Abstract
Phototropism, the bending response of plant organs to or away from a directional light source, is one of the best studied blue light responses in plants. Although phototropism has been studied for more than a century, recent advances have improved our understanding of the underlying signaling mechanisms involved. The NPH1 gene of Arabidopsis thaliana encodes a blue light-dependent autophosphorylating protein kinase with the properties of a photoreceptor for phototropism. NPH1 apoprotein noncovalently binds FMN to form the holoprotein nph1. The N-terminal region of the protein contains two LOV (light, oxygen, or voltage) domains that share homology with sensor proteins from a diverse group of organisms. These include the bacterial proteins NIFL and AER, both of which bind FAD, and the phy3 photoreceptor from Adiantium capillus-veneris. The LOV domain has therefore been proposed to reflect a flavin-binding site, regulating nph1 kinase activity in response to blue light-induced redox changes. Herein we demonstrate that the LOV domains of two nph1 proteins and phy3 bind stoichiometric amounts of FMN when expressed in Escherichia coli. The spectral properties of the chromopeptides are similar to the action spectrum for phototropism, implying that the LOV domain binds FMN to function as a light sensor. Thus, our findings support the earlier model that nph1 is a dual-chromophoric flavoprotein photoreceptor regulating phototropic responses in higher plants. We therefore propose the name phototropin to designate the nph1 holoprotein.
Environmental factors have an extensive regulatory influence on plant growth and development. Perhaps the most important environmental factor is light. Light is not only a substrate for photosynthesis but a stimulus that regulates a wide range of metabolic and developmental processes, including seed germination, leaf development, stem extension, floral induction, and phototropism (1).
In higher plants, the effects of light on plant development are controlled by several classes of photoreceptors (2). These include the phytochromes (3), which monitor the red and far-red regions of the electromagnetic spectrum. UV-A/blue light perception is mediated by the cryptochromes (4), a photoreceptor regulating stomatal aperture in response to blue light (5), and the chromoprotein encoded by the NPH1 locus (6). Recent physiological studies indicate that these UV-A/blue light photoreceptors activate genetically separable signal transduction pathways in Arabidopsis, although the pathways may interact to influence the overall magnitude of a given response (7).
For a number of years, phototropism has been hypothesized to involve the activity of a 120-kDa plasma membrane-associated protein kinase that is rapidly phosphorylated in response to blue light irradiation (8, 9). In vitro studies indicate that the 120-kDa protein undergoes autophosphorylation after blue light treatment and is possibly a blue light-regulated protein kinase (10). Unilateral irradiation of etiolated oat coleoptiles results in a gradient of protein phosphorylation, consistent with the hypothesis that the 120-kDa protein is directly involved in the phototropic response (11–13). The 120-kDa phosphoprotein was subsequently shown to be the product of the NPH1 locus in Arabidopsis (14). Mutants at this locus, designated nph1 (nonphototropic hypocotyl), are deficient in all known phototropic responses, including the negative phototropic response of roots (15). The nph1 protein was therefore hypothesized to function as a photoreceptor for phototropism in higher plants (15). The NPH1 gene of Arabidopsis was recently isolated and found to encode a plasma membrane-associated protein of 996 amino acids (14). Homologues of Arabidopsis nph1 have been identified in a number of plant species, including oat (Avena sativa) nph1 (16) and the phy3 from the fern Adiantum (17). The C-terminal region of the nph1 protein has high similarity with serine/threonine protein kinases and contains all of the 11 conserved signature sequences (18). The N-terminal region of the protein contains two similar domains of about 110 amino acids (LOV1 and LOV2) present in a number of sensor proteins that are regulated by environmental factors that affect their redox status: light, oxygen, or voltage (hence, LOV) (14). Oxygen sensors containing the LOV domain include the bacterial regulator of nitrogen fixation NIFL (19) and the aerotaxis protein AER from Escherichia coli (20). Both AER and NIFL have been identified as flavoproteins binding FAD (21–23). The FAD-binding site of NIFL has recently been shown to reside within the N-terminal 284 amino acids of the protein, a region spanning the NIFL LOV domain (24). Therefore, the LOV domain has been proposed to function as a flavin-binding site, with the bound flavin acting as a sensor for environmental stimuli such as light or oxygen (14). Consistent with this hypothesis are studies involving the putative blue light photoreceptor WC-1 from the filamentous fungus Neurospora crassa. Mutations in the LOV domain of WC-1 result in mutant strains that are blind to blue light, demonstrating the importance of this domain in blue light sensing (25).
Alignment of the LOV1 and LOV2 domains from oat nph1, Arabidopsis nph1, and Adiantum phytochrome 3 (phy3) shows that the LOV domains are highly conserved between diverse plant species (Table 1). A high degree of homology is observed intra- or intermolecularly between the LOV1 and LOV2 domains of the oat nph1, Arabidopsis nph1, and Adiantum phy3 (40–44% identity), suggesting that the functions of the two LOV domains may also be evolutionarily conserved. Adiantum phy3 is an unusual protein with features of both phytochrome and nph1 photoreceptors (17). The N-terminal region of phy3 shows sequence homology to the chromophore-binding domain of phytochrome and includes all of the signature residues required for covalent attachment of the linear tetrapyrrole prosthetic group. Indeed, recombinant phy3 covalently binds phycocyanobilin to yield to a photochromic holoprotein phytochrome species (17). In contrast, the C-terminal region of the phy3 protein bears striking homology to nph1, including both LOV domains followed by the complete serine/threonine kinase domain. Given the complexity of fern responses to red and blue light, phy3 is hypothesized to function as a dual photosensor for red and blue light. In contrast to other plant species, phototropism in Adiantum can be induced by red light in addition to blue (26–28). Therefore, we speculate that Adiantum phy3 is a photoreceptor regulating both red and blue light-induced phototropism in this organism.
Table 1.
Sequence homology between the LOV domains of Avena sativa (As) nph1, Arabidopsis thaliana (At) nph1, and Adiantum capillus-veneris (Ac) phy3
| % identity
|
|||||
|---|---|---|---|---|---|
| As NPH1 LOV1 | At NPH1 LOV1 | Ac PHY3 LOV1 | As NPH1 LOV2 | At NPH1 LOV2 | |
| As NPH1LOV1 | |||||
| At PHY1LOV1 | 85.0 | ||||
| Ac PHY3LOV1 | 64.8 | 64.8 | |||
| As NPH1LOV2 | 42.1 | 41.1 | 42.6 | ||
| At NPH1LOV2 | 42.1 | 42.1 | 43.5 | 91.6 | |
| Ac PHY3LOV2 | 43.6 | 41.8 | 40.5 | 69.1 | 66.4 |
Amino acid sequence identities of the LOV1 and LOV2 domains are shown.
We recently reported that nph1 is a chromoprotein that functions as a photoreceptor for phototropism (29). When expressed in insect cells, nph1 is autophosphorylated in response to blue light irradiation, indicating that the NPH1 gene encodes a blue light-activated kinase. Moreover, recombinant nph1 noncovalently binds FMN, a likely chromophore for this light-dependent autophosphorylation. Indeed, the biochemical and photochemical properties of the recombinant protein are similar to those of the plant-derived protein, suggesting that FMN is the chromophore mediating photoactivation of nph1 in plants.
Herein we directly investigate whether the LOV domains of nph1 function as binding sites for the FMN chromophore. To address this question, we expressed a range of peptide fragments containing LOV domains from oat nph1, Arabidopsis nph1, and Adiantum phy3 in E. coli. Each of the fusion proteins generated were found to bind stoichiometric amounts of FMN, confirming the previous hypothesis that the nph1 LOV domains are flavin-binding motifs. Spectral analysis of the purified LOV domains indicates that the bound FMN most likely acts as a UV-A/blue light absorbing chromophore mediating phototropic curvature in higher plants.
MATERIALS AND METHODS
Sequence Alignment.
LOV domain sequences of Arabidopsis thaliana nonphototropic hypocotyl 1 (nph1), the nph1 homologue from Avena sativa, and Adiantum capillus-veneris phytochrome 3 (phy3) were obtained from the National Center of Biotechnology Information Entrez Web service (http://www.ncbi.nlm.nih.gov/Entrez/). Sequence alignment and determination of sequence identities of the LOV domains were performed with the program macboxshade.
Generation of Calmodulin Binding Peptide (CBP)-LOV Fusion Proteins.
Using the cDNAs of A. sativa NPH1, A. thaliana NPH1 and A. capillus-veneris PHY3 as templates, we synthesized DNA fragments corresponding to the various LOV domains with the PCR and oligonucleotides that contained the appropriate restriction sites and bordered the domain(s) of interest (Fig. 1A). The amplified DNA was isolated, digested, and cloned into the T7 RNA polymerase-based (30) pCAL bacterial expression vector (Stratagene) as a translational fusion to the calmodulin binding peptide (CBP). The following CBP N-terminal fusion proteins were prepared: AsNPH1LOV1 (amino acids 134–256), AsNPH1LOV2 (amino acids 404–559), AsNPH1LOV1/2 (amino acids 134–517), AtNPH1LOV1/2 (amino acids 7–628), and AcPHY3LOV2 (amino acids 924-1051). AsNPH1LOV1 and AsNPH1LOV1/2 sequences were cloned into the NcoI–HindIII site of the vector pCAL-n. AsNPH1LOV2, AtNPH1LOV1/2, and AcPHY3LOV2 sequences were cloned into the EcoRI–NcoI site of the vector pCAL-n-EK. Recombinant plasmids containing the AsNPH1LOV1 and AsNPH1LOV2 sequences were used to transform the E. coli host strain BL21(DE3) (Novagen). E. coli BL21(DE3)pLysS (Novagen) was transformed with recombinant plasmids containing the AsNPH1LOV1/2, AtNPH1LOV1/2, and AcPHY3LOV2 sequences. Large-scale cultures were grown at 37°C to an OD600 of approximately 0.5. Protein expression was carried out in complete darkness for 3 h at 30°C in the presence of 1 mM isopropyl β-d-galactopyranoside. Cultures were harvested and the CBP fusion proteins purified under dim red light on calmodulin resin in accordance to the instructions of Stratagene.
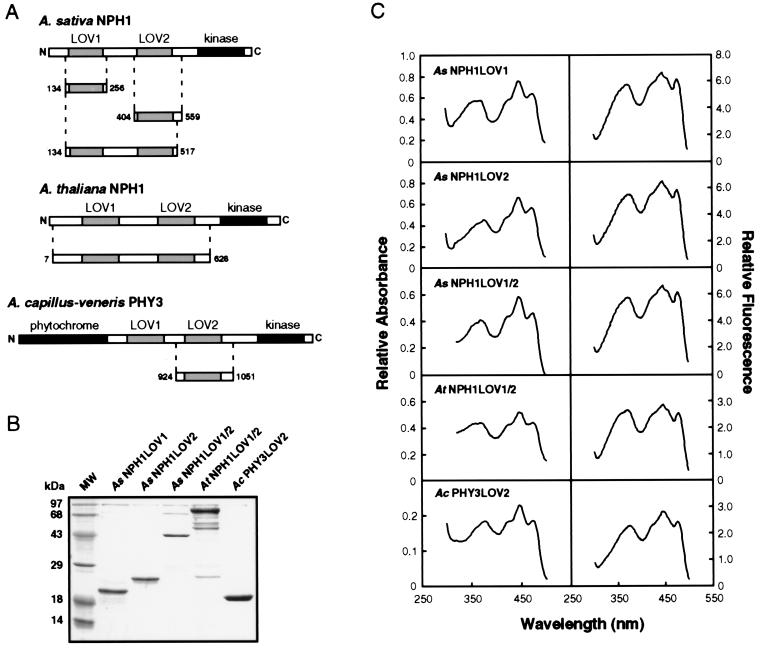
Figure 1.
Purification and spectral analysis of the CBP-LOV fusion proteins. (A) Schematic representation of the CBP-LOV domain fusion proteins. The various regions of Avena sativa nph1, Arabidopsis thaliana nph1, and Adiantum capillus-veneris phy3 used to generate the CBP fusion proteins are shown. Structural features of the nph1 and phy3 proteins are also indicated. LOV domains are shown as shaded boxes and other features are shown as solid boxes. Abbreviations: kinase, serine/threonine kinase domain; phytochrome, phytochrome-related region. The CBP region of the fusion proteins is not shown. (B) SDS/PAGE analysis of the purified LOV domain fusion proteins. Coomassie blue-stained SDS/12.5% polyacrylamide gel showing each of the purified CBP-LOV domain fusion proteins. The molecular masses of the marker proteins (MW) are indicated on the left in kilodaltons (kDa). (C) Spectral analysis of the LOV domain fusions, AsNPH1LOV1, AsNPH1LOV2, AsNPH1LOV1/2, AtNPH1LOV1/2, and AcPHY3LOV2. Absorption and fluorescence excitation spectra obtained for each of the CBP-LOV fusions are shown.
Spectral Analysis.
Absorption and fluorescence spectra of the LOV domain fusion proteins (approximately 10 μM) were obtained with a Beckman DU-70 spectrophotometer and a Photon Technology International Alphascan spectrofluorometer, respectively. Fluorescence excitation spectra were obtained by monitoring the emission at 535 nm. Fluorescence emission spectra were measured by using an excitation wavelength of 390 nm. For chromophore analysis, protein samples (150 μg) were treated with 10% trichloroacetic acid and centrifuged at 16,000 × g for 10 min. Supernatants were then examined for the presence of free chromophore.
TLC.
Protein samples (150 μg) were dialyzed against distilled water for 16 h before analysis. The chromophore was released by boiling in 70% ethanol for 2 min. Samples were then chilled on ice and centrifuged at 16,000 × g for 10 min. The flavin-containing supernatants were lyophilized to dryness and resuspended in 35% ethanol (5–10 μl). TLC was performed as described (31) with n-butanol/acetic acid/water, 3:1:1 (vol/vol), as solvent.
Calculation of FMN/protein Ratios.
Molecular weights of the CBP-LOV fusions were calculated with expasy proteomics tools (http://expasy.hcuge.ch/). The following sizes were predicted: AsNPH1LOV1 (18.7 kDa), AsNPH1LOV2 (23.8 kDa), AsNPH1LOV1/2 (49.2 kDa), AtNPH1LOV1/2 (75.1 kDa), and AcPHY3LOV2 (20.9 kDa). Purified AcPHY3LOV2 appeared to migrate faster during electrophoresis than expected from its predicted size (Fig. 1B). The presence of six proline residues at the extreme C-terminal end of this fusion protein may account for this discrepancy. Protein concentrations were determined by the Bradford protein assay (Bio-Rad) using BSA as standard. To determine the concentration of FMN associated with each of the CBP fusion proteins, chromophore was released by treatment with 10% trichloroacetic acid. Samples were centrifuged at 16,000 × g for 10 min, and the supernatant diluted 1:5 with distilled water. Fluorescence emission at 520 nm of the diluted sample was determined by using an excitation wavelength of 390 nm. The concentration of FMN in each sample was calculated by using a series of FMN standards containing an equivalent amount of trichloroacetic acid. The molar ratio of FMN to protein was expressed as FMN concentration against protein concentration. Ratios were calculated at least three times and varied from 0.6 to 1.0 for AsNPH1LOV1, AsNPH1LOV2, and AcPHY3LOV2; from 1.6 to 2.1 for AsNPH1LOV1/2; and from 1.2 to 1.6 for AtNPH1LOV1/2, depending on the protein preparation. The data presented (Table 2) are the ratios obtained for the CBP fusion proteins shown in Fig. 1B.
Table 2.
Molar ratios of FMN to each of the CBP-LOV fusion proteins
| Peptide | FMN/protein |
|---|---|
| AsNPH1LOV1 | 1.0 |
| AsNPH1LOV2 | 1.0 |
| AsNPH1LOV1/2 | 2.1 |
| AtNPH1LOV1/2 | 1.6 |
| AcPHY3LOV2 | 0.9 |
Calculated FMN/protein molar ratios for the CBP fusions, AsNPH1LOV1, AsNPH1LOV2, AsNPH1LOV1/2, AtNPH1LOV1/2, and AcPHY3LOV2 are shown.
RESULTS AND DISCUSSION
To characterize the molecular properties of the LOV domains, we constructed a range of bacterial expression vectors designed to express LOV domain-containing peptides in E. coli as fusions with CBP (Fig. 1A). Three LOV domain fusions were generated for oat nph1 containing the LOV1 domain, the LOV2 domain, or both LOV1 and LOV2 domains (designated AsNPH1LOV1, AsNPH1LOV2, and AsNPH1LOV1/2, respectively). A single fusion peptide was produced for the Arabidopsis nph1 protein that included both the LOV1 and LOV2 domains (designated AtNPH1LOV1/2). Finally, a smaller peptide containing sequences spanning the LOV2 domain was generated for Adiantum phy3 (designated AcPHY3LOV2). In each case, the fusion proteins were found to be partially soluble and could be purified to near homogeneity by affinity chromatography with calmodulin resin (Fig. 1B). Additional peptides were routinely observed to copurify with AtNPH1LOV1/2 fusion protein. These lower molecular weight peptides likely result from AtNPH1LOV1/2 degradation because they are recognized by specific polyclonal nph1 antiserum (data not shown). Nph1 is associated with the plasma membrane on isolation from Arabidopsis and other plant species (10). Although the mechanism of association remains to be determined, the soluble nature of CBP-LOV fusions in E. coli suggests that the LOV domains do not mediate this process.
We previously demonstrated that full-length nph1 expressed in insect cells is a chromoprotein that noncovalently binds FMN, with spectral properties similar to the action spectrum for phototropism (29). The action spectrum for phototropism typically shows maximal activity between 400 and 500 nm and shows a substantial degree of fine structure with a major band at 450 nm, a subsidiary shoulder at 430 nm, and a sharp peak at 470 nm (32). An additional broad less-effective peak is typically observed at 380 nm. As for the holoprotein expressed in insect cells (29), the absorption spectrum for each of the CBP-LOV fusions closely resembles the action spectrum for phototropism (Fig. 1C). Similar spectral properties were also detectable in the fluorescence excitation spectra for each of the LOV domain fusions (Fig. 1C). These findings are consistent with the earlier hypothesis that the LOV domains of nph1 function as chromophore-binding sites (14). CBP fusions AsNPH1LOV1 and AsNPH1LOV2 exhibit similar spectral properties, suggesting that both these regions of nph1 are involved in chromophore binding.
The fluorescence excitation spectrum for each of the CBP fusion proteins is not identical to the corresponding absorption spectrum (Fig. 1C). Although the fluorescence excitation and absorption maxima are similar, their relative peak intensities are somewhat different, especially in the UV-A region of the spectrum. It is possible that the observed spectral differences result from the photochemical properties of the chromophore associated with the LOV domain. Indeed, preliminary analysis suggests that the fluorescence of the LOV domain fusion proteins is light-intensity-dependent, with bleaching at high-excitation fluences (data not shown). Therefore, although the results presented herein demonstrate that the LOV domains are chromophore-binding sites, a more detailed photochemical and spectroscopic analysis of the isolated domains is required to gain a better understanding of the reaction mechanism of nph1 photoexcitation. One important question to address is whether the photochemical properties of LOV1 differ from those of LOV2. Because the spectral properties of both AsNPH1LOV1 and AsNPH1LOV2 closely resemble the action spectrum for phototropism (Fig. 1C), it is likely that both LOV domains and their associated chromophores play a role in light perception.
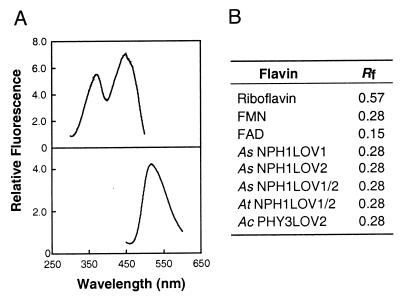
Recombinant nph1 binds FMN, a likely chromophore for light-dependent nph1 autophosphorylation (29). We therefore investigated whether the CBP-LOV fusion proteins expressed in E. coli also bind FMN. The cofactor for each fusion protein was found to be noncovalently bound and released by acid denaturation. For all CBP fusions, the released chromophore exhibited the fluorescence properties characteristic of free flavins (Fig. 2A). The flavin associated with each of the LOV domains was identified as FMN by thin-layer chromatography, according to its mobility relative to FAD, FMN, and riboflavin standards (Fig. 2B). These observations are consistent with the FMN-binding properties of full-length nph1 expressed in insect cells (29) and demonstrate that the LOV domains are binding sites for the FMN chromophore. Because the spectra shown in Fig. 1C resemble the action spectrum for phototropism, it seems likely that FMN is the chromophore mediating phototropism in higher plants. An extinction coefficient at 450 nm of 11.2 mM−1⋅cm−1 was estimated for AcPHY3LOV2 using the concentration of FMN released from the CBP-LOV fusion. A fusion protein containing the LOV1 domain of Adiantum phy3 was also found to bind FMN (data not shown). However, poor solubility significantly reduced the purification yield of this particular CBP fusion. Nevertheless, our results demonstrate that phy3 from Adiantum exhibits the properties of a UV-A/blue light photoreceptor. Further studies are required to establish whether phy3, like nph1, undergoes light-dependent autophosphorylation.
Figure 2.
Spectrofluorometric and TLC analysis of the chromophore associated with each of the LOV domain fusion proteins. (A) A typical fluorescence excitation spectrum (Upper) and fluorescence emission spectrum (Lower) of the chromophore released from each of the CBP fusion proteins. (B) Identification of the chromophore as FMN by TLC. The mobility of the chromophore released from each of the CBP-LOV fusions, relative to the solvent front (Rf), is indicated. Rf values for riboflavin, FAD, and FMN standards are also shown.
The molar ratio of FMN to each of the CBP fusion proteins appears to be stoichiometric (Table 2): fusion proteins containing one LOV domain bind one molecule of FMN, whereas fusions containing both LOV1 and LOV2 bind two molecules of FMN. Protein instability most likely accounts for the lower FMN/protein ratio obtained for the AtNPH1LOV1/2 fusion (Fig. 1B). These findings are consistent with the earlier proposal that NPH1 encodes the apoprotein of a dual-chromophoric photoreceptor regulating phototropic responses in higher plants (15). The spectral properties of full-length nph1 (29) and the LOV domains are not typically characteristic of free flavins (6). In general, flavins do not exhibit the fine structure observed in the blue region of the absorption spectrum (Fig. 1C). Indeed, these spectral qualities are more indicative of carotenoids rather than flavins (33). Evidently, binding of the FMN chromophore to the LOV domain provides a restricted hydrophobic environment that results in an absorption spectrum characteristic of the action spectrum for phototropism. Nph1 therefore represents a unique class of flavoprotein photoreceptors, unrelated to the cryptochromes, that binds FMN as a light-sensing chromophore to regulate phototropic responses in higher plants. As a result, we now introduce the name phototropin to designate the nph1 chromoprotein.
The LOV domain is also related to the well-characterized PAS domain, found in a number of proteins associated with light perception and signaling, including the phytochromes (34). One function of the PAS domain is to mediate protein–protein interactions. A second function of the PAS domain is to mediate cofactor binding, as is the case for two bacterial proteins, photoactive yellow protein (PYP) (35) and the oxygen-sensor FixL (36). With regard to cofactor binding, our results demonstrate that the LOV domains of phototropin have a similar functional role. Moreover, recent studies have shown that the LOV domain of Neurospora WC-1 is able to dimerize in vitro (25). Thus, the PAS/LOV domain appears to represent a highly conserved function in evolution. Indeed, PAS/LOV domains have been identified in a number of sensor proteins from an extremely diverse group of organisms, including archea, eubacteria, and eukaryotes (14, 34). Molecular evolution therefore seems to have created a functionally flexible motif that can adapt to serve a wide range of sensory roles.
Crystal structures of the PYP and FixL PAS domains have recently been determined and are shown to contain a β-sheet core, the site of cofactor binding, flanked by α-helices (35, 37, 38). The PAS-related domain of the human potassium channel HERG, designated eag, has also been reported to form a similar structure (39). Although HERG is oxygen-regulated (40), it is unclear whether this region, like the PAS domains in PYP and FixL, binds a prosthetic group. It will be of interest to determine whether the LOV domains of phototropin form a similar three-dimensional structure to accommodate the binding of FMN. Furthermore, detailed spectral studies are required to determine whether the LOV domain undergoes a self-contained photocycle as is the case for PYP (37). Such studies will provide insights into the mechanisms of phototropin photoexcitation and help identify the primary signaling events associated with the phototropic signal transduction pathway.
Acknowledgments
We thank Margaret A. Olney for critical reading of the manuscript. This work was supported by a grant (Ru 108/31-2) of the Deutsche Forschungsgemeinschaft; North Atlantic Treaty Organization Travel Grant CRG. 930555; Grant-in Aid for International Research (Joint Research 10044214) from the Ministry of Education, Science, Sports and Culture in Japan; PROBAIN and NOVARTIS Foundation Grant to M.W.; and National Science Foundation Grant IBN-9601164 to W.R.B. This is Carnegie Institution of Washington Department of Plant Biology Publication No. 1423.
ABBREVIATIONS
- LOV
light, oxygen, or voltage
- CBP
calmodulin binding peptide
References
- 1.Kendrick R E, Kronenberg G H M. In: Photomorphogenesis in Plants. Mohr H, editor. Dordrecht, the Netherlands: Kluwer Academic; 1994. pp. 353–373. [Google Scholar]
- 2.Batschauer A. Planta. 1998;206:479–492. doi: 10.1007/s004250050425. [DOI] [PubMed] [Google Scholar]
- 3.Fankhauser C, Chory J. Curr Biol. 1999;9:123–126. doi: 10.1016/s0960-9822(99)80078-5. [DOI] [PubMed] [Google Scholar]
- 4.Cashmore A R, Jarillo J A, Wu Y-J, Liu D. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- 5.Zeiger E, Zhu J. J Exp Bot. 1998;49:433–442. [Google Scholar]
- 6.Briggs, W. R. & Huala, E. (1999) Annu. Rev. Cell Dev., in press. [DOI] [PubMed]
- 7.Lascève G, Leymarie J, Olney M A, Liscum E, Christie J M, Vavasseur A, Briggs W R. Plant Physiol. 1999;120:605–614. doi: 10.1104/pp.120.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Short T W, Briggs W R. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:143–171. [Google Scholar]
- 9.Briggs W R, Liscum E. Plant Cell Environ. 1997;20:768–771. doi: 10.1046/j.1365-3040.1997.d01-116.x. [DOI] [PubMed] [Google Scholar]
- 10.Reymond P, Short T W, Briggs W R. Plant Physiol. 1992;100:655–661. doi: 10.1104/pp.100.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salomon M, Zacherl M, Luff L, Rüdiger W. Plant Physiol. 1997;115:493–500. doi: 10.1104/pp.115.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salomon M, Zacherl M, Rüdiger W. Bot Acta. 1997;110:214–216. [Google Scholar]
- 13.Salomon M, Zacherl M, Rüdiger W. Plant Physiol. 1997;115:485–491. doi: 10.1104/pp.115.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huala E, Oeller P W, Liscum E, Han I-S, Larsen E, Briggs W R. Science. 1997;278:2120–2130. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 15.Liscum E, Briggs W R. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zacherl M, Huala E, Rüdiger W, Briggs W R, Salomon M. Plant Physiol. 1998;116:869. [Google Scholar]
- 17.Nozue K, Kanegae T, Imaizumi T, Fukada S, Okamoto H, Yeh K-C, Lagarias J C, Wada M. Proc Natl Acad Sci USA. 1998;95:15826–15830. doi: 10.1073/pnas.95.26.15826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanks S K, Hunter T. FASEB J. 1995;9:576–610. [PubMed] [Google Scholar]
- 19.Dixon R. Arch Microbiol. 1998;169:371–380. doi: 10.1007/s002030050585. [DOI] [PubMed] [Google Scholar]
- 20.Bibikov S I, Biran R, Rudd K E, Parkinson J S. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill S, Austin S, Eydmann T, Jones T, Dixon R. Proc Natl Acad Sci USA. 1996;93:2143–2148. doi: 10.1073/pnas.93.5.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grishanin R N, Bibikov S I. Biosci Reports. 1997;17:77–83. doi: 10.1023/a:1027391402753. [DOI] [PubMed] [Google Scholar]
- 23.Rebbapragada A, Johnson M S, Harding G P, Zucarelli A J, Fletcher H M, Taylor B L. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Söderbäck E, Reyes-Ramirez F, Eydmann T, Austin S, Hill S, Dixon R. Mol Microbiol. 1998;28:179–192. doi: 10.1046/j.1365-2958.1998.00788.x. [DOI] [PubMed] [Google Scholar]
- 25.Ballario P, Talora C, Galli D, Linden H, Macino G. Mol Microbiol. 1998;29:719–729. doi: 10.1046/j.1365-2958.1998.00955.x. [DOI] [PubMed] [Google Scholar]
- 26.Kadota A, Koyama M, Wada M, Furuya M. Physiol Plant. 1984;107:181–186. [Google Scholar]
- 27.Hayami J, Kadota A, Wada M. Plant Cell Physiol. 1986;27:1571–1577. [Google Scholar]
- 28.Wada M, Sei H. J Plant Res. 1994;107:181–186. [Google Scholar]
- 29.Christie J M, Reymond P, Powell G K, Bernasconi P, Raibekas A A, Liscum E, Briggs W R. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 30.Studier W F, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 31.Raibekas A A. J Biolumin Chemilumin. 1991;6:169–176. doi: 10.1002/bio.1170060306. [DOI] [PubMed] [Google Scholar]
- 32.Baskin T I, Iino M. Photochem Photobiol. 1987;46:127–136. [Google Scholar]
- 33.Quiñones M A, Lu Z, Zeiger E. Proc Natl Acad Sci USA. 1996;93:2224–2228. doi: 10.1073/pnas.93.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhulin I B, Taylor B L. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 35.Pellequer J I, Wager-Smith K A, Kay S A, Getzoff E D. Proc Natl Acad Sci USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilles-Gonzalez M A, Ditta G, Helinski D R. Nature (London) 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 37.Genick U K, Borgstahl G E O, Ng K, Ren Z, Pradervand C, Burke P M, Srajer V, Teng T-Y, Schildkamp W, McRee D E, et al. Science. 1997;275:1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- 38.Gong W, Hao B, Mansy S S, Gonzalez G, Gilles-Gonzalez M A, Chan M K. Proc Natl Acad Sci USA. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabral J H M, Lee A, Cohen S L, Chait B T, Li M, Mackinnon R. Cell. 1998;95:649–655. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- 40.Taglialatela M, Castaldo P, Iossa S, Pannaccione A, Fresi A, Ficker E, Annunziato L. Proc Natl Acad Sci USA. 1997;94:11698–11703. doi: 10.1073/pnas.94.21.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]