Abstract
Huntington’s disease (HD) is a hereditary neurodegenerative disorder presenting with chorea, dementia, and extensive striatal neuronal death. The mechanism through which the widely expressed mutant HD gene mediates a slowly progressing striatal neurotoxicity is unknown. Glutamate receptor-mediated excitotoxicity has been hypothesized to contribute to the pathogenesis of HD. Here we show that transgenic HD mice expressing exon 1 of a human HD gene with an expanded number of CAG repeats (line R6/1) are strongly protected from acute striatal excitotoxic lesions. Intrastriatal infusions of the N-methyl-d-aspartate (NMDA) receptor agonist quinolinic acid caused massive striatal neuronal death in wild-type mice, but no damage in transgenic HD littermates. The remarkable neuroprotection in transgenic HD mice occurred at a stage when they had not developed any neurological symptoms caused by the mutant HD gene. At this stage there was no change in the number of striatal neurons and astrocytes in untreated R6/1 mice, although the striatal volume was decreased by 17%. Moreover, transgenic HD mice had normal striatal levels of NMDA receptors, calbindin D28k (calcium buffer), superoxide dismutase activity (antioxidant enzyme), Bcl-2 (anti-apoptotic protein), heat shock protein 70 (stress-induced anti-apoptotic protein), and citrate synthase activity (mitochondrial enzyme). We propose that the presence of exon 1 of the mutant HD gene induces profound changes in striatal neurons that render these cells resistant to excessive NMDA receptor activation.
Huntington’s disease (HD) is an autosomal dominant, progressive neurodegenerative disorder. Typically, symptoms appear in midlife and are characterized by involuntary choreiformic movements, cognitive impairment, and emotional disturbances (1). Although there is some atrophy of the neocortex, the major site of pathology is the striatum, where up to 90% of the neurons may be lost (2, 3). The disease mutation consists of an expanded CAG trinucleotide repeat in exon 1 of a gene coding for a protein with unknown function called huntingtin (4). Whereas unaffected individuals typically display 6–34 CAG repeats in this gene, patients with HD usually exhibit between 36 and 120 CAG repeats (5). The mechanism through which mutant huntingtin mediates neurotoxicity is unknown. It has been suggested that a mutant huntingtin exhibits a conformational change (6), which may cause abnormal interactions with other proteins or between mutant huntingtin molecules (7–11). Such interactions may explain the intranuclear inclusions containing huntingtin that are found in brains of human HD patients (12). Huntingtin is ubiquitously expressed in peripheral tissues and in the central nervous system, with no specific enrichment in the striatum (13–16). Therefore it is particularly intriguing that the neuropathology is focused to the striatum.
The striatum receives dense excitatory glutamatergic input from the corticostriatal pathway. For more than 20 years, glutamate receptor-mediated cell death—i.e., excitotoxicity—has been hypothesized to be involved in the pathogenesis of HD (17–21). Excitotoxic striatal lesions produced by local injections of quinolinic acid, an agonist of the N-methyl-d-aspartate (NMDA) receptor subtype of glutamate receptors, mimic many of the neurochemical and neuropathological characteristics of HD (22–24). For example, there is a selective loss of medium-sized spiny striatal projection neurons (the predominant striatal cell type), relative sparing of striatal aspiny interneurons containing somatostatin, neuropeptide Y, and NADPH-diaphorase (22–24), and sparing of afferents to the striatum and fibers of passage (25). Importantly, NMDA receptors are preferentially lost in the striatum of postmortem brains from HD patients, suggesting that neurons with NMDA receptors degenerate (26).
A recently developed transgenic mouse model that expresses exon 1 of the human HD gene with an expanded number of CAG trinucleotide repeats has provided an important advance in HD research (27). Three different lines [R6/1, (CAG)115; R6/2, (CAG)145; and R6/5 (CAG)128–156] that display progressive neurological symptoms have been created. Importantly, striatal neurons of all affected transgenic lines exhibit intranuclear inclusions containing huntingtin prior to onset of symptoms (28). In addition, the R6/2 line has been reported to exhibit metabolic dysfunction in the brain (29).
In the present report, we investigated whether the chronic expression of mutant huntingtin changes the reaction of striatal neurons to excessive NMDA receptor activation. We studied striatal excitotoxic lesions produced by infusion of quinolinic acid, an endogenous NMDA receptor agonist, in 18-week-old presymptomatic transgenic (line R6/1) and wild-type littermate mice.
MATERIALS AND METHODS
Animals and Quinolinic Acid Lesion.
Heterozygous transgenic R6/1 males of the CBA × C57BL/6 strain were purchased from The Jackson Laboratory and maintained by crossing carrier males with CBA females. Mice were genotyped by using a PCR assay. Blood samples for determination of glucose concentration were obtained from the tail and analyzed with a Glucometer II (model 5529; Ames, Miles Laboratories). Quinolinic acid (Sigma) was dissolved in 0.1 M phosphate buffered-saline (pH 7.4). Under halothane anesthesia, the mice received intrastriatal injections of 30 nmol (1 μl) of quinolinic acid from a 2-μl Hamilton syringe at the following stereotaxic coordinates: 0.7 mm rostral to bregma; 1.9 mm lateral to midline; 2.5 mm ventral from the dural surface; with the tooth-bar set at zero. The toxin was injected over 2 min, and afterwards the cannula was left in place for an additional 3 min. Body temperature was controlled with a heating pad set at 37°C. Mortality after quinolinic acid injection of transgenic HD mice was 4/18 and of wild-type littermate mice, 3/15.
Perfusion and Immunohistochemistry.
Mice were anesthetized and perfusion fixed (4% paraformaldehyde). Brains were postfixed and dehydrated in 20% sucrose/0.1 M phosphate buffer. Coronal sections were cut (40 μm), and every third section was processed for immunohistochemistry. For dopamine- and cAMP-regulated phosphoprotein of a molecular mass of 32 kDa (DARPP-32) and glial fibrillary acidic protein (GFAP) immunohistochemistry, endogenous peroxidase was quenched with 3% hydrogen peroxide. Free-floating sections were incubated in 10% horse serum/0.2% Triton X-100/0.1 M phosphate-buffered saline for 1 h at room temperature, followed by a reaction overnight with an antibody against GFAP (1:200; Dakopatts, Glostrup, Denmark) or DARPP-32 (1:20 000; donated by P. Greengard and H. Hemmings, Weill Medical College, New York). Sections were incubated with the corresponding biotinylated secondary antibody (1:200) for 1 h, and bound antibody was visualized by using the ABC system (Vectastain ABC Kit, Vector Laboratories), with 3,3′-diaminobenzidine as chromogen. GFAP-stained sections were counterstained with cresyl violet.
Terminal Deoxynucleotidyltransferase-Mediated dUTP-Biotin Nick End Labeling (TUNEL).
In 10-μm-thick cryostat sections, the number of TUNEL-positive cells was measured with a commercially available kit according to the enclosed protocol (Apoptag; Oncor, Gaithersburg, MD). After staining, sections were incubated with Hoechst 33342 (2 μg/ml for 5 min; Molecular Probes) to label all nuclei. Positive controls were carried out by treating sections with DNase I (1 μg/ml; Sigma) before the assay.
Fluoro-Jade.
Fluoro-Jade staining was performed as described by Schmued et al. (30). Briefly, 10-μm-thick brain sections were cut, mounted, dried, and immersed in 100% ethanol, followed by 70% ethanol. Sections were then treated with 0.06% potassium permanganate for 15 min. After rinsing, sections were immersed in Fluoro-Jade (0.001% Fluoro-Jade/0.1% acetic acid; Histo-Chem, Jefferson, AR) for 30 min, followed by a 5-min incubation with Hoechst 33342 (2 μg/ml).
Immunoblotting.
Striata were excised and snap-frozen in liquid nitrogen. They were homogenized in 100 μl of buffer containing 150 mM NaCl, 50 mM Tris⋅HCl (pH 7.6), 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mM EGTA, and protease inhibitors. Protein content of the homogenates was determined by the bicinchoninic acid method (Bio-Rad). Equal amounts of protein were separated by SDS/polyacrylamide gel electrophoresis (12% for calbindin D28k and Bcl-2; 8% for NMDA-R1 and heat shock protein 70), and electroblotted onto nitrocellulose membranes (Hybond ECLTM, Amersham–Buchler). Protein content was routinely controlled by redeveloping the membranes with an anti-actin antibody (1:100,000 Chemicon, clone C4). The membranes were blocked with 5% (wt/vol) low-fat milk and 1% fetal calf serum in a buffer containing 10 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, and 0.5% Tween 20, and then incubated with the primary antibodies (calbindin D28, Sigma, 1:200; Bcl-2: clone3F11, PharMingen, 1:200; NMDA-R1, Chemicon, 1:100; heat shock protein 70, clone C92F3A-5, Stressgen, Victoria, BC, 1:1000). Blots were developed with IgG-horseradish peroxidase followed by enhanced chemiluminescence detection (ECL, Amersham).
Citrate Synthase and Superoxide Dismutase Activity.
Homogenization and protein determination of striatal tissue were performed essentially as described for immunoblotting. Citrate synthase activity was measured spectrophotometrically in striatal tissue homogenates as previously described (31). The activity of superoxide dismutase was quantified spectrophotometrically at 340 nm in striatal homogenates according to Paoletti et al. (32). One unit of activity is defined as the amount of enzyme that inhibits 50% of the oxidation of NADH induced by superoxide. When validating the method we found a 98% inhibition of NADH oxidation with purified superoxide dismutase (100 units/ml; Sigma).
Cell Counting and Data Analysis.
The number of cells positive for DARPP-32, GFAP, cresyl violet, TUNEL, Fluoro-Jade, or Hoechst 33342 was assessed on blind-coded slides with a semi-automated stereological system [Olympus C.A.S.T. Grid system (version 1.10), composed of an Olympus BX50 microscope and a X–Y–Z step motor stage run by a computer). The area of the striatum was delineated and a counting frame was randomly placed within the striatum to mark the first area to be sampled. The frame was then systematically moved through the striatum. The number of positive cells was then extrapolated according to a stereological algorithm (33). The lesion volume was determined by using the same equipment as described above. Cell counts in untreated mice and all volume measurements were performed on sections from the whole striatum. When assessing the number of dying and surviving cells after quinolinic acid infusion, the number of cells was investigated on five serial sections (120 μm apart) surrounding the cannula track. All data were analyzed by unpaired two-tailed Student’s t test and presented as mean ± SD.
RESULTS
Histological and Phenotypic Characteristics of Transgenic HD Mice.
In our colony of R6/1 HD mice, the transgenic mice began to exhibit behavioral changes after 22–26 weeks. These were initially subtle and included hunched posture, tremor, and poor grooming. The histological features of the intact striatum of 18-week-old presymptomatic transgenic HD mice were compared with wild-type littermate controls. The total number of cell bodies (including both neurons and glia) in the striatum of transgenic HD mice was unchanged compared with wild-type littermates, when assessed in cresyl violet-stained sections (Table 1). The number of medium-sized spiny striatal projection neurons, which is the most affected neuronal population in HD (34, 35), was investigated with immunohistochemistry for DARPP-32. In transgenic mice, the number of DARPP-32-expressing neurons was not different from that seen in wild-type littermates (Table 1). Also, regarding the number of GFAP-positive astrocytes, there was no difference between R6/1 transgenic and wild-type mice (Table 1). In addition, we observed no dying cells within the striatum of untreated transgenic mice when sections were stained for TUNEL or Fluoro-Jade (data not shown). Even though there were no changes in cell number, the volume of striatum in transgenic mice was reduced by 17% compared with wild-type littermates (Table 1).
Table 1.
Cell numbers and striatal volume in wild-type and transgenic animals
| Mice | No. of cells × 10−5
|
Striatal volume, mm3 | ||
|---|---|---|---|---|
| Total | DARPP-32+ | GFAP+ | ||
| Wild-type | 8.1 ± 1.1 | 3.9 ± 0.4 | 0.33 ± 0.11 | 6.0 ± 0.15 |
| Transgenic | 8.0 ± 0.4 | 4.0 ± 0.4 | 0.38 ± 0.10 | 5.0 ± 0.19* |
Data are given as mean ± SD; n = 3 wild-type mice; n = 4 transgenic mice. ∗, P < 0.01.
Cell Death After Intrastriatal Infusion of Quinolinic Acid.
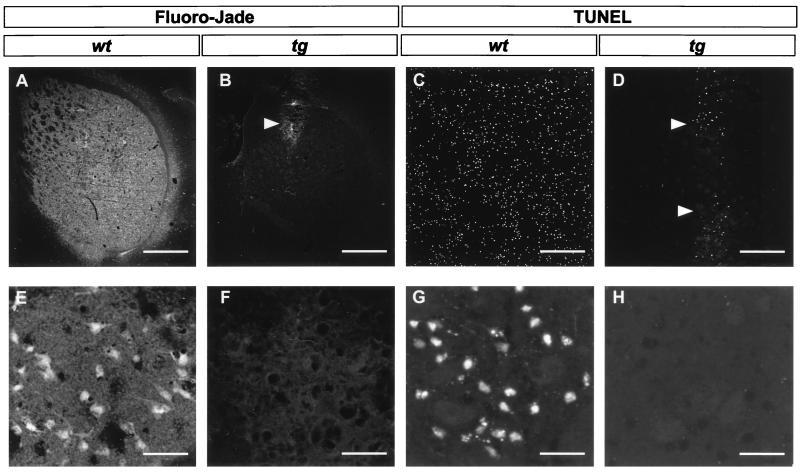
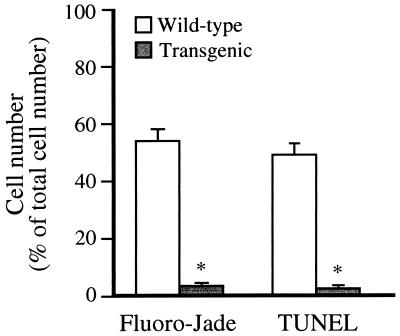
To investigate whether the chronic expression of mutant HD gene changes the reaction of striatal neurons to excessive NMDA receptor activation, we performed intrastriatal infusions of quinolinic acid in transgenic HD and wild-type littermate mice. The pattern of cell death 48 h after infusion of quinolinic acid was studied with Fluoro-Jade (Fig. 1 A, B, E, and F) and TUNEL (Fig. 1 C, D, G, and H) staining. Fluoro-Jade, a fluorescent marker for dying neurons (29), stained 54% of the striatal cells in the five coronal sections closest to the site of the injection in wild-type littermate controls (Fig. 1 A and E and Fig. 2). In contrast, the transgenic HD mice were almost totally resistant to intrastriatal injections of quinolinic acid. Only a very small number of cells along the cannula track were found to be labeled with Fluoro-Jade (Fig. 1 B and F and Fig. 2). On adjacent sections we used the TUNEL method, which labels nuclei with double-strand DNA breaks as an indication of apoptosis. This technique also showed vast labeling throughout the striatum of wild-type littermate mice (49%) and labeling of only a very small number of cells (3%) along the track of the cannula used to deliver quinolinic acid into the brains of transgenic HD mice (Fig. 1 C, D, G, and H and Fig. 2). The numbers of cells labeled by Fluoro-Jade or TUNEL in the transgenic HD mice were similar to the numbers of labeled cells after sham injection of saline into the striatum (data not shown). Therefore the dying neurons in the transgenic HD brains may be the result of mechanical damage caused by the cannula, rather than an effect of quinolinic acid. Control experiments showed that intrastriatal quinolinic acid injections in normal female CBA mice result in a size of lesion, as evaluated with the Fluoro-Jade staining on coronal brain sections, similar to that observed in wild-type littermate mice (data not shown).
Figure 1.
Micrographs of striatal sections prepared from brains 48 h after intrastriatal quinolinic acid injection, labeled with the two fluorescent cell death markers Fluoro-Jade (A, B, E, and F) or TUNEL (C, D, G, and H). Sections from wild-type mice (wt; A, C, E, and G) contain numerous stained cells. In sections from transgenic mice (tg; B, D, F, and H) only very few cells are labeled along the cannula track (arrowheads). [Bar = 720 μm (A and B), 250 μm (C and D), and 30 μm (E–H).]
Figure 2.
Quantification of the number of striatal Fluoro-Jade- and TUNEL-positive cells 48 h after intrastriatal quinolinic acid injection. The number of positive cells was counted in the five coronal sections closest to the site of injection and is expressed as a mean (error bar = SD) percentage of the total number of cells in the same sections (n = 5 per group). ∗, P < 0.0001.
Cell Survival After Intrastriatal Infusion of Quinolinic Acid.
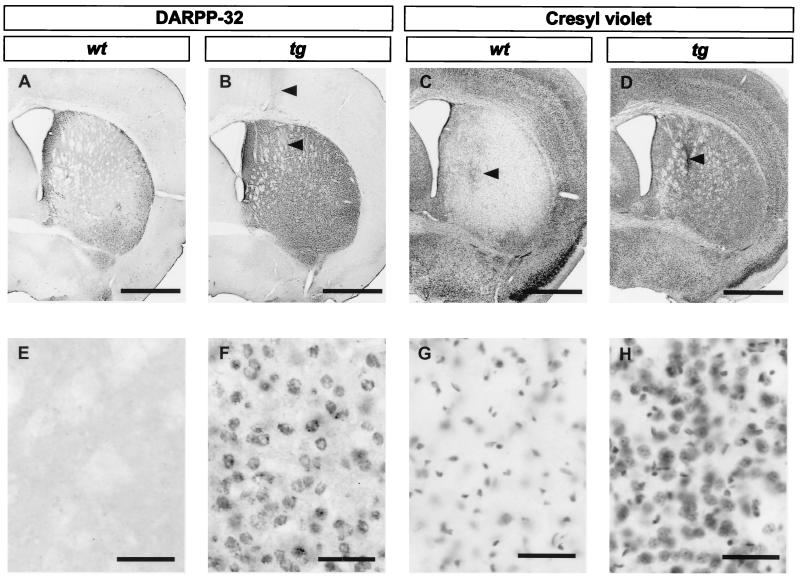
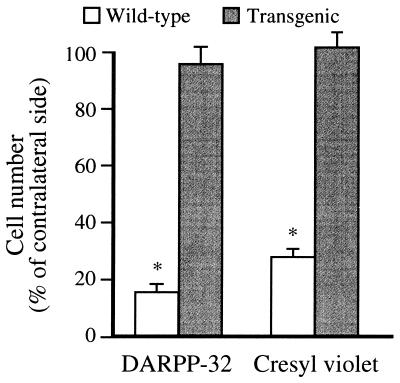
We employed DARPP-32 immunohistochemistry and cresyl violet staining to study the surviving neurons 14 days after intrastriatal quinolinic acid injection in both transgenic HD and wild-type littermate mice (Fig. 3). Consistent with the observation of a high degree of cell death in wild-type littermates at 48 h after quinolinic acid injection, 85% of the DARPP-32-immunostained striatal projection neurons in the five sections closest to the stereotactic injection were lost 14 days after quinolinic acid-induced lesion (Fig. 3 A and E, and Fig. 4). In contrast, only 3% of the DARPP-32-labeled neurons had disappeared in the transgenic HD mice (Fig. 3 B and F and Fig. 4). Since the decreased number of DARPP-32 immunopositive neurons could be caused by down-regulation of DARPP-32, we also assessed the total number of nonpyknotic cells (both neurons and glia) in cresyl violet-stained sections (Fig. 3 C, D, G, and H). In wild-type littermates, a 72% decrease in number of cresyl violet-stained cells was observed (Fig. 3 C and G and Fig. 4). In contrast, in the brains of transgenic HD mice the number of striatal cells was not decreased, but there was an increase in cell density close to the cannula tract, possibly caused by cell proliferation/infiltration (Fig. 3 D and H and Fig. 4). In addition, we measured lesion volume on cresyl violet-stained sections. The lesion volume in wild-type mice was 3.90 ± 0.13 mm3 and in transgenic HD mice only 0.04 ± 0.01 mm3.
Figure 3.
Micrographs of striatal sections prepared from brains 14 days after intrastriatal quinolinic acid injection, processed for DARPP-32 immunohistochemistry (A, B, E, and F) or cresyl violet staining (C, D, G, and H). Sections from wild-type mice (wt; A, C, E, and G) show massive loss of DARPP-32- and cresyl violet-stained cells. Sections from transgenic mice (tg; B, D, F, and H) reveal only a minor loss of DARPP-32- positive neurons (B and F) and an increased number of cresyl violet-stained cells (D and H) along the cannula track (arrowheads). [Bar = 1.0 mm (A–D) and 50 μm (E–H).]
Figure 4.
Quantification of the number of striatal DARPP-32- and cresyl violet- labeled cells 14 days after intrastriatal quinolinic acid injection. The number of positive cells was counted in the five coronal sections closest to the site of injection and is expressed as a mean (error bar = SD) percentage of the number of positive cells on the contralateral unlesioned side (n = 9 transgenic mice; n = 7 wild-type mice). ∗, P < 0.0001.
Biochemical Markers.
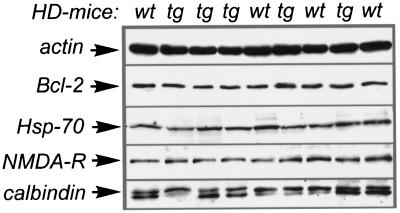
We addressed possible mechanisms underlying the marked reduction in sensitivity of the striatum of R6/1 transgenic HD mice to quinolinic acid-induced toxicity. Thus we examined several proteins that are of relevance to NMDA receptor-mediated toxicity in striatal tissue from untreated R6/1 transgenic HD mice and wild-type littermates. First, we observed normal levels of the NMDA-NR1 subunit of the NMDA receptor in transgenic HD mice when we used immunoblotting, indicating that NMDA receptor level is unchanged (Fig. 5). Similarly, we saw no changes in the levels of the calcium-binding protein calbindin D28k or in the levels of the two antiapoptotic proteins bcl-2 and heat shock protein 70 (Fig. 5). In addition, we measured the activity of the antioxidant enzyme superoxide dismutase and found no significant increase in transgenic HD mice (wild-type = 86.7 ± 9.8 units/mg of protein; transgenic = 99.2 ± 6.3 units/mg of protein; P = 0.06; n = 5 per group). Citrate synthase activity (a general marker for mitochondrial density) was also measured. Again there was no difference in enzymatic activity in striatal tissue from transgenic and wild-type littermates (wild-type = 208 ± 12 units/mg of protein; transgenic = 200 ± 8 units/mg of protein; P = 0.28; n = 5 per group). Because it has been reported that mice of the R6/2 line may develop diabetes (36), we measured blood glucose levels in both fasting and nonfasting mice. However, transgenic R6/1 mice have normal blood glucose levels (fasting blood glucose in wild-type mice = 6.5 ± 0.1 mM, transgenic = 6.5 ± 0.4 mM; nonfasting blood glucose in wild-type mice = 11.0 ± 0.7 mM, transgenic = 10.2 ± 0.3 mM; n = 5 per group).
Figure 5.
Quantification of proteins possibly regulating susceptibility to excitotoxic damage. Striata of randomly chosen wild-type (wt) or transgenic (tg) mice were excised and homogenized. Proteins were separated by SDS/polyacrylamide gel electrophoresis and detected by immunoblot (Hsp-70, heat shock protein 70; NMDA-R, NMDA receptor subunit NMDA-NR1). The anti-calbindin D28k antibody recognized a nonspecific band at 26 kDa in addition to the 28-kDa calbindin band (arrow).
DISCUSSION
We have shown that presymptomatic R6/1 transgenic HD mice are strongly protected from NMDA receptor-mediated excitotoxicity induced by intrastriatal injections of quinolinic acid, indicating that the presence of exon 1 of the mutant HD gene induces profound changes in striatal neurons. After intrastriatal injection of quinolinic acid the transgenic HD mice exhibited total protection of striatal projection neurons stained by DARPP-32 immunohistochemistry and an almost complete absence of cells stained by the fluorescent cell death markers Fluoro-Jade and TUNEL.
The vast majority of previous work regarding the natural history of transgenic mice expressing exon 1 of a human HD gene with an expanded number of CAG trinucleotide repeats has focused on the R6/2 line (27, 28, 37). The number of CAG repeats in the R6/2 mice is 145, whereas it is 115 in the R6/1 mice used in this study. As a result R6/2 mice develop symptoms already at 9–11 weeks of age, including resting tremor, irregular gait, stereotypic and abrupt movements, epileptic seizures, and diabetes (27, 36). In our colony of R6/1 transgenic HD mice, the transgenic mice begin to exhibit neurological symptoms after 22–26 weeks. Presymptomatic 18-week-old mice of the R6/1 line exhibited a reduced striatal volume (17%), but the numbers of cresyl violet-stained cells, DARPP-32-positive projection neurons, and GFAP-stained astrocytes were still not reduced and no ongoing cell death was observed. Taken together, these observations suggest that cell size and/or extracellular matrix is reduced because of the HD mutation. Brain size is also reduced by 19% in the R6/2 line (27), but no quantification of cell number has hitherto been reported.
The observed dramatic reduction in excitotoxic striatal damage in presymptomatic transgenic R6/1 mice was not caused by a decreased expression of NMDA receptors, since we found normal levels of striatal NMDA receptors when we used immunoblotting. These data are in agreement with earlier findings by Cha et al. (37), who studied the expression of glutamate receptors in the R6/2 line of mice. The calcium-binding protein calbindin D28k has been shown to protect against excitotoxicity (38), and there is evidence that calbindin D28k is increased in a subpopulation of striatal neurons in HD patients (39), maybe a result of excitotoxic stress (40). However, immunoblotting revealed that the level of calbindin D28k was unchanged in transgenic HD mice. In addition, the formation of oxygen free radicals plays an important role in excitotoxicity (41), and the antioxidant enzyme superoxide dismutase protects against quinolinic acid-induced striatal lesions (42). Nevertheless, the superoxide dismutase activity in the R6/1 transgenic HD mice was not significantly increased.
Widespread neuronal apoptosis occurs after excitotoxic brain damage (43). This observation was well illustrated by the abundance of TUNEL-positive neurons in the quinolinic acid-injected striatum in the present study. Therefore, it seemed valid to hypothesize that there is an up-regulation of antiapoptotic defenses in the brains of R6/1 mice. However, we did not observe any changes in basal striatal levels of Bcl-2, a well known antiapoptotic protein (44), or in the levels of heat shock protein 70, a stress-induced protein that has been shown to counteract apoptosis (45). Similarly, there were no alterations of citrate synthase, suggesting that there was no major change in the mitochondrial density in R6/1 transgenic mice. This observation is important because mitochondria play a pivotal role in excitotoxic damage (46).
The reduced sensitivity to NMDA receptor-mediated cell death in the transgenic HD mice is particularly interesting in view of the longstanding hypothesis that excitotoxicity plays an important role in HD pathogenesis (19–21, 47). One can speculate whether the transgene in the R6/1 mice induces a protracted, sublethal grade of excitotoxicity in the striatal neurons, which results in a response with increased recruitment of cellular defenses against excitotoxic demise—e.g., by means of up-regulation of growth factors and their receptors (48). A sublethal grade of excitotoxicity could be caused either by changes in glutamate transmission or by a secondary mechanism involving an impaired metabolic function (for review, see refs. 20 and 47). Interestingly, in vitro studies have shown that incubation with sublethal concentrations of NMDA results in a protein synthesis-dependent neuroprotection against excitotoxicity (49). An analogous form of neuroprotection, called ischemic tolerance, is known to develop in the brain after minor ischemic insults in experimental animals. After the priming sublethal ischemic insult the neurons can survive when subjected to a marked ischemia that would normally kill them (for review see ref. 50). The mechanism of ischemic tolerance is still unknown, but NMDA receptor activation and protein synthesis are required (50).
The changes we observed were very marked and robust, indicating that the presence of exon 1 of the mutant HD gene induces profound changes in the physiology of striatal neurons. In HD, a reduced sensitivity to NMDA receptor-mediated cell death could offer a possible explanation for the relatively slow and protracted development of neuropathology. Further studies in the R6/1 mouse are required to determine whether the reduced sensitivity to NMDA receptor-mediated cell death is also apparent in neocortex, which displays some degeneration in clinical HD, or other regions that are unaffected in the clinical human disorder. Moreover, it will be of interest to establish whether the transgenic neurons are also less sensitive to injury induced by excessive stimulation of other glutamate receptor subtypes. Ultimately, the unraveling of mechanisms underlying the marked reduction of striatal sensitivity to excessive NMDA receptor activation in R6/1 transgenic HD mice may help us to understand the slow pathogenic process in HD.
Acknowledgments
We acknowledge the technical assistance of T. Björklund, H. Naumann, B. Lindberg, and B. Mattsson. This study was supported by grants from the Hereditary Disease Foundation, the Swedish Medical Research Council, and the Deutsche Forschungsgemeinschaft. O.H. and R.F.C. are supported by the National Network in Neuroscience and the Thorsten and Elsa Segerfalk Foundation, respectively.
ABBREVIATIONS
- HD
Huntington’s disease
- DARPP-32
dopamine- and cAMP-regulated phosphoprotein of a molecular mass of 32 kDa
- GFAP
glial fibrillary acidic protein
- NMDA
N-methyl-d-aspartate
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling
Note Added in Proof
Since this work was submitted, we have obtained similar results when performing intrastriatal injections of quinolinic acid (30 nmol) in 6-week-old presymptomatic transgenic HD mice of the R6/2 line (27).
References
- 1.Harper P S. Huntington’s Disease. London: Saunders; 1996. [Google Scholar]
- 2.Vonsattel J P, Myers R H, Stevens T J, Ferrante R J, Bird E D, Richardson E P., Jr J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Hedreen J C, Folstein S E. J Neuropathol Exp Neurol. 1995;54:105–120. doi: 10.1097/00005072-199501000-00013. [DOI] [PubMed] [Google Scholar]
- 4.The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald M E, Gusella J F. Curr Opin Neurobiol. 1996;6:638–643. doi: 10.1016/s0959-4388(96)80097-3. [DOI] [PubMed] [Google Scholar]
- 6.Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, Weber C, David G, Tora L, et al. Nature (London) 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- 7.Li X J, Li S H, Sharp A H, Nucifora F C, Jr, Schilling G, Lanahan A, Worley P, Snyder S H, Ross C A. Nature (London) 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- 8.Burke J R, Enghild J J, Martin M E, Jou Y S, Myers R M, Roses A D, Vance J M, Strittmatter W J. Nat Med. 1996;2:347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- 9.Bao J, Sharp A H, Wagster M V, Becher M, Schilling G, Ross C A, Dawson V L, Dawson T M. Proc Natl Acad Sci USA. 1996;93:5037–5042. doi: 10.1073/pnas.93.10.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalchman M A, Koide H B, McCutcheon K, Graham R K, Nichol K, Nishiyama K, Kazemi-Esfarjani P, Lynn F C, Wellington C, Metzler M, et al. Nat Genet. 1997;16:44–53. doi: 10.1038/ng0597-44. [DOI] [PubMed] [Google Scholar]
- 11.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 12.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 13.Strong T V, Tagle D A, Valdes J M, Elmer L W, Boehm K, Swaroop M, Kaatz K W, Collins F S, Albin R L. Nat Genet. 1993;5:259–265. doi: 10.1038/ng1193-259. [DOI] [PubMed] [Google Scholar]
- 14.Gutekunst C A, Levey A I, Heilman C J, Whaley W L, Yi H, Nash N R, Rees H D, Madden J J, Hersch S M. Proc Natl Acad Sci USA. 1995;92:8710–8714. doi: 10.1073/pnas.92.19.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharp A H, Loev S J, Schilling G, Li S H, Li X J, Bao J, Wagster M V, Kotzuk J A, Steiner J P, Lo A, et al. Neuron. 1995;14:1065–1074. doi: 10.1016/0896-6273(95)90345-3. [DOI] [PubMed] [Google Scholar]
- 16.Trottier Y, Devys D, Imbert G, Saudou F, An I, Lutz Y, Weber C, Agid Y, Hirsch E C, Mandel J L. Nat Genet. 1995;10:104–110. doi: 10.1038/ng0595-104. [DOI] [PubMed] [Google Scholar]
- 17.Coyle J T, Schwarcz R. Nature (London) 1976;263:244–246. doi: 10.1038/263244a0. [DOI] [PubMed] [Google Scholar]
- 18.McGeer E G, McGeer P L. Nature (London) 1976;263:517–519. doi: 10.1038/263517a0. [DOI] [PubMed] [Google Scholar]
- 19.DiFiglia M. Trends Neurosci. 1990;13:286–289. doi: 10.1016/0166-2236(90)90111-m. [DOI] [PubMed] [Google Scholar]
- 20.Albin R L, Greenamyre J T. Neurology. 1992;42:733–738. doi: 10.1212/wnl.42.4.733. [DOI] [PubMed] [Google Scholar]
- 21.Sharp A H, Ross C A. Neurobiol Dis. 1996;3:3–15. doi: 10.1006/nbdi.1996.0002. [DOI] [PubMed] [Google Scholar]
- 22.Beal M F, Kowall N W, Ellison D W, Mazurek M F, Swartz K J, Martin J B. Nature (London) 1986;321:168–171. doi: 10.1038/321168a0. [DOI] [PubMed] [Google Scholar]
- 23.Beal M F, Ferrante R J, Swartz K J, Kowall N W. J Neurosci. 1991;11:1649–1659. doi: 10.1523/JNEUROSCI.11-06-01649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrante R J, Kowall N W, Cipolloni P B, Storey E, Beal M F. Exp Neurol. 1993;119:46–71. doi: 10.1006/exnr.1993.1006. [DOI] [PubMed] [Google Scholar]
- 25.Schwarcz R, Whetsell W O, Jr, Mangano R M. Science. 1983;219:316–318. doi: 10.1126/science.6849138. [DOI] [PubMed] [Google Scholar]
- 26.Young A B, Greenamyre J T, Hollingsworth Z, Albin R, D’Amato C, Shoulson I, Penney J B. Science. 1988;241:981–983. doi: 10.1126/science.2841762. [DOI] [PubMed] [Google Scholar]
- 27.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies S W, Bates G P. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 28.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 29.Bogdanov M B, Ferrante R J, Kuemmerle S, Klivenyi P, Beal M F. J Neurochem. 1998;71:2642–2644. doi: 10.1046/j.1471-4159.1998.71062642.x. [DOI] [PubMed] [Google Scholar]
- 30.Schmued L C, Albertson C, Slikker W., Jr Brain Res. 1997;751:37–46. doi: 10.1016/s0006-8993(96)01387-x. [DOI] [PubMed] [Google Scholar]
- 31.Shepherd D, Garland P B. Methods Enzymol. 1969;13:11–16. [Google Scholar]
- 32.Paoletti F, Aldinucci D, Mocali A, Caparrini A. Anal Biochem. 1986;154:536–541. doi: 10.1016/0003-2697(86)90026-6. [DOI] [PubMed] [Google Scholar]
- 33.West M J, Slomianka L, Gundersen H J. Anat Rec. 1991;231:482–497. doi: 10.1002/ar.1092310411. [DOI] [PubMed] [Google Scholar]
- 34.Graveland G A, Williams R S, DiFiglia M. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 35.Reiner A, Albin R L, Anderson K D, D’Amato C J, Penney J B, Young A B. Proc Natl Acad Sci USA. 1988;85:5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurlbert M S, Zhou W, Wasmeier C, Kaddis F G, Hutton J C, Freed C R. Diabetes. 1999;48:649–651. doi: 10.2337/diabetes.48.3.649. [DOI] [PubMed] [Google Scholar]
- 37.Cha J H, Kosinski C M, Kerner J A, Alsdorf S A, Mangiarini L, Davies S W, Penney J B, Bates G P, Young A B. Proc Natl Acad Sci USA. 1998;95:6480–6485. doi: 10.1073/pnas.95.11.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meier T J, Ho D Y, Sapolsky R M. J Neurochem. 1998;69:1039–1047. doi: 10.1046/j.1471-4159.1997.69031039.x. [DOI] [PubMed] [Google Scholar]
- 39.Ferrante R J, Kowall N W, Richardson E P., Jr J Neurosci. 1991;11:3877–3887. doi: 10.1523/JNEUROSCI.11-12-03877.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang Q, Zhou D, Sapp E, Aizawa H, Ge P, Bird E D, Vonsattel J P, DiFiglia M. Neuroscience. 1995;65:397–407. doi: 10.1016/0306-4522(94)00494-p. [DOI] [PubMed] [Google Scholar]
- 41.Coyle J T, Puttfarcken P. Science. 1993;262:689–695. doi: 10.1126/science.7901908. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz P J, Reaume A, Scott R, Coyle J T. Brain Res. 1998;789:32–39. doi: 10.1016/s0006-8993(97)01469-8. [DOI] [PubMed] [Google Scholar]
- 43.Portera-Cailliau C, Hedreen J C, Price D L, Koliatsos V E. J Neurosci. 1995;15:3775–3787. doi: 10.1523/JNEUROSCI.15-05-03775.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams J M, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 45.Jaattela M, Wissing D, Kokholm K, Kallunki T, Egeblad M. EMBO J. 1998;17:6124–6134. doi: 10.1093/emboj/17.21.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castilho R F, Hansson O, Ward M W, Budd S L, Nicholls D G. J Neurosci. 1998;18:10277–10286. doi: 10.1523/JNEUROSCI.18-24-10277.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beal M F. Curr Opin Neurobiol. 1992;2:657–662. doi: 10.1016/0959-4388(92)90035-j. [DOI] [PubMed] [Google Scholar]
- 48.Lindvall O, Kokaia Z, Bengzon J, Elmer E, Kokaia M. Trends Neurosci. 1994;17:490–496. doi: 10.1016/0166-2236(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 49.Marini A M, Paul S M. Proc Natl Acad Sci USA. 1992;89:6555–6559. doi: 10.1073/pnas.89.14.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J, Simon R. Neurology. 1997;48:306–311. doi: 10.1212/wnl.48.2.306. [DOI] [PubMed] [Google Scholar]