedented structural feature: the presence of a glutamine, Gln163, as a stacking residue for the PLP ring. In all three-dimensional structures of PLP-dependent enzymes studied to date, the amino acid residue interacting with the pyridine ring of PLP is generally Tyr, Trp, Phe or His. Another peculiar feature of the GadB active site is the binding mode of the γ-carboxylate of glutamate, as deduced from the binding of acetate in form A. The carboxylate is held in place by hydrogen bonding interactions not only with the protein main chain (Phe63) and with a threonine side chain (Thr62), but also with the side-chain carboxylate of an aspartate from the neighbouring subunit (Asp86*). This fits with the observation that the maximum in the GadB activity profile lies at low pH, where at least one of the two carboxylates of Glu and of Asp86* must be protonated. A similar substrate binding mode may also occur in plant GADs, which are maximally active at pH 5.8 (Snedden et al., 1996) even though they contain a residue equivalent to Asp86*. The protein environment might affect the pKa of this residue so that its side-chain carboxylate remains protonated at a higher pH; a classical example for this is Asp26 in E.coli thioredoxin, for which a pKa value of 7.5 was experimentally determined (Langsetmo et al., 1991). In E.coli GadB, the side chains of Asp86* and Glu89* are not far away from each other in the low-pH structure, while in the neutral-pH structure they point in opposite directions.
Another interesting feature of the GadB active site is that it possesses a residue, Arg422, corresponding in sequence and spatial position to the arginine which binds the α-carboxylate of the substrate in many PLP-dependent enzymes. In GadB the side chain of Arg422 is prevented from binding the α-carboxylate of glutamate by an interaction with the side-chain ring of Phe63, which keeps the guanidinium group of Arg422 away so that it cannot interfere with the α-decarboxylation of the substrate. A similar arrangement is observed in the active site of DDC.
In the neutral-pH GadB structure (crystal form C, where the internal aldimine was reduced with NaBH3CN, see Materials and methods) there appears to be a gap between the electron density of the cofactor and that of Lys276. After ruling out biochemical problems, the possibility of crystallographic artefacts was also excluded by using simulated annealing omit maps (starting at 5000 K). It is possible that the reduced Schiff base in the GadB active site may be in a particularly constrained (tense) conformation, which makes it labile to radiation damage, especially after reduction. Evidence for radiation damage comes from the data scaling output: in the last few data collection frames the I/σ value is approximately half compared to that of the initial frames. Comparison with crystal form A (internal aldimine, non-reduced) shows that in the reduced form the cofactor is rotated by ∼13° away from Lys276 (rotation along the P–O5 axis).
The two crystal structures at neutral and low pH also allow unambiguous assignment of the species absorbing at 420 and 340 nm; in fact, the less hydrated environment of the neutral-pH structure, where the active site is locked by the C-terminal tail, favours the 340-nm-absorbing enolimine tautomer of the cofactor. Instead, the low-pH structure, with an increased active site polarity, favours the 420-nm-absorbing ketoenamine PLP tautomer (Tramonti et al., 2002a).
The combination of crystallographic and biochemical data presented here shed light on how the glutamate-dependent acid resistance system is likely to work. An extremely low external pH renders the bacterial membrane leaky to H+ and increases the influx of volatile fatty acids (Meng and Bennett, 1992; Foster, 2000). Both events, independently or in combination, lead to acidification of the cytoplasm, deleterious for many biological intracellular functions. In our model, incoming protons are expected to be more abundant on the cytoplasmic side of the inner membrane. A pH-dependent chemical equilibrium is responsible for recruiting and anchoring (upon the conformational changes described in the present work) active GadB in this part of the cell. Membrane-localized GadB counteracts the low pH in two ways: by organicating protons (irreversibly incorporated into GABA, which is then exported) and by acting as a biological buffer, because of its ability to uptake at least 30 protons/hexamer.
Because of the impact of the acid resistance trait in pathogenesis and food microbiology (Foster, 2000), we expect the structural elucidation of GadB to be the first step towards a detailed mechanistic understanding of the glutamate-based acid resistance system. Moreover, knowing how the system functions might be relevant for both the design of novel antimicrobial agents and vaccine development.
Materials and methods
Analytical reagents
Vent polymerase was from New England Biolabs. Restriction enzymes were from Roche. DEAE–Sepharose and the DNA ligation kit were from Amersham-Pharmacia Biotech. Ingredients for bacterial growth were from Difco. Oligonucleotide synthesis and DNA sequencing were from the custom services of MWG Biotech. Gabase was from Sigma. Other chemicals were from Merck.
Structure determination
Recombinant E.coli wild-type GadB was purified in the holo form (100% PLP bound) as described previously (De Biase et al., 1996). Both native GadB and reduced GadB (GadBred) were used for crystallization. To prepare GadBred, 15 mg (280 nmol) of native GadB in 2 ml of 0.1 M sodium acetate pH 4.6, containing 0.1 mM DTT, were treated with 350 nmol of sodium cyanoborohydride (NaBH3CN). Reduction was controlled spectrophotometrically by following both the decrease in absorbance at 422 nm (λmax for the Schiff base), and the increase in absorbance at 334 nm (λmax for the reduced Schiff base). After extensive dialysis against 0.1 M sodium acetate pH 4.6, containing 0.1 mM DTT, GadBred was concentrated to 20 mg/ml using Centricon-30 devices (Amicon). Three different crystal forms were obtained using the vapour diffusion method. They belong to space groups P1 (form A), twinned P3 (form B, used only for structure solution) and P32 (form C) (Table I). Form A was obtained by mixing 1 µl of protein solution containing 5 mg/ml GadB, 100 mM Na acetate pH 4.6 with 1 µl of reservoir, containing 135 mM Na acetate pH 4.6, 700 mM Na formate, 13% PEG 4000. Forms B and C were obtained by mixing 1 µl of protein solution containing 20 mg/ml GadBred with 1 µl of reservoir solution containing 1.9–2.0 M (NH4)2SO4 and 100 mM Tris pH 7.6. These conditions follow, with some modification, those described in Malashkevich et al. (1998) for non-reduced GadB. Crystals of both forms occurred in the same drop. Data were collected at 100 K using synchrotron radiation (Table I). Form A was cryoprotected by adding 17% ethylene glycol to the reservoir solution, forms B and C were first transferred to a stabilizing solution containing 1.9 M (NH4)2SO4 and 100 mM Tris pH 7.6, then cryoprotected using a stabilizing solution like the previous one but also containing 33% (v/v) glycerol. All data were processed with DENZO (Otwinowski and Minor, 1997) and programs of the CCP4 suite (CCP4, 1994). Form B turned out to be merohedrally twinned. Data collection statistics are summarized in Table I.
The structure of GadB was solved using crystal forms B and C, with a combination of the MIRAS and MAD methods. An initial set of MIRAS phases was obtained using a Ta6Br12 cluster as heavy atom derivative. The resolution of the data did not allow the determination of the Ta cluster orientation, and the phasing power of the Ta cluster, treated as a pseudo-atom, fell rapidly beyond ∼6 Å. A three-wavelength MAD experiment was then carried out at beamline BW7A (DESY) on a poorly isomorphous mercury acetate derivative. Bijvoet difference Fourier analysis using the low resolution phases from the Ta cluster yielded six Hg positions, which were input into SHARP (de La Fortelle and Bricogne, 1997). Acceptable phasing power to 3.25 Å was obtained. Density modification yielded an interpretable electron density map. A preliminary model for the GadB monomer could be built and duplicated into a dimer using NCS deduced from some of the heavy atom positions. The dimer was used as a search model in AMoRe (Navaza, 1994) and three molecular replacement solutions for crystal form B were determined. Multiple crystal averaging with DMMULTI (with one dimer in form B and three in form C) lead to a high-quality electron density map, where a nearly complete hexameric GadBred model could be easily built in. The low-pH crystal form A (one hexamer per a.u.) was solved by molecular replacement with AMoRe. Crystal forms A and C were refined using CNS (Brünger et al., 1998). NCS restraints were imposed during refinement and released in the final stage for form A. The refinement statistics are given in Table II.
Table II. Refinement statistics.
| Crystal form | A (native low pH) | C (native reducedneutral pH) |
|---|---|---|
| Space group | P1 | P32 |
| Resolution range (Å) | 25–2.0 | 20–2.3 |
| Reflections used in refinement | 178 480 | 136 306 |
| R-factor (%)a | 18.3 | 20.0 |
| Rfree (%) (no. of reflections) | 21.3 (5238) | 23.3 (2749) |
| Number of atoms | 23 453 | 23 202 |
| Protein | 21 497 | 21 799 |
| Cofactor | 90 | 90 |
| Ligand | 24 | 0 |
| Solvent and buffer |
1842 |
1313 |
| R.m.s.ds | ||
| Bond lengths (Å) | 0.005 | 0.0065 |
| Bond angles (°) | 1.23 | 1.26 |
| NCS Cα atoms (Å) | 0.19 | 0.23 |
| R.m.s. ΔB NCS Cα atoms (Å2) | 2.2 | 2.6 |
| Wilson B-factor (Å2) | 15.9 | 39.1 |
| Mean B factor (all atoms, Å2) |
15.8 |
28.0 |
| Ramachandran plot regions (%) | ||
| Most favoured | 90.52 | 89.6 |
| Additionally allowed | 9.43 | 9.9 |
| Generously allowed | 0.05 | 0.5 |
| Disallowed | 0.0 | 0.0 |
aR-factor = (Σhkl||Fo| – |Fc||)/Σhkl|Fo|, where |Fo| and |Fc| are the observed and scaled calculated structure factor amplitudes, respectively.
The atomic coordinates and structure factors of GadB have been deposited in the Protein Data Bank, with entry codes 1PMM (low pH) and 1PMO (neutral pH).
Modelling of the PLP–glutamate complex (external aldimine) into the GadB active site
An atomic model and its corresponding topology and parameter files for phosphopyridoxyl-l-glutamate were obtained from the HIC-UP server (Kleywegt and Jones, 1998), and modified to obtain the Schiff base adduct (external aldimine) between PLP and Glu. The PLP–Glu model was first energy-minimized with X-PLOR (Brünger, 1992), then manually positioned into the GadB active site using the program O. The interactions between the protein and the external aldimine adduct were optimized manually by modifying torsion angles in the adduct. The orientation of the cofactor ring was changed by modifying the torsion angles θ, ψ and φ, as defined in McPhalen et al. (1992). This resulted in a rotation of ∼20° for the cofactor ring along the C5A–C2A axis, with the phosphate moiety essentially immobilized. The C5A–C2A axis was also slightly tilted. The resulting atomic model was energy-minimized with X-PLOR (300 cycles of Powell dynamics), with a linear distance-dependent dielectric function for electrostatic interactions. The topology and parameter files TOPH19.PRO and PARAM19.PRO were used. The minimization was carried out under harmonic restraints for the protein Cα positions.
Construction, expression and purification of GadBΔ1–14
Construction of the E.coli gadB deletion mutant GadBΔ1–14, where the first 14 residues of the mature polypeptide chain would be missing, was performed by PCR amplification on the entire gadB gene as cloned in pQgadB (De Biase et al., 1996). Two primers were used. The first was a mutagenic primer annealing from the 15th codon and containing an upstream additional nucleotide sequence was used to introduce an NcoI restriction site at the 5′-end, which provided the necessary ATG start codon (5′-GGCCATGGGTTCACGTTTTGGTGCG-3′). A second primer annealing over the C-terminus of wild-type gadB (5′-GGAAGC TTACCGTTAAACGTTATCAGGT-3′) was employed. The italicized sequences indicate the NcoI and HindIII restriction sites used for directional cloning of the PCR product into pQE60, following an already described cloning strategy (De Biase et al., 1996), while the underlined sequences are the start and stop codons. An A→G point mutation (double underlined) was introduced at the 15th codon (Asp→Gly) to allow efficient removal of the Met residue at the N-terminus by the bacterial methionine aminopeptidase. The newly generated plasmid pQgadBΔ1–14 was fully sequenced on both strands within the gadBΔ1–14 coding sequence to confirm the mutation, and was used to transform the E.coli host strain JM109/pREP4 (De Biase et al., 1996). The conditions used for expression and purification of GadBΔ1–14 are essentially as described for wild-type GadB (De Biase et al., 1996) except that expression of the mutant was prolonged for an additional hour following IPTG induction compared with the wild type. This was done to compensate for both reduced bacterial growth rate and decreased levels of expression (∼50%). The final yield of GadBΔ1–14 from a 2 l bacterial culture was ∼50% (35 mg) of that normally achieved with the wild-type enzyme (De Biase et al., 1996). Purity of the mutant, assessed by 12.5% SDS–PAGE, was judged to be 90% and the specific activity was ∼70% of that of wild-type GadB. The enzyme concentration and activity, and the PLP content were determined as previously described (Tramonti et al., 2002a).
Spectroscopic analyses
Absorption spectra were recorded on a Hewlett-Packard model 8452 diode-array spectophotometer. Curve fitting and statistical analyses were carried out using the data manipulation software Scientist (Micromath, Salt Lake City, UT). The pH-dependent absorbance variation of GadBΔ1–14 was analysed using the following equation:
where AbsHnE and AbsE are the absorbances of the completely protonated and unprotonated forms of the enzyme, K is the intrinsic dissociation constant, and n is the number of protons involved in the titration.
Cell fractionation
Cytoplasmic and membrane fractions from E.coli strain JM109/pREP4 containing either pQgadB or pQgadBΔ1–14 were obtained as follows. After IPTG induction, bacteria from a 1 l culture were harvested by centrifugation at 4200 g for 30 min at 4°C. The bacterial pellet was initially resuspended in 40 ml of 50 mM Tris–HCl pH 7.5, containing 1 mM DTT and a protease inhibitor cocktail (Complete, Roche). The resuspension was then divided into two 20 ml aliquots. One of the aliquots was brought to pH 5.6 by dropwise (few microlitres) addition of 8 N HCl. The two samples were then sonicated to disrupt cells and adjusted to the desired pH values (7.5 and 5.6, respectively), where necessary. An additional centrifugation at 4100 g for 10 min at 4°C was performed to remove cell debris and unbroken cells, thus generating the cell supernatants. The cytoplasmic and membrane fractions from each cell supernatant were separated via ultracentrifugation at 150 000 g for 1 h at 10°C. The pellet from the ultracentrifugation, representing the membrane fraction, was resuspended using 2 ml of 0.1 M Tris–HCl pH 8.0, containing 0.15 M NaCl, 5 mM EDTA and 0.5% lauroyl sarcosine. Solubilized proteins were separated from the insoluble material by ultracentrifugation at 150 000 g for 40 min at 10°C. Total protein content of each sample was determined using the detergent-compatible protein assay Micro BCA™ protein assay (Pierce). Immunoblot analysis for GadB detection was carried out using affinity-purified anti-GadB rabbit polyclonal antibodies (De Biase et al., 1999) and horseradish peroxidase-labelled secondary antibody provided with the BM chemiluminescence western blotting kit (Roche).
Acknowledgments
Acknowledgements
We wish to thank the staff of beamlines ID14-1, ESRF, Grenoble, France, X31 and BW7A, DESY, Hamburg, Germany and PX, SLS, Villigen, Switzerland. Financial support from the Swiss NCCR (Structural Biology) to M.G.G., from the Istituto Pasteur-Fondazione Cenci Bolognetti to D.D.B. and from the Italian MIUR (PRIN) to F.B. are gratefully acknowledged.
References
- Baum G., Lev-Yadun,S., Fridmann,Y., Arazi,T., Katsnelson,H., Zik,M. and Fromm,H. (1996) Calmodulin binding to glutamate decarboxylase is required for regulation of glutamate and GABA metabolism and normal development in plants. EMBO J., 15, 2988–2996. [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenhorn D., Phillips,J. and Slonczewski,J.L. (1999) Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol., 181, 2209–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F.R. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- Booth I.R. (1999) The regulation of intracellular pH in bacteria. In Derek,J.C. and Gardner,G. (eds), Bacterial Response to pH. John Wiley and Sons, Chichester, UK. [Google Scholar]
- Brünger A.T. (1992) X-PLOR Manual, Version 3.1. Yale University, New Haven, CT. [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Dominici,P., Borri-Voltattorni,C., Jansonius,J.N. and Malashkevich,V.N. (2001) Structural insight into Parkinson’s disease treatment from drug-inhibited DOPA decarboxylase. Nat. Struct. Biol., 8, 963–967. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cherrington C.A., Hinton,M., Mead,G.P. and Chopra,I. (1991) Organic acids: chemistry, antibacterial activity and practical applications. Adv. Microb. Physiol., 32, 249–259. [DOI] [PubMed] [Google Scholar]
- Coleman S.T., Fang,T.K., Rovinsky,S.A., Turano,F.J. and Moye-Rowley,W.S. (2001) Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J. Biol. Chem., 276, 244–250. [DOI] [PubMed] [Google Scholar]
- Cotter P.D., Gahan,C.G. and Hill,C. (2001) A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol., 40, 465–475. [DOI] [PubMed] [Google Scholar]
- De Biase D., Tramonti,A., John,R.A. and Bossa,F. (1996) Isolation, overexpression and biochemical characterization of the two isoforms of glutamic acid decarboxylase from Escherichia coli. Protein Expr. Purif., 8, 430–438. [DOI] [PubMed] [Google Scholar]
- De Biase D., Tramonti,A., Bossa,F. and Visca,P. (1999) The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol., 32, 1198–1211. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Foster J.W. (2000) Microbial response to acid stress. In Storz,G. and Hengge-Aronis,R. (eds), Bacterial Stress Responses. ASM Press, Washington, DC, pp. 99–115. [Google Scholar]
- Giannella R.A., Broitmann,S.A. and Zamcheck,N. (1972) Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro. Gut, 13, 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh B.M., Farooq,F.T., Barstad,D.N., Blankenhorn,D.L. and Slonczewski,J.L. (1996) A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol., 178, 3978–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Janin J. (1997) Specific versus non-specific contacts in protein crystals. Nat. Struct. Biol., 4, 973–974. [DOI] [PubMed] [Google Scholar]
- Jansonius J.N. (1998) Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol., 8, 759–769. [DOI] [PubMed] [Google Scholar]
- Jansonius J.N. and Vincent,M.G. (1987) Structural basis for catalysis by aspartate aminotransferase. In Jurnak,F.A. and McPherson,A. (eds), Biological Macromolecules & Assemblies: Active Sites of Enzymes, Vol. 3. J. Wiley & Sons, New York, NY, pp. 187–285. [Google Scholar]
- Kaiser J.T., Clausen,T., Bourenkow,G.P., Bartunik,H.D., Steinbacher,S. and Huber,R. (2000) Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly. J. Mol. Biol., 297, 451–464. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1998) Databases in protein crystallography. Acta Crystallogr. D., 54, 1119–1131. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Langsetmo K., Fuchs,J.A. and Woodward,C. (1991) The conserved, buried aspartic acid in oxidized Escherichia coli thioredoxin has a pKa of 7.5. Its titration produces a related shift in global stability. Biochemistry, 30, 7603–7609. [DOI] [PubMed] [Google Scholar]
- Lin J., Lee,I.S., Frey,J., Slonczewski,J.L. and Foster,J.W. (1995) Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri and Escherichia coli. J. Bacteriol., 177, 4097–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Smith,M.P., Chapin,K.C., Baik,H.S., Bennett,G.N. and Foster,J.W. (1996) Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol., 62, 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malashkevich V.N., De Biase,D., Markovic-Housley,Z., Schlunegger,M.P., Bossa,F. and Jansonius,J.N. (1998) Crystallization and preliminary X-ray analysis of the beta-isoform of glutamate decarboxylase from Escherichia coli. Acta Crystallogr. D, 54, 1020–1022. [DOI] [PubMed] [Google Scholar]
- McPhalen C.A., Vincent,M.G. and Jansonius,J.N. (1992) X-ray structure refinement and comparison of three forms of mitochondrial aspartate aminotransferase. J. Mol. Biol., 225, 495–517. [DOI] [PubMed] [Google Scholar]
- Meng S.Y. and Bennett,G.N. (1992) Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J. Bacteriol., 174, 2659–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D Photorealistic Molecular Graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 450, 157. [Google Scholar]
- O’Leary M.H. and Brummund,W.,Jr (1974) pH jump studies of glutamate decarboxylase. Evidence for a pH-dependent conformation change. J. Biol. Chem., 249, 3737–3745. [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. In Carter,C.W. and Sweet,R.M. (eds), Methods Enzymol., Vol. 276. Academic Press, pp. 307–328. [DOI] [PubMed] [Google Scholar]
- Rhee S., Silva,M.M., Hyde,C.C., Rogers,P.H., Metzler,C.M., Metzler,D.E. and Arnone,A. (1997) Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate. J. Biol. Chem., 272, 17293–17302. [PubMed] [Google Scholar]
- Sanner M.F., Spehner,J.-C. and Olson,A.J. (1996) Reduced surface: an efficient way to compute molecular surfaces. Biopolymers, 38, 305–320. [DOI] [PubMed] [Google Scholar]
- Shukuya R. and Schwert,G.W. (1960a) Glutamic acid decarboxylase. 1. Isolation procedures and properties of the enzyme. J. Biol. Chem., 235, 1649–1652. [PubMed] [Google Scholar]
- Shukuya R. and Schwert,G.W. (1960b) Glutamic acid decarboxylase. 2. Spectrum of the enzyme. J. Biol. Chem., 235, 1653–1657. [PubMed] [Google Scholar]
- Slonczewski J.L. and Foster,J.W. (1996) pH-regulated genes and survival at extreme pH. In Neidhardt,F.C., Ingraham,J.L. and Curtiss,R.,III (eds), Escherichia coli and Salmonella typhimurium., Vol. 1. ASM Press, Washington, DC, pp. 1539–1552. [Google Scholar]
- Smith D.K., Kassam,T., Singh,B. and Elliott,J.F. (1992) Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol., 174, 5820–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden W.A., Koutsia,N., Baum,G. and Fromm,H. (1996) Activation of a recombinant petunia glutamate decarboxylase by calcium/calmodulin or by a monoclonal antibody which recognizes the calmodulin binding domain. J. Biol. Chem., 271, 4148–4153. [DOI] [PubMed] [Google Scholar]
- Soghomonian J.J. and Martin,D.L. (1998) Two isoforms of glutamate decarboxylase: why? Trends Pharmacol. Sci., 19, 500–505. [DOI] [PubMed] [Google Scholar]
- Sukhareva B.S., Tikhonenko,A.S. and Darii,E.L. (1994) Study of the quaternary structure of glutamate carboxylase from Escherichia coli. Mol. Biol. (Mosk.), 28, 1407–1411. [PubMed] [Google Scholar]
- To C.M. (1971) Quaternary structure of glutamate decarboxylase of Escherichia coli as revealed by electron microscopy. J. Mol. Biol., 59, 215–217. [DOI] [PubMed] [Google Scholar]
- Tramonti A., John,R.A., Bossa,F. and De Biase,D. (2002a) Contribution of Lys276 to the conformational flexibility of the active site of glutamate decarboxylase from Escherichia coli. Eur. J. Biochem., 269, 4913–4920. [DOI] [PubMed] [Google Scholar]
- Tramonti A., Visca,P., De Canio,M., Falconi,M. and De Biase,D. (2002b) Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol., 184, 2603–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman S.R. and Small,P.L. (1996) Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol. Microbiol., 21, 925–940. [DOI] [PubMed] [Google Scholar]
- Zik M., Arazi,T., Snedden,W.A. and Fromm,H. (1998) Two isoforms of glutamate decarboxylase in Arabidopsis are regulated by calcium/calmodulin and differ in organ distribution. Plant Mol. Biol., 37, 967–975. [DOI] [PubMed] [Google Scholar]