Abstract
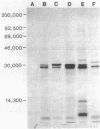
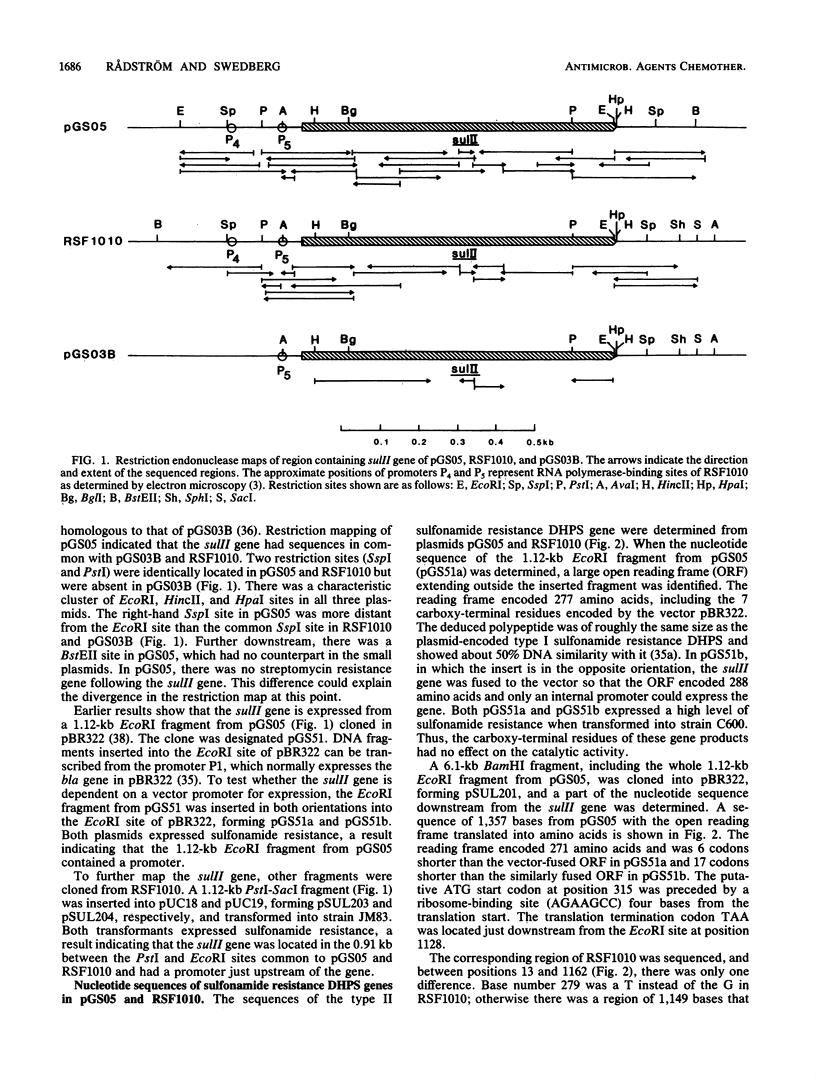
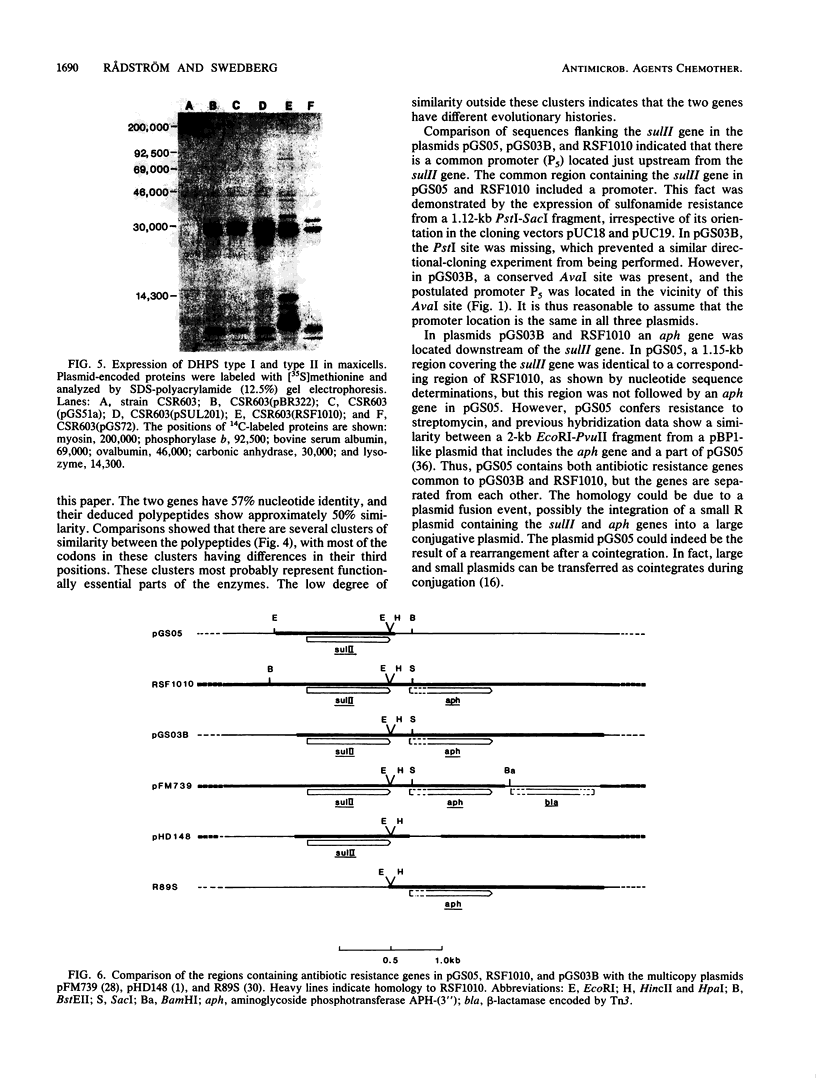
The nucleotide sequence of the type II sulfonamide resistance dihydropteroate synthase (sulII) gene was determined. The molecular weight determined by maxicells was 30,000, and the predicted molecular weight for the polypeptide was 28,469. Comparison with the sulI gene encoded by Tn21 showed 57% DNA similarity. The sulII-encoded polypeptide has 138 of 271 amino acids in common with the polypeptide encoded by sulI. The sulII gene is located on various IncQ (broad-host-range) plasmids and other small nonconjugative resistance plasmids. Detailed restriction maps were constructed to compare the different plasmids in which sulII is found. The large conjugative plasmid pGS05 and the IncQ plasmid RSF1010 contained identical nucleotide sequences for the sulII gene. This type of sulfonamide resistance is very frequently found among gram-negative bacteria because of its efficient spread to various plasmids.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albritton W. L., Brunton J. L., Slaney L., MacLean I. Plasmid-mediated sulfonamide resistance in Haemophilus ducreyi. Antimicrob Agents Chemother. 1982 Jan;21(1):159–165. doi: 10.1128/aac.21.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Balbás P., Soberón X., Merino E., Zurita M., Lomeli H., Valle F., Flores N., Bolivar F. Plasmid vector pBR322 and its special-purpose derivatives--a review. Gene. 1986;50(1-3):3–40. doi: 10.1016/0378-1119(86)90307-0. [DOI] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz R., Staudenbauer W. L. Replication of the broad host range plasmid RSF1010 in cell-free extracts of Escherichia coli and Pseudomonas aeruginosa. Nucleic Acids Res. 1982 Aug 11;10(15):4687–4702. doi: 10.1093/nar/10.15.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan G., Saul M., Twigg A., Gill R., Sherratt D. Polypeptides expressed in Escherichia coli K-12 minicells by transposition elements Tn1 and Tn3. J Bacteriol. 1979 Apr;138(1):48–54. doi: 10.1128/jb.138.1.48-54.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Adam S. A., Choi Y. D. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol Cell Biol. 1984 Mar;4(3):415–423. doi: 10.1128/mcb.4.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EDLIN G. GENE REGULATION DURING BACTERIOPHAGE T4 DEVLOPMENT. I. PHENOTYPIC REVERSION OF T4 AMBER MUTANTS BY 5-FLUOROURACIL. J Mol Biol. 1965 Jun;12:363–374. doi: 10.1016/s0022-2836(65)80260-1. [DOI] [PubMed] [Google Scholar]
- Grinter N. J., Barth P. T. Characterization of SmSu plasmids by restriction endonuclease cleavage and compatibility testing. J Bacteriol. 1976 Oct;128(1):394–400. doi: 10.1128/jb.128.1.394-400.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., van Embden J., Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974 Feb;117(2):619–630. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Heffron F., Rubens C., Falkow S. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3623–3627. doi: 10.1073/pnas.72.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe H., Tanaka T., Mitsuhashi S. Streptomycin and Spectinomycin resistance mediated by plasmids. Antimicrob Agents Chemother. 1978 Jun;13(6):1031–1035. doi: 10.1128/aac.13.6.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. D., Selsing E., Wells R. D. A rapid microscale technique for isolation of recombinant plasmid DNA suitable for restriction enzyme analysis. Plasmid. 1980 Jan;3(1):88–91. doi: 10.1016/s0147-619x(80)90037-2. [DOI] [PubMed] [Google Scholar]
- Korfmann G., Lüdtke W., van Treeck U., Wiedemann B. Dissemination of streptomycin and sulfonamide resistance by plasmid pBP1 in Escherichia coli. Eur J Clin Microbiol. 1983 Oct;2(5):463–468. doi: 10.1007/BF02013905. [DOI] [PubMed] [Google Scholar]
- Lopez P., Espinosa M., Greenberg B., Lacks S. A. Sulfonamide resistance in Streptococcus pneumoniae: DNA sequence of the gene encoding dihydropteroate synthase and characterization of the enzyme. J Bacteriol. 1987 Sep;169(9):4320–4326. doi: 10.1128/jb.169.9.4320-4326.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Hill B., Crapo L., Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci U S A. 1968 Apr;59(4):1259–1264. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofverstedt L. G., Hammarström K., Balgobin N., Hjertén S., Pettersson U., Chattopadhyaya J. Rapid and quantitative recovery of DNA fragments from gels by displacement electrophoresis (isotachophoresis). Biochim Biophys Acta. 1984 Jun 16;782(2):120–126. doi: 10.1016/0167-4781(84)90014-9. [DOI] [PubMed] [Google Scholar]
- Rubens C., Heffron F., Falkow S. Transposition of a plasmid deoxyribonucleic acid sequence that mediates ampicillin resistance: independence from host rec functions and orientation of insertion. J Bacteriol. 1976 Oct;128(1):425–434. doi: 10.1128/jb.128.1.425-434.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saano A. K., Zinchenko V. V. A new IncQ plasmid R89S: properties and genetic organization. Plasmid. 1987 May;17(3):191–201. doi: 10.1016/0147-619x(87)90027-8. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupert C. S. Determination of plasmid molecular weights from ultraviolet sensitivities. Nature. 1978 Mar 30;272(5652):471–472. doi: 10.1038/272471a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sköld O. R-factor-mediated resistance to sulfonamides by a plasmid-borne, drug-resistant dihydropteroate synthase. Antimicrob Agents Chemother. 1976 Jan;9(1):49–54. doi: 10.1128/aac.9.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedberg G. Organization of two sulfonamide resistance genes on plasmids of gram-negative bacteria. Antimicrob Agents Chemother. 1987 Feb;31(2):306–311. doi: 10.1128/aac.31.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedberg G., Sköld O. Characterization of different plasmid-borne dihydropteroate synthases mediating bacterial resistance to sulfonamides. J Bacteriol. 1980 Apr;142(1):1–7. doi: 10.1128/jb.142.1.1-7.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedberg G., Sköld O. Plasmid-borne sulfonamide resistance determinants studied by restriction enzyme analysis. J Bacteriol. 1983 Mar;153(3):1228–1237. doi: 10.1128/jb.153.3.1228-1237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise E. M., Jr, Abou-Donia M. M. Sulfonamide resistance mechanism in Escherichia coli: R plasmids can determine sulfonamide-resistant dihydropteroate synthases. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2621–2625. doi: 10.1073/pnas.72.7.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Treeck U., Schmidt F., Wiedemann B. Molecular nature of a streptomycin and sulfonamide resistance plasmid (pBP1) prevalent in clinical Escherichia coli strains and integration of an ampicillin resistance transposon (TnA). Antimicrob Agents Chemother. 1981 Mar;19(3):371–380. doi: 10.1128/aac.19.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]