Summary
Although notable progress has been made in the treatment of non-small-cell lung cancer (NSCLC) in recent years, this disease is still associated with a poor prognosis for most patients. Modern techniques have facilitated the identification of specific genetic factors that may play a role in disease progression and patient response to therapy, prompting research efforts to identify the clinical predictors of outcome for NSCLC. Recent evidence suggests that the application of a pharmacogenomic approach has the potential to greatly improve survival in certain subpopulations of patients with NSCLC, which could profoundly influence the decision-making process used in evolving treatment strategies for this malignancy.
Keywords: Chromosome 11p15.5, ERCC1, gemcitabine, immunohistochemistry, NSCLC, pharmacogenomics, RRM1
Genomic abnormalities in lung cancer
The genome of cancer cells, as identified by modern, sophisticated techniques that can pinpoint chromosome breaks and rearrangements, is extremely complex. The predominant type of genome instability in cancer is structural aberration of chromosomes, such as deletions, translocations, and insertions, which may arise due to impaired repair of DNA double-strand breaks [1,2] [Rouet 1994, Liang 1998].
Loss-of-heterozygosity (LOH) analysis is the most frequently used technique to assess genomic aberrations [3] [Bepler 2002]. The identification of allele loss with this technique has led to the discovery of numerous genes with key functions in tumour development and progression [4] [Pitterle 1998].
Molecular genetic studies have detected many chromosomal regions with frequent LOH in lung cancer, including areas on chromosomes 3, 5, 8, 9, 11 and 17 [4–8] [Bepler 1994, Whang-Peng 1982, Takahashi 1989, Naylor 1987, Pitterle 1998]. The identification of frequent allele loss on chromosome 11p15.5 in NSCLC prompted investigations to identify and characterise tumour-suppressor genes with potential involvement in the development and progression of this disease [5] [Bepler 1994]. LOH in this region has since been linked to patient outcome in NSCLC, and is highly predictive of poor survival [3] [Bepler 2002].
RRM1, a gene in the 11p LOH region
Following the identification of its potential role in NSCLC, the centromeric part of the 11p15.5 chromosome segment, known as LOH11A, was mapped and sequenced [9–10] [Bepler 1999, Zhao 2001], and positional cloning studies identified the putative tumour suppressor gene, RRM1, within this region [11] [Pitterle 1999].
Early genetic complementation studies with chromosome 11 strongly suggested that a gene, or genes, in the LOH11A region inhibited tumourigenicity in nude mice and growth in liquid culture [12] [O’Briant]. Subsequently, it was demonstrated that the RRM1 gene suppresses invasion, migration and in vivo metastasis formation through up-regulation of the PTEN tumour suppressor gene when overexpressed in human and mouse lung cancer cell lines [13] [Gautam 2003]. In a recent transgenic mouse study, mice continuously overexpressing RRM1 were found to be less susceptible to carcinogen-induced lung cancer formation and displayed improved survival compared with control mice [14]. [Gautam A, Bepler G 2006] Splenocyte assays revealed that transgenic RRM1 overexpressing mice had a higher capacity to repair DNA damage, which could explain the observed reduction in susceptibility to lung tumour induction.
These in vitro and in vivo observations suggest that overexpression of RRM1 results in a more ‘benign’ phenotype and could, therefore, be a significant predictor of survival in NSCLC. This hypothesis was investigated through the study of retrospective and prospective datasets of patients with resectable NSCLC [15] [Bepler 2004]. This analysis concluded that RRM1 is a biologically and clinically important determinant of malignant behaviour in NSCLC and represents a strong predictor of outcome in patients with resectable disease. It was also suggested that future randomised trials of NSCLC should stratify patients based on RRM1 expression since tumours with high levels of expression have an intrinsically less malignant phenotype.
RRM1 is the molecular target of gemcitabine
The inherent or induced resistance of tumours to cytotoxic agents represents a major clinical problem. Several recent publications have highlighted a possible link between RRM1 expression and increased resistance to the antimetabolite gemcitabine in NSCLC [16–19] [Bergman 2002, Davidson 2004, Bergman 2005, Bepler 2006 Clin Oncol 2006.
Ribonucleotide reductase is the rate-limiting enzyme in DNA synthesis, and it is the only known enzyme that converts ribonucleotides to deoxyribonucleotides, which are required for DNA synthesis and repair. The ribonucleotide reductase holoenzyme consists of two dimerised subunits (RRM1 and RRM2), the pairing of which is essential for deoxynucleotide synthesis. Although the physical relationship between gemcitabine and mammalian ribonucleotide reductase has not been well characterised, data support the hypothesis that the RRM1 subunit is the most likely intracellular target for gemcitabine diphosphate [17,20] [Fan 1997, Davidson 2004]
Davidson et al. identified increased expression of RRM1 as the major determinant of gemcitabine resistance [17] [Davidson 2004] In 2005, Bergman et al. developed the first in vivo model of resistance to gemcitabine as a result of repetitive treatment using a clinically relevant schedule. In line with previous in vitro studies, microarray profiling revealed a marked increase in RMM1 expression, and, therefore, identified this gene as a key target for acquired in vivo gemcitabine resistance [18] [Bergman 2005]. These observations clearly indicate that RMM1 could play a role in the prediction of patient outcome, and draw attention to the fact that response to gemcitabine represents an area of research where the application of pharmacogenomics could be of vital importance. The challenges faced by this emerging model include determination of whether its application improves treatment response and survival, while reducing the toxicity experienced by patients.
Current approaches to lung cancer therapy
Platinum-based combination therapy is the established standard of care for the first-line treatment of advanced NSCLC. Although the current practice for treating patients with metastatic disease includes the addition of newer generation agents such as vinorelbine, gemcitabine, paclitaxel or docetaxel to a platinum agent, no combination has emerged as a gold standard [21 [Fossella 2003]. Recent, large Phase III trials comparing modern platinum-based regimens in the first-line treatment of advanced NSCLC found no clear advantage for any regimen [22–25] [Schiller 2002, Kelly 2001, Belani 2005, Rosell 2002]. Similarly, studies investigating non-platinum doublets versus platinum doublets were unable to demonstrate significant differences in outcome [26–29] [Georgoulias 2001, Kosmidis 2002, Gridelli 2003, Smit 2003].
Most recently, trials have investigated the addition of molecularly targeted agents such as gefitinib (INTACT-1 and -2 trials) [30–31] [Giaccone 2004, Herbst 2004] and erlotinib (TALENT and TRIBUTE trials) [32–33] [Gatzemeier 2005, Herbst 2005] to cytotoxic chemotherapy regimens in untreated patients with advanced NSCLC, but no benefit in terms of increased response rate, time to progression or overall survival has been demonstrated. The one exception is bevacizumab, which recently became the first targeted therapy to demonstrate superior efficacy combined with standard doublet chemotherapy over chemotherapy alone in the treatment of NSCLC, although this is based on preliminary data [34] [Sandler 2005]
Therefore, although it remains the key treatment modality, the one size fits all approach to first-line chemotherapy of NSCLC appears to have reached a therapeutic plateau. Currently, subgroups of NSCLC patients that might have different responses to treatment are primarily defined on the basis of clinical parameters such as performance status, personal preference, convenience, central nervous system metastases, histology, bleeding disorders, gender and smoking status. However, pharmacogenomics has the potential to allow the selection of specific patients on a genetic basis. It is hypothesised that this specific tailoring of therapy, guided by individual patient genetics, could lead to unequivocally superior responses following chemotherapy treatment.
RRM1- and ERCC1-based chemotherapy selection
As outlined earlier in this article, the available evidence indicates that if RRM1 is highly expressed (within physiological range) in genetically modified cell lines, resistance to gemcitabine increases, and, if RRM1 is not highly expressed, cell lines become sensitive to gemcitabine (Figure 1). [19] [Bepler 2006]
Figure 1.
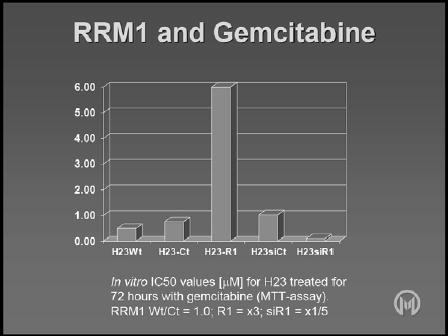
Effect of RRM1 levels on gemcitabine in vitro.
Since it has been demonstrated that RRM1 has an impact on DNA damage and repair, it would be expected to have an influence on the activity of other drugs, particularly the platinum agents. In vitro studies have identified a minor but consistent impact of RRM1 on platinum chemosensitivity, with increased RRM1 levels making cells slightly more resistant to carboplatin (Figure 2) [19] [Bepler 2006]. Evidently, this could have significant implications for platinum combination chemotherapy.
Figure 2.
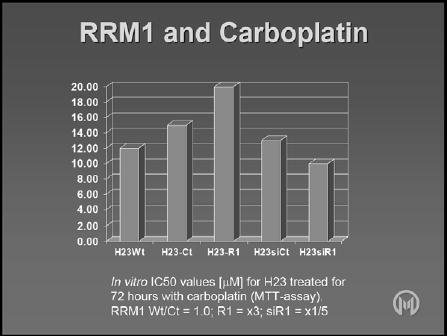
Effect of RRM1 levels on carboplatin in vitro.
Two exploratory retrospective datasets investigating this hypothesis in patients with stage IV NSCLC have provided evidence that RRM1 mRNA expression is a crucial predictive marker of survival in patients treated with gemcitabine plus cisplatin [35–36] [Rosell 2003, Rosell 2004]. Although these studies have certain limitations due to their retrospective nature and size, and the fact that no assessable impact on disease response was documented, the observed effects on survival suggest that genetic testing of RRM1 mRNA expression levels can and should be used to personalise platinum-based chemotherapy [36] [Rosell 2004]
The second of these studies also showed that the excision-repair cross-complementing group 1 (ERCC1) gene is related to cisplatin activity, [Rosell 2004] and other studies have confirmed that lung cancer is a malignancy in which the expression of this excision nuclease is directly related to the outcome of DNA-damaging therapy [37–38] [Reed 2005, Lord 2002]
Based on the evidence that RRM1 and ERCC1 may result in chemoresistance, two prospective Phase II trials were initiated, the results of which will be published in the near future. The goal of the first study (MCC-13240) was to obtain tumour biopsies under optimal conditions and measure RRM1 and ERCC1 expression to determine whether there is a direct correlation with response to gemcitabine plus carboplatin in patients with locally advanced stage IIIa and IIIb NSCLC [39]. The aim of the second trial (MCC-13208), referred to as the Molecular Analysis-Directed Individualized Treatment for Advanced NSCLC (MADeIT) trial, was to tailor chemotherapy based on the expression of these genes.
Following two cycles of chemotherapy with gemcitabine plus carboplatin, data from 35 patients in the MCC-13240 trial revealed a highly significant correlation between high levels of RRM1 expression and poor treatment response. Similarly, low levels of RRM1 expression were associated with an increased likelihood of patient response. The same trend was observed for ERCC1expression, although statistical significance was not reached.
In the MADeIT study, chemotherapy was administered based on the level of expression of RRM1 and ERCC1. Patients with stage III/IV disease had dedicated tumour biopsies, and, if high levels of RRM1 expression were identified, the patients received a chemotherapy doublet not containing gemcitabine, while those with low levels of RRM1 expression were given a doublet that included gemcitabine (see Figure 3).
Figure 3.
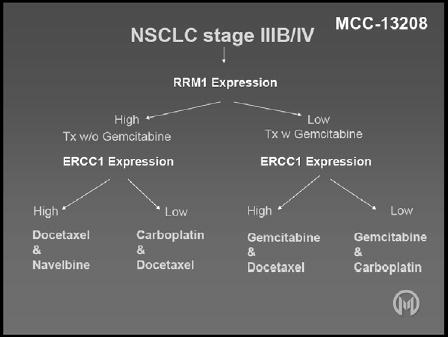
Study design for the MCC-13208 (MADeIT) trial.
The goal of this study was not to compare different treatments, but to demonstrate that upfront patient selection can lead to the administration of the most suitable treatment, which should, theoretically, result in the best possible outcome. This was found to be the case, with data indicating an unprecedented 12-month survival rate of 62%. Comparison with another study (MCC-12621) conducted at the same institution by the same physicians and with matching referral patterns, staging and enrolment criteria revealed that results obtained in the MADeIT study were close to 50% better (Table 1) [40]. Similarly, results from another study (E1594) revealed that the tailored treatment used in the MADeIT trial produced 12-month survival values that were almost double those previously observed [22].
Table 1.
Comparison of results from the MCC-13208 (MADeIT), 12621 and E1594 trials.

Development of immunohistochemistry for determination of RRM1 and ERCC1 expression
Clearly, the method of treatment selection used in the above-mentioned studies holds tremendous promise. However, this approach currently represents a boutique therapy since the expression analysis techniques used require a substantial infrastructure and, therefore, may not be readily accessible to the vast majority of patients.
However, recent evidence suggests that ERCC1 and RRM1 expression can be determined using immunohistochemistry, and preliminary results indicate that data obtained with this technology are consistent with values produced by real-time quantitative polymerase chain reaction. Soria et al. recently used a standard protocol of immunohistochemistry to confirm that patients with completely resected NSCLC and ERCC1-negative tumours derive a substantial benefit from adjuvant cisplatin-based chemotherapy. [Soria 2006] [41] Since this powerful antibody staining technique is simple to carry out and results can be easily interpreted, it could be widely used in the clinic. In addition, triple-staining automated systems, which eliminate potential errors due to subjectivity, have recently been developed to analyse RRM1, highlighting that immunohistochemistry has a vital role to play in the prediction of pharmacogenomic responses.
Conclusion
Due to the extremely complex nature of the genome of cancer cells, new discoveries are continually being made in this exciting field of research. Pharmacogenomics centres on the principle that these molecular genetic findings have the potential to ultimately affect therapeutic decisions in the clinic and greatly improve the treatment benefits experienced by patients. The available evidence clearly indicates that the identification and exploitation of genetic markers, predictive of response to specific cytotoxic drugs, is an achievable goal, and should, therefore, become a priority of current cancer research and future trials. Early results indicate that the application of pharmacogenomics in the field of NSCLC has the potential to profoundly influence outcomes and greatly improve survival in this fatal malignancy.
Acknowledgments
Funding for these studies was provided by NIH grants R01-CA102726 and R21-CA106616 and from Eli Lilly & Company and Sanofi-Aventis.
References
- 1.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bepler G, Gautam A, McIntyre LM, Beck AF, Chervinsky DS, et al. Prognostic significance of molecular genetic aberrations on chromosome segment 11p15.5 in non-small-cell lung cancer. J Clin Oncol. 2002;20:1353–1360. doi: 10.1200/JCO.2002.20.5.1353. [DOI] [PubMed] [Google Scholar]
- 4.Pitterle DM, Jolicoeur EC, Bepler G. Hot spots for molecular genetic alterations in lung cancer. In Vivo. 1998;12:643–658. [PubMed] [Google Scholar]
- 5.Bepler G, Garcia-Blanco MA. Three tumor-suppressor regions on chromosome 11p identified by high-resolution deletion mapping in human non-small-cell lung cancer. Proc Natl Acad Sci USA. 1994;91:5513–5517. doi: 10.1073/pnas.91.12.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whang-Peng J, Kao-Shan CS, Lee EC, Bunn PA, Carney DN, et al. Specific chromosome defect associated with human small-cell lung cancer: Deletion 3p(14–23) Science. 1982;215:181–182. doi: 10.1126/science.6274023. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, et al. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246:491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 8.Naylor SL, Johnson BE, Minna JD, Sakaguchi AY. Loss of heterozygosity of chromosome 3p markers in small-cell lung cancer. Nature. 1987;329:451–454. doi: 10.1038/329451a0. [DOI] [PubMed] [Google Scholar]
- 9.Bepler G, O’Briant K, Kim Y, Schreiber G, Pitterle D. A 1.4-Mb high-resolution physical map and contig of chromosome segment 11p15.5 and genes in the LOH11A metastasis suppressor region. Genomics. 1999;55:164–175. doi: 10.1006/geno.1998.5659. [DOI] [PubMed] [Google Scholar]
- 10.Zhao B, Bepler G. Transcript map and complete genomic sequence for the 310 kb region of minimal allele loss on chromosome segment 11p15.5 in non-small-cell lung cancer. Oncogene. 2001;20:8154–8164. doi: 10.1038/sj.onc.1205027. [DOI] [PubMed] [Google Scholar]
- 11.Pitterle DM, Kim YC, Jolicoeur EMC, Cao Y, O’Briant KC, et al. Lung cancer and the human gene for ribonucleotide reductase subunit M1 (RRM1) Mamm Genome. 1999;10:916–922. doi: 10.1007/s003359901114. [DOI] [PubMed] [Google Scholar]
- 12.O’Briant K, Jolicoeur E, Garst J, Campa M, Schreiber G, et al. Growth inhibition of a human lung adenocarcinoma cell line by genetic complementation with chromosome 11. Anticancer Res. 1997;17:3243–3252. [PubMed] [Google Scholar]
- 13.Gautam A, Li ZR, Bepler G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene. 2003;22:2135–2142. doi: 10.1038/sj.onc.1206232. [DOI] [PubMed] [Google Scholar]
- 14.Gautam A, Bepler G. Suppression of tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res. 2006;66:6497–6502. doi: 10.1158/0008-5472.CAN-05-4462. [DOI] [PubMed] [Google Scholar]
- 15.Bepler G, Sharma S, Cantor A, Gautam A, Haura E, et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:1878–1885. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Bergman AM, Pinedo HM, Peters GJ. Determinants of resistance to 2′,2′-difluorodeoxycytidine (gemcitabine) Drug Resist Updat. 2002;5:19–33. doi: 10.1016/s1368-7646(02)00002-x. [DOI] [PubMed] [Google Scholar]
- 17.Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, et al. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64:3761–3766. doi: 10.1158/0008-5472.CAN-03-3363. [DOI] [PubMed] [Google Scholar]
- 18.Bergman A, Eijk P, van Haperen V, Smid K, Veerman G, et al. In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as a major determinant. Cancer Res. 2005;65:9510–9516. doi: 10.1158/0008-5472.CAN-05-0989. [DOI] [PubMed] [Google Scholar]
- 19.Bepler G, Kusmartseva I, Sharma S, Gautam A, Cantor A, et al. RRM1-modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small cell lung cancer. J Clin Oncol. 2006;(26) doi: 10.1200/JCO.2006.06.1101. (will be printed before publication of this supplement – please ask author for page numbers) [DOI] [PubMed] [Google Scholar]
- 20.Fan H, Huang A, Villegas C, Wright JA. The R1 component of mammalian ribonucleotide reductase has malignancy-suppressing activity as demonstrated by gene transfer experiments. Proc Natl Acad Sci USA. 1997;94:13181–13186. doi: 10.1073/pnas.94.24.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fossella F, Pereira JR, von Pawel J, Pluzanska A, Gorbounova V, et al. Randomized, multinational, phase III study of docetaxel plus platinum combinations versus vinorelbine plus cisplatin for advanced non-small-cell lung cancer: the TAX 326 study group. J Clin Oncol. 2003;21:1–9. doi: 10.1200/JCO.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 22.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 23.Kelly K, Crowley J, Bunn PA, Jr, Presant CA, Grevstad PK, et al. Randomized phase III trial of paclitaxel plus carboplatin versus vinorelbine plus cisplatin in the treatment of patients with advanced non-small-cell lung cancer: a Southwest Oncology Group trial. J Clin Oncol. 2001;19:321–328. doi: 10.1200/JCO.2001.19.13.3210. [DOI] [PubMed] [Google Scholar]
- 24.Belani C, Lee JS, Socinski MA, Robert F, Waterhouse D, et al. Randomized phase III trial comparing cisplatin-etoposide to carboplatin-paclitaxel in advanced or metastatic non-small cell lung cancer. Ann Oncol. 2005;16:1069–1075. doi: 10.1093/annonc/mdi216. [DOI] [PubMed] [Google Scholar]
- 25.Rosell R, Gatzemeier U, Betticher DC, Keppler U, Macha HN, et al. Phase III randomized trial comparing paclitaxel/carboplatin with paclitaxel/cisplatin in patients with advanced non-small-cell lung cancer: a cooperative multinational trial. Ann Oncol. 2002;13:1539–1549. doi: 10.1093/annonc/mdf332. [DOI] [PubMed] [Google Scholar]
- 26.Georgoulias V, Papadakis E, Alexopoulos A, Tsiafaki X, Rapti A, et al. Platinum-based and non-platinum-based chemotherapy in advanced non-small-cell lung cancer: a randomized multicentre trial. Lancet. 2001;357:1478–1484. doi: 10.1016/S0140-6736(00)04644-4. [DOI] [PubMed] [Google Scholar]
- 27.Kosmidis P, Mylonakis N, Nicolaides C, Kalophonos C, Samantas E, et al. Paclitaxel plus carboplatin versus gemcitabine plus paclitaxel in advanced non-small-cell lung cancer: a phase III randomized trial. J Clin Oncol. 2002;20:3578–3585. doi: 10.1200/JCO.2002.12.112. [DOI] [PubMed] [Google Scholar]
- 28.Gridelli C, Gallo C, Shepherd FA, Illiano A, Piantedosi F, et al. Gemcitabine plus vinorelbine compared with cisplatin plus vinorelbine or cisplatin plus gemcitabine for advanced non-small-cell lung cancer: a phase III trial of the Italian GEMVIN Investigators and the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2003;21:3025–3034. doi: 10.1200/JCO.2003.06.099. [DOI] [PubMed] [Google Scholar]
- 29.Smit EF, van Meerbeeck JP, Lianes P, Debruyne C, Legrand C, et al. European Organization for Research and Treatment of Cancer Lung Cancer Group. Three-arm randomized study of two cisplatin-based regimens and paclitaxel plus gemcitabine in advanced non-small-cell lung cancer: a phase III trial of the European Organization for Research and Treatment of Cancer Lung Cancer Group - EORTC 08975. J Clin Oncol. 2003;21:3909–3917. doi: 10.1200/JCO.2003.03.195. [DOI] [PubMed] [Google Scholar]
- 30.Giaccone G, Herbst RS, Manegold C, Scagliotti G, Rosell R, et al. Gefitinib in combination with gemcitabine and cisplatin in advanced non-small-cell lung cancer: a phase III trial--INTACT 1. J Clin Oncol. 2004;22:777–784. doi: 10.1200/JCO.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Herbst RS, Giaccone G, Schiller JH, Natale RB, Miller V, et al. Gefitinib in combination with paclitaxel and carboplatin in advanced non-small-cell lung cancer: a phase III trial – INTACT 2. J Clin Oncol. 2004;22:785–794. doi: 10.1200/JCO.2004.07.215. [DOI] [PubMed] [Google Scholar]
- 32.Gatzemeier U, Heller A, Foernzler D, Moecks J, Ward C, et al. Exploratory analyses EGFR, kRAS mutations and other molecular markers in tumors of NSCLC patients (pts) treated with chemotherapy +/− erlotinib (TALENT) J Clinl Oncol. 2005;23(16S):7028. [Google Scholar]
- 33.Herbst RS, Prager D, Hermann R, Fehrenbacher L, Johnson BE, et al. TRIBUTE: a phase III trial of erlotinib. J Clin Oncol. 2005;23:5892–5899. doi: 10.1200/JCO.2005.02.840. [DOI] [PubMed] [Google Scholar]
- 34.Sandler AB, Gray R, Brahmer J, Dowlati A, Schiller JH, et al. Randomised phase II/III trial of paclitaxel plus carboplatin with or without bevacizumab in patients with advanced non-squamous non-small-cell lung cancer (NSCLC): An Easter Cooperative Oncology Group (ECOG) trial – E4599. J Clin Oncol. 2005;23(16):LBA4. [Google Scholar]
- 35.Rosell R, Scagliotti G, Danenberg KD, Lord RV, Bepler G, et al. Transcripts in pretreatment biopsies from a three-arm randomized trial in metastatic non-small-cell lung cancer. Oncogene. 2003;22:3548–3553. doi: 10.1038/sj.onc.1206419. [DOI] [PubMed] [Google Scholar]
- 36.Rosell R, Danenberg K, Alberola V, Bepler G, Sanchez JJ, et al. Ribonucleotide reductase mRNA expression and survival in gemcitabine/cisplatin-treated advanced non-small-cell lung cancer patients. Clin Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 37.Reed E. ERCC1 and clinical resistance to platinum-based therapy. Clin Cancer Res. 2005;11:6100–6102. doi: 10.1158/1078-0432.CCR-05-1083. [DOI] [PubMed] [Google Scholar]
- 38.Lord RV, Brabender J, Gandara D, Alberola V, Camps C, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res. 2002;8:2286–2291. [PubMed] [Google Scholar]
- 39.Williams CC, Wagner H, Greenberg H, Sharma A, Hazelton T, et al. Phase II Study of Induction Chemotherapy with Gemcitabine and Carboplatin (IndGC) followed by Paclitaxel and Carboplatin with Concurrent Thoracic Radiation (PCRT) for Patients with Unresectable Stage III Non-Small-Cell Lung Cancer (NSCLC): MCC-13240. J Clinl Oncol. 2005;23(16S):7307. [Google Scholar]
- 40.Chiappori A, Simon G, Williams C, Haura E, Rocha-Lima C, et al. Phase II study of first-line sequential chemotherapy with gemcitabine-carboplatin followed by docetaxel in patients with advanced non-small-cell lung cancer. Oncology. 2005;68:382–390. doi: 10.1159/000086979. [DOI] [PubMed] [Google Scholar]
- 41.Soria J, Haddad V, Olaussen KA, Fouret P, Dunant A, et al. Immunohistochemical staining of the Excision Repair Cross-Complementing 1 (ERCC1) protein as predictor for benefit of adjuvant chemotherapy (CT) in the International Lung Cancer Trial (IALT) J Clin Oncol. 2006;24(18S):7010. [Google Scholar]


