Abstract
Background
Depending on their larval environment, female honeybees develop into either queens or workers. As in other polyphenisms, this developmental switch depends not on genomic differences between queens and workers but on the differential expression of entire suites of genes involved with larval fate. As such, this and other polyphenic systems can provide a novel tool for understanding how genomes and environmental conditions interact to produce different developmental trajectories. Here we use gene-expression profiles during honeybee caste determination to present the first genomic view of polyphenic development.
Results
Larvae raised as queens or workers differed greatly in their gene-expression patterns. Workers remained more faithful than queens to the expression profiles of younger, bipotential, larvae. Queens appeared to both downregulate many of the genes expressed by bipotential larvae and turn on a distinct set of caste-related genes. Queens overexpressed several metabolic enzymes, workers showed increased expression of a member of the cytochrome P450 family, hexameric storage proteins and dihydrodiol dehydrogenase, and young larvae overexpressed two putative heat-shock proteins (70 and 90 kDa), and several proteins related to RNA processing and translation.
Conclusions
Large differences in gene expression between queens and workers indicate that social insect castes have faced strong directional selection pressures. Overexpression of metabolic enzymes by queen-destined larvae appears to reflect the enhanced growth rate of queens during late larval development. Many of the differently expressed genes we identified have been tied to metabolic rates and cellular responses to hormones, a result consistent with known physiological differences between queen and worker larvae.
Background
Social insect colonies are defined in part by reproductive division of labor, whereby some colony members are considerably more fecund than others. The reproductive roles of colony members often are fixed by events that occur during larval development. An understanding of the mechanisms behind role, or caste, determination allows tests of several models for the evolution and maintenance of social life [1,2,3]. Female honeybees (Apis mellifera) are fated to become queens or workers during the first few days of larval development. Despite detailed knowledge of the physiological differences between workers and queens during development [4], relatively little is known of the underlying genetic machinery that drives these changes. Although differences in honeybee larval transcription had been inferred [5], caste-biased genes were identified only recently [3,6,7]. On the basis of inferred homology with known genes [3], these genes appear to be involved with diverse metabolic, nutritional and signaling processes during development.
Gene-expression arrays provide a powerful new tool to clarify the timing and nature of genetic events during insect development [8]. Here we use this technique to explore the genetic mechanisms of divergence between social insect castes. We contrast the gene-expression patterns of worker-destined and queen-destined larvae and then compare both castes with younger, bipotential, individuals. These comparisons allow tests of hypotheses concerning the evolutionary relationship between queen- or worker-destined developmental pathways in social insects (see, for example, [2]). More generally, changes in gene expression during insect caste determination can shed light on diverse developmental events such as differences in hormonal titers, the development or apoptosis of reproductive tissues, and developmental rates and metabolism [9]. We present evidence for widespread divergence in gene expression between workers and queens, and identify particular genes that appear to be integral to the production of these distinct adult phenotypes.
Results
We assayed gene expression using an array of gene fragments derived from reciprocal queen and worker larval sub-tractive cDNA libraries. Of 158 unique, consistently amplified clones isolated from these libraries, 63 showed a significant match to genes with known function. The functions assigned to these genes span approximately half of the major functional groups described by Adams et al. [10] for Drosophila melanogaster, suggesting that we have captured a fairly broad cross-section of the expressed genes in larval bees. Nevertheless, the clones derived from our libraries were biased toward particular functional groups, in a fashion that suggests a relationship between gene function and caste (Table 1). For example, we found an apparent over-representation of both ribosomal proteins and hexameric storage proteins in our libraries, relative to the expected frequencies of these gene families based on the Drosophila sequence. Of the 63 characterized clones, 15 (24%) showed significant similarity to known ribosomal protein genes. In contrast, 128 of the 6,537 annotated genes (2%) in the Drosophila melanogaster genome encode ribosomal proteins [10]. Gene knock-out experiments have implicated several of these as regulators of development rate and body size in Drosophila [11]. Assuming that ribosomal proteins help modulate ribosome activity, they may be indicators of the developmental tempo of larvae, and therefore could help pinpoint the initiation of caste determination or other major developmental events. We also cloned six apparently distinct hexameric storage proteins from the libraries, indicating a strong bias toward this family given the frequencies of hexamerins in the Drosophila genome of <<1% [10].
Table 1.
Normalized gene expression by functional group and larval class
| Functional group | Number of clones | Young larvae mean (SE) | Worker larvae mean (SE) | Queen larvae mean (SE) |
| Binding protein | 10 | 0.028 (0.015) | 0.025 (0.012) | -0.017 (0.014) |
| Enzyme | 16 | 0.006 (0.009) | 0.023 (0.011) | -0.001 (0.015) |
| Hexamerin | 6 | -0.042 (0.009) | 0.048 (0.017) | 0.005 (0.012) |
| Metabolic enzyme | 3 | -0.018 (0.028) | -0.012 (0.014) | 0.053 (0.004) |
| Ribosomal protein | 15 | 0.033 (0.009) | -0.013 (0.006) | -0.009 (0.013) |
| Structural protein | 6 | 0.015 (0.012) | -0.020 (0.007) | 0.044 (0.016) |
| RNA processing | 7 | 0.050 (0.013) | 0.001 (0.011) | -0.030 (0.017) |
| Unknown or other | 95 | 0.008 (0.004) | 0.005 (0.004) | 0.008 (0.005) |
Numbers in bold represent groups overexpressed in a particular larval class. Binding proteins were defined as non-metabolic proteins known to bind to other proteins or nucleic acids. SE, standard error.
We raised female honeybee larvae using standard methods, then measured gene-expression patterns for larvae collected at six 24-hour intervals from the embryo stage to the middle of the fifth and final instar. A subset (n = 13) of these larvae was raised as queens, while the others (n = 14) were raised workers. Analyses of covariance in gene expression indicate three primary clusters of genes with shared expression patterns across larvae (Figure 1). These clusters define genes upregulated primarily in queen-destined larvae relative to the other larvae screened; genes downregulated in queens; and genes upregulated in young, bipotential larvae. Genes within these clusters are more similar to each other in function than expected by chance (likelihood-ratio analysis based on functional group, df = 14, X = 38.2, p < 0.001).
Figure 1.
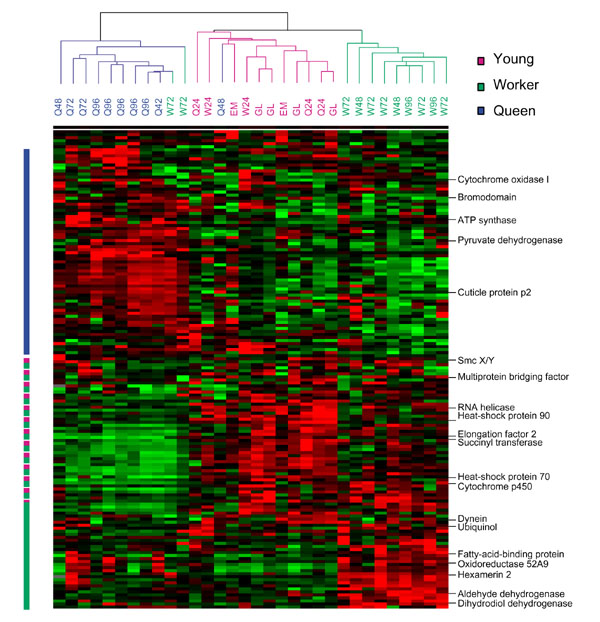
Cluster analyses showing expression patterns by gene and larval group. Pink, young bipotential larvae; green, worker-destined larvae; blue, queen-destined larvae. Green cells represent relatively low levels of expression, dark gray cells represent moderate expression levels, and red cells represent higher expression. Bars to the left of the array reflect boundaries of genes with relatively high expression in queens (blue), workers and young larvae (pink and green) and workers alone (green).
Genes related to RNA processing and translation showed higher rates of expression in young larvae (first and second instars) than in older larvae (Table 1). RNA helicase was strongly expressed by young larvae, as was an RNA-binding protein and translation elongation factor 2. Interestingly, two heat-shock proteins also were overexpressed in the youngest larvae. One shows a significant match to the Hsp70 family, whereas the second matches Hsp90. Heat-shock proteins are named for their ubiquitous presence following hyperthermy and other stresses [12], but members of this group have a more general role as molecular chaperones, and are involved in the assembly of newly translated proteins [13]. Hsp90 appears to be involved in the assembly of steroid hormone receptors [13]. Expression analyses on a more precise time scale, and at the level of specific tissues, should help determine whether the covariance between the expression of Hsps and agents involved in RNA processing reflects a joint role in producing essential proteins during early larval development. Queen-destined and worker-destined larvae showed broadly similar expression profiles late in the second instar, hours after queens and workers receive differential treatment in colonies. This suggests that most gene-expression changes associated with the caste programs occur after this point. Nevertheless, several genes were expressed differently at this early stage, including dihydrodiol dehydrogenase, which was expressed in lower levels by second-instar queens (normalized x = -0.067, SE = 0.025) than by either bipotential larvae (x = -0.026, SE = 0.009) or second-instar workers (x = 0.001, SE = 0.007). Dihydrodiol dehydrogenase has a general role in deactivating steroid hormones in mammals [14]. Consequently, it is intriguing that higher levels of expression of this enzyme by workers coincide with a period of decreased steroid titers in workers relative to queens. A putative lipid-binding protein also showed higher expression in workers than in queens from the second instar onward.
By the third instar, queen and worker larvae showed many differentially expressed genes (Figures 1,2) and, in each case, retained these different levels of expression through the remaining two instars. A cytochrome P450 (from the CYP4 subfamily) was more strongly expressed by workers than by queens. Members of the CYP4 subfamily have been implicated in the downregulation of steroid levels [15,16] although this subfamily is involved in a diversity of additional functions related to the metabolism of endogenous and exogenous substrates [17]. As a group, hexameric storage proteins were expressed at significantly higher levels in workers than in queens (Table 1), although a single hexamerin, from clone 1CB6 [3], showed nearly equal levels of expression in queens and workers. Hexamerins have a nutrient storage role during insect development, by bundling amino acids that accumulate during larval development for use during metamorphosis or by the adult insects. The accelerated growth rates of queens may limit their ability to allocate resources to hexamerin storage during the larval stages we assayed, as shown for Manduca sexta larvae [18]. Other genes consistently overexpressed by workers include an oxidoreductase, a fatty-acid-binding protein, aldehyde dehydrogenase and a second lipid-binding protein.
Figure 2.
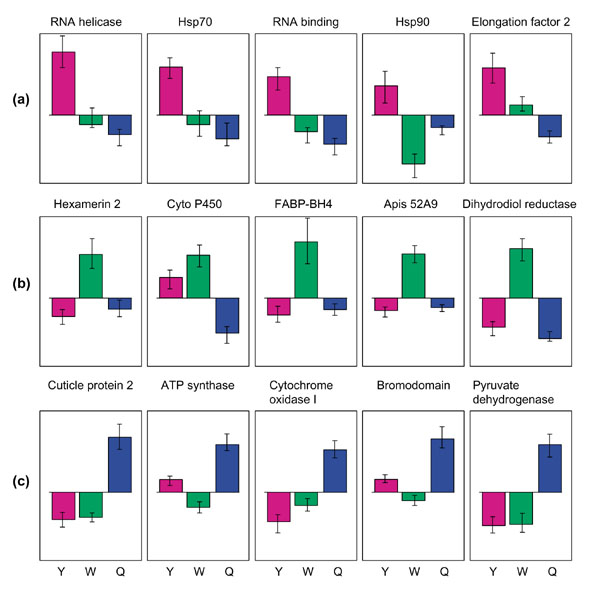
Proportional expression levels of specific genes. (a) Genes with high expression in young larvae (Y). (b) Genes with high expression in worker larvae (W). (c) Genes with high expression in queen larvae (Q). Normalized expression levels are shown, with standard error bars. FABP is a fatty-acid-binding protein and 52A9 is an oxidoreductase.
Queens overexpressed two genes directly linked to increased metabolic rates - the nuclear-encoded mitochondrial protein ATP synthase and the mitochondrial gene cytochrome oxidase I. Higher expression of two mitochondrial genes was reported previously [6], and was suggested to be an indicator of higher metabolic respiration in queen versus worker larvae. Interestingly, increased expression of these genes by queen larvae in our study was not apparent until the fifth instar in the bees we followed, a fact consistent with a role in caste-biased respiration. During the final instar, queens show the most extreme differences in growth rates, relative to workers. Queens also showed relatively high expression of a set of structural (cuticular) proteins, a histone acetyltransferase bromodomain, an ortholog to the Smcx/y mammalian sex-differentiation gene, and pyruvate dehydrogenase (Figures 1,2).
Discussion
As well as providing expression information for specific genes, the analyses presented here provide the first look at genome-level processes during the divergence of two social insect castes. Caste- and age-based biases in gene expression were indicated by a strong tendency for independent samples to group together in the clustering analyses (Figure 1). The castes differed widely in gene-expression patterns, supporting the idea that social insect castes have faced strong directional selection pressures [2]. Workers remained more faithful than queens to the expression profiles of younger, bipotential, larvae. In contrast, queens appeared simultaneously to downregulate many of the genes expressed by bipotential larvae and turn on a distinct set of caste-related genes. This difference could reflect, in part, a common impact of regulatory hormones on gene expression in young larvae and in larvae destined to become workers. Juvenile hormone and ecdysteroids are both much reduced in these larvae, relative to queen-destined larvae [4], and ecdysteroids have been shown to downregulate several genes expressed in the ovaries of developing workers [7].
Honeybees and other social insects provide a novel opportunity to measure the impacts of insect hormones on gene expression [9]. Specifically, hormone levels in different social insect castes are partially decoupled from molting and metamorphosis [19]. This fact can be used to retrieve expression differences caused by hormones per se from differences caused by other developmental events such as metamorphosis [8]. The identification of common transcriptional regulators for these genes, as shown in Drosophila embryos [20], would point to hormonal control mechanisms and unite caste-related genes with genes involved in insect development more generally.
Materials and methods
Array development
To generate the genetic arrays, cDNA clones were isolated from four subtractive libraries derived from worker- and queen-destined larvae collected in the third and fourth instars. We chose 144 clones from worker-biased libraries and 144 clones from queen-biased libraries. These clones were amplified by the polymerase chain reaction (PCR) using endogenous adaptor primers (primers Nest1 and Nest2r, described in [3]), then separated and verified by agarose gel electrophoresis. After being denatured for 5 min at 95°C in a final molarity 0.2 M NaOH, approximately 5 μg DNA from each amplified clone was fixed onto nylon membranes (Hybond N+, Amersham) using a 5 μl slot-pin library replicator (V and P Scientific) followed by UV irradiation. Each of 96 quadrants in a 384-cell printing contained three samples and a negative control. An aliquot of each cDNA clone was purified then sequenced using fluorescent-dye labeling and an ABI Prism 373 DNA analysis machine (Applied Biosystems).
Sample collection
Embryos, along with worker- and queen-destined larvae, were harvested from several colonies of A. mellifera ligustica of the same genetic stock maintained in the Bee Research Lab apiary. Two groups of 50 late-stage (2-day-old) embryos were harvested from standard (worker) cells and immediately frozen at -80°C. Four groups of 20 bipotential larvae aged 24 h post-hatching also were pooled and frozen. Fourteen worker-destined larvae aged 48, 72, 96, and 120 h post-hatching were also collected, along with 13 queen-destined larvae from the same age groups, raised in natural queen-rearing (swarm) cells. The ages of these queen larvae were determined by wet weight and body size comparisons with larvae of known age raised in artificial queen cells. For both worker- and queen-destined larvae, ages were likely to be accurate within 8-12 h, on the basis of wet-weight comparisons with larvae of known age [3]. Total RNA was extracted from these samples using the RNAqueous protocol (Ambion).
Labeling and hybridization
DNA probes were generated from 5 μg total RNA by reverse transcriptase synthesis (Superscript II enzyme, Life Technologies), using oligo-dT primers and biotin-labeled dUTP and dATP (in a 1:10 molar ratio to unmodified dTTP and DATP, respectively). Probes were cleaned by spin filtration, denatured, and then incubated in hybridization buffer (50% formamide, 6 × SSPE, 0.5% SDS, 5 × Denhardt's solution) with 0.1 pmol poly-(A)25 for 2 h at 42°C. Hybridization was carried out at 42°C, followed by a series of stringency washes, as in [3]. Membranes were washed, bathed in the Avidx alkaline-phosphatase conjugate then exposed to the chemiluminescent substrate CDP-star, according to the manufacturer's instructions (Tropix, Applied Biosystems). Membranes were next exposed to autoradiographic film for between 10 min and 8 h. Each membrane was used only once, giving an independent hybridization for 2 embryo replicates, 4 bipotential larvae, 13 queen, and 13 worker larval samples.
Data analysis
Cloned sequences were aligned using the CLUSTALW algorithm (Omiga 2.0, Oxford Molecular) to assess sequence quality and identify multiple captures of the same sequence. Each unique clone (167 out of 288) was compared against the GenBank database using the BLAST-X and BLAST-N algorithms at the site maintained by the US National Center for Biotechnology Information [21]. Matches showing a probability score of < 1.0 × 10-3 were treated as significant matches. Of the 167 unique clones, nine were dropped from the analyses as a result of evidence of multiple bands or inconsistent PCR amplification. Of the remaining clones, 64 showed a match to one or more sequences in GenBank and, of these, 63 could be placed into one of the gene function groups defined in [10] and the Gene Ontology database [22]. All gene fragment sequences used are available, along with their BLAST search results, at the Beenome Project website [23], and in the NCBI dbEST database [24].
Scanned images of exposed films were scored using the software program Zero Dscan (Scanalytics), giving a densitometry score for each cell in the membrane's 384-cell matrix. Spreadsheet macros (Microsoft Excel) were then used to generate an average intensity value for each of 158 unique clones that consistently showed a single band in the PCR amplifications. These values were normalized as a proportion of the strongest signal on each membrane, then were further normalized and centered on unity across genes and samples [18]. Centroid clustering was then carried out and visualized using the programs Cluster 1.0 and TreeView 1.0, respectively [25].
Additional data
The following additional data are included as Excel files: raw expression data and normalized data used in cluster analyses.
Supplementary Material
Raw expression data.
Normalized expression data.
Acknowledgments
Acknowledgements
We thank D. Lopez and D. Pham for technical assistance, and A. Collins and B. Tabashnik for comments on the manuscript. G. Hoffman, J. Martin, and colleagues in the Beltsville Bee Research Lab shared insights into honeybees and queen production.
References
- West-Eberhard M. Phenotypic plasticity and the origins of diversity. Annu Rev Ecol Syst. 1989;20:249–278. [Google Scholar]
- Gadagkar R. The evolution of caste polymorphism in social insects: genetic release followed by diversifying evolution. J Genet. 1997;76:167–179. [Google Scholar]
- Evans J, Wheeler D. Differential gene expression between developing queens and workers in the honeybee, Apis mellifera. Proc Natl Acad Sci USA. 1999;96:5575–5580. doi: 10.1073/pnas.96.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfelder K, Engels W. Social insect polymorphism: hormonal regulation of plasticity in development and reproduction in the honeybee. Curr Top Dev Biol. 1998;40:45–77. doi: 10.1016/s0070-2153(08)60364-6. [DOI] [PubMed] [Google Scholar]
- Severson D, Williamson J, Aiken J. Caste-specific transcription in the female honeybee. Insect Biochem. 1989;19:215–220. [Google Scholar]
- Corona M, Estrada E, Zurita M. Differential expression of mito-chondrial genes between queens and workers during caste determination in the honeybee Apis mellifera. J Exp Biol. 1999;202:929–938. doi: 10.1242/jeb.202.8.929. [DOI] [PubMed] [Google Scholar]
- Hepperle C, Hartfelder K. Differential display PCR reveals ecdysteroid-responsive genes in the ovary of honeybee worker larvae. In Proceedings of the International Congress of Entomology Edited by Gazzoni D Londrina, Brazil: Embrapa Press. 2000. p. 552.
- White K, Rifkin S, Hurban P, Hogness D. Microarray analysis of Drosophila development during metamorphosis. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- Evans J, Wheeler D. Gene expression and the evolution of insect polyphenisms. BioEssays. 2001;23:1–7. doi: 10.1002/1521-1878(200101)23:1<1::AID-BIES1000>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Torok I. Down-regulation of RpS21, a putative translation initiation factor interacting with p40, produces viable minute imagos and larval lethality with overgrown hematopoietic organs and imaginal discs. Mol Cell Biol. 1999;19:2308–2321. doi: 10.1128/mcb.19.3.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder M. Organismal, ecological, and evolutionary aspects of heat-shock proteins and the stress response: established conclusions and unresolved issues. Am Zool. 1999;39:857–864. [Google Scholar]
- Caplan A. Hsp90's secrets unfold: new insights from structural and functional studies. Trends Cell Biol. 1999;9:262–268. doi: 10.1016/s0962-8924(99)01580-9. [DOI] [PubMed] [Google Scholar]
- Hung C, Penning T. Members of the nuclear factor 1 transcription factor family regulate rat 3alpha-hydroxysteroid/dihydrodiol dehydrogenase (3 alpha-HSD/DD AKR1C9) gene expression: a member of the aldo-keto reductase superfamily. Mol Endocr. 1999;13:1704–1717. doi: 10.1210/mend.13.10.0363. [DOI] [PubMed] [Google Scholar]
- Dauphin-Villemant C, Bocking D, Tom M, Maibeche M, Lafont R. Cloning of a novel cytochrome p450 (CYPAC15) differently expressed in the steroidogenic glands of an arthropod. Biochem Biophys Res Commun. 1999;264:413–418. doi: 10.1006/bbrc.1999.1363. [DOI] [PubMed] [Google Scholar]
- Sutherland T, Unnithan G, Feyereisen R. Terpenoid w-hydroxylase (CYP4C7) messenger RNA levels in the corpora allata: a marker for ovarian control of juvenile hormone in Diploptera punctata. J Insect Physiol. 2000;46:1219–1227. doi: 10.1016/s0022-1910(00)00042-1. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. Insect p450 enzymes. Annu Rev Entomol. 1999;44:507–533. doi: 10.1146/annurev.ento.44.1.507. [DOI] [PubMed] [Google Scholar]
- Webb B, Riddiford L. Synthesis of two storage proteins during larval development of the tobacco hornworm, Manduca sexta. Dev Biol. 1988;130:671–681. doi: 10.1016/0012-1606(88)90359-4. [DOI] [PubMed] [Google Scholar]
- Wheeler D. Developmental and physiological determinants of caste in social Hymenoptera: Evolutionary implications. Am Nat. 1986;128:13–34. [Google Scholar]
- Liang Z, Biggin M. Eve and ftz regulate a wide array of genes in blastoderm embryos: the selector homeoproteins directly or indirectly regulate most genes in Drosophila. Development. 1998;125:4471–4482. doi: 10.1242/dev.125.22.4471. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (NCBI) http://www.ncbi.nlm.nih.gov/blast
- Gene Ontology database http://www.flybase.indiana.edu
- Beenome Project website http://www.barc.usda.gov/psi/brl/beenome.html
- NCBI dbEST database http://www.ncbi.nlm.nih.gov/dbEST/index.html
- Cluster 1.0 and TreeView http://rana.stanford.edu/software/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw expression data.
Normalized expression data.


