Abstract
Durability of plant disease resistance (R) genes may be predicted if the cost of pathogen adaptation to overcome resistance is understood. Adaptation of the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae (Xoo), to virulence in rice is the result of the loss of pathogen avirulence gene function, but little is known about its effect on aggressiveness under field conditions. We evaluated the cost in pathogenic fitness (aggressiveness and persistence) associated with adaptation of Xoo to virulence on near-isogenic rice lines with single R genes (Xa7, Xa10, and Xa4) at two field sites endemic for bacterial blight. Disease severity was high in all 3 years on all lines except the line with Xa7. Of two Xoo lineages (groups of strains inferred to be clonally related based on DNA fingerprinting) detected, one, lineage C, dominated the pathogen population at both sites. All Xoo strains were virulent to Xa4, whereas only lineage C strains were virulent to Xa10. Only a few strains of lineage C were virulent to Xa7. Adaptation to virulence on Xa7 occurred through at least four different pathways and was associated with a reduction in aggressiveness. Loss of avirulence and reduced aggressiveness were associated with mutations at the 3′ terminus of the avrXa7 allele. Strains most aggressive to Xa7 were not detected after the second year, suggesting they were less persistent than less aggressive strains. These experiments support the prediction that Xa7 would be a durable R gene because of a fitness penalty in Xoo associated with adaptation to Xa7.
Host plant resistance has been used extensively for disease control in many crop species. Many types of resistance, however, are not long lasting as a result of rapid changes in pathogen populations. Durability of resistance (R) genes is thus central to sustainable disease management. To date, it has been possible to assess the durability of an R gene only after its deployment for a long time over a large area (1). The capacity to predict the durability of R genes would be highly desirable for making sound investments in breeding and genetic engineering.
The R genes used in plant disease management are largely single dominant genes that direct the recognition of pathogen components encoded by avirulence (avr) genes; this relationship is referred to as a gene-for-gene interaction (2). The mechanisms by which these plant and pathogen genes interact to initiate resistance are becoming clearer because of the isolation and characterization of several plant R genes and pathogen avr genes (reviewed in refs. 3 and 4). The majority of cloned R genes encode leucine-rich repeat domains, which mediate specific protein–protein interactions or ligand binding. The R proteins are predicted to act as receptors to bind specifically to a pathogen-produced ligand, which is produced directly or indirectly by the avr gene. This direct interaction of the R protein and Avr ligand results in activation of the plant defense response, which often involves a hypersensitive response (HR). Therefore, the function of the R gene depends on the presence of a recognizable Avr ligand in the pathogen.
Functional analyses of pathogen avr genes have provided evidence that prediction of the durability of disease R genes may lie in an understanding of the fitness cost in pathogen evolution to overcome the resistance. In addition to their role in avirulence, several avr genes, such as avrBs2 (5), PthA (6), avrA and avrE (7), avrB (8), avrRPM1 (9), avrPto (10), and nip1 (11), possess a function in aggressiveness (amount of disease) or disease symptom expression, both of which are components of pathogenic fitness. The dual functions (avirulence and fitness) associated with these genes has led to speculation that the R genes corresponding to these avr genes may be more durable in the field (12, 13). Currently, two observations are generally taken as supportive of this hypothesis: (i) the measurable fitness functions of specific avr genes, and (ii) the prevalence of the avr genes in the genomes of pathogens (reviewed in refs. 14 and 15). No field experiments have been reported that are designed to directly test whether an assessment of the fitness penalty associated with pathogen adaptation in the field can be used to predict durability of R genes.
The interaction between rice and Xanthomonas oryzae pv. oryzae (Xoo) follows the classic gene-for-gene model. The three characterized avr genes (avrxa5, avrXa7, and avrXa10) from Xoo that correspond to the rice bacterial blight R genes xa5, Xa7, and Xa10 are members of a highly conserved gene family that contains a central domain composed of a near-perfect repeated sequence of 34 aa (16). The number of repeats varies among genes and the sequences of the repeats are almost identical, with the significant variation occurring at codons corresponding to amino acids 12 and 13 with respect to each repeat (reviewed in ref. 14). The order and type of repeats in the central domain govern resistance specificity of a particular gene family member (17–20). We recently demonstrated that insertional inactivation of the Xoo avr genes avrXa7 and avrxa5 reduced pathogenic aggressiveness of Xoo, whereas inactivation of another gene family member, avrXa10, did not affect aggressiveness (21). Based on these laboratory studies, we would predict that the corresponding R genes, Xa7 and xa5, would be more durable than Xa10 under field conditions.
In the rice bacterial blight pathosystem, adaptation to rice varieties containing a single dominant R gene often results from the loss of function of a corresponding pathogen avr gene. For example, in the early 1970s, the pathogen population was dominated by strains that could not cause disease on rice with the Xa4 R gene (22). After deployment of rice varieties carrying Xa4, strains virulent to Xa4 became predominant. Many more Xa R genes are being introduced into commercial rice lines, but the durability of these genes is uncertain. The well-developed genetic tools for analysis of Xoo (23, 24), together with the availability of rice near-isogenic lines carrying single R genes (25), provided a unique opportunity to test whether a detailed assessment of the genetic changes in the pathogen and associated epidemiological consequences can lead to a prediction of the inherent value of a R gene. Here, we provide evidence that the fitness penalty (reduced aggressiveness and lack of persistence) resulting from mutation of avrXa7 is sufficiently high to reduce the potential for disease epidemics on a rice line with Xa7. In contrast, there was no apparent penalty to loss of avrXa10 function (21), and epidemics occurred on the rice line with Xa10. Our results support the hypothesis that the inherent quality of an R gene is a function of the amount of fitness penalty imposed on the pathogen. This concept could be of general applicability to breeding for durable resistance against other diseases.
Materials and Methods
Field Experiments and Disease Assessment.
Field experiments were conducted in six continuous dry and wet seasons during 1993, 1994, and 1995 at two sites (Calauan and Mabitac) in the Philippines where bacterial blight is endemic. Three near-isogenic rice lines, IRBB4, IRBB7, and IRBB10, containing the Xa4, Xa7, and Xa10 genes for bacterial blight resistance, respectively, and the recurrent parent IR24 were planted in a randomized complete block design (Calauan) or a Latin Square Design (Mabitac) as described (26). Percentage of diseased leaf area (% DLA), a measure of disease severity, was assessed at the late epidemic stage (91–97 days after transplanting). To compare severity among the lines, ANOVA was performed for all lines per year at each site (SAS Institute, Cary, NC).
Collection of Infected Leaves and Bacterial Isolation.
Leaf samples were collected at the late epidemic stage during wet season crops. Samples were collected from seven sampling areas in a W-pattern at each treatment plot. At each site, each treatment was replicated four times. In Calauan, the sampling area consisted of 2 × 2-m (100 hills), whereas in Mabitac, sampling areas were 4 × 4-m (400 hills). From each sampling area, 7–10 symptomatic leaf samples were collected, totaling at least 49 samples per rice line per replication. Bacteria isolated from a minimum of seven symptomatic leaves were cultured on modified Wakimoto's medium (27). Of the 3,136 leaves sampled, only 1,417 yielded Xoo because some lesions resulted from physiological stress or other pathogens.
Genomic and Virulence Analyses of Bacterial Populations.
Bacterial strains were analyzed by PCR assays using DNA from pure cultures, whole cells from plates or bacterial leachate from leaves (28). Repetitive sequence-based PCR (29), as modified for Xoo (30), or amplification of DNA between sequences of a high-copy insertion element from Xoo (31), were used to distinguish Xoo populations. Restriction fragment length polymorphism analysis of bacterial genomic DNA after digestion with BamHI, EcoRI, or PstI was performed as described (28) by using the 3.1-kb BamHI fragment of avr gene avrXa10 as the probe (16). The presence or absence of the XorI restriction-modification system was assessed by digestion of Xoo DNA with PstI, an isoschizomer of XorI (32).
To determine race grouping, strains (n = 124) selected randomly from IR24 plots and strains (n = 62) from IRBB7 plots at both locations were inoculated in greenhouses to three seedlings per replication of each of the rice differential lines IR24, IRBB4, IRBB7, IRBB10, and IRBB5 (33). Lesion lengths from two replications of each line were scored at 14 days. Race designations were assigned based on lesion lengths (34).
Data Analyses.
DNA fingerprints generated by combined insertion element-based PCR and repetitive sequence-based PCR profile for each strain were grouped according to polymorphic banding patterns as described (30, 31). A total of 55 positions represented the final haplotype designation for each strain. A pairwise comparison of banding patterns for strains, cluster analysis, and bootstrap analysis using 2,000 iterations were performed as described (30, 35).
Differences in lineage distribution among years, sites, and lines were tested by using a weighted mixed-model analysis. The frequencies for each lineage were analyzed separately by using a split-plot model in which site is the whole-plot effect, line is the subplot effect, and year is a repeated measure effect that is stripped across sites, blocks, and lines. Pairwise comparisons between levels of significant effects were performed by using the least significant differences. All computations were performed by using proc mixed in sas (SAS Institute).
Cloning and Characterization of Wild-Type and Mutant avrXa7 Alleles.
Genomic DNA from field isolates PXO1865 (wild type, race 3), PXO0314 (race 9b), and PXO2684 (race 9c) were digested with BamHI and separated in agarose gels. Fragments of about 4.2 kb were extracted from the gel, ligated into plasmid pBluescript II KS+, and transformed into Escherichia coli DH5α. Inserts of 4.2 kb that hybridized to an avrXa10 probe were sequenced. To assist in accurate assembly of the repeat region, clones were linearized by digestion at a SmaI site in the polylinker, and then were partially digested with MscI (each repeat has only one MscI site). Sequences were obtained for recircularized clones with different-sized deletions and compared by using a bcm multiple comparison program (36).
For analysis of avirulence and aggressiveness functions, the 4.2-kb BamHI fragments from field Xoo strains PXO1865, PXO2684, and PXO0314 were cloned into plasmid pK117 (J.F.B., unpublished work), which contains a 170-bp 3′ terminus of the wild-type avrXa7 gene in pHM1, a vector that replicates in Xoo (R. Innes, personal communication). These plasmids, pB1865 (from the functional avrXa7 in PXO1865), pB2684 (mutant avrXa7 from PXO2684), and pB0314 (mutant avrXa7 from PXO0314) were introduced by electroporation (37) into strain PXO99A (16), which is virulent to IRBB7 or into KL7M, which contains a Tn5 insertion that inactivates avrXa7 (21). To confirm that regions of genes showing sequence differences between the wild-type and mutant avrXa7 alleles were responsible for loss of avrXa7 activity, HincII/BamHI fragments in the 3′ end of each gene were exchanged, and the resulting constructs were introduced into strain PXO99A. To assay gene function, bacterial suspensions (5 × 108 colony-forming unit/ml) were infiltrated into 14-day-old rice plants (38), and leaves were scored for an HR or water-soaked response at 3 days after inoculation. To assess aggressiveness, lesion lengths were measured at 15 days after clip-inoculation of 21-day-old plants.
Results
Disease Severity.
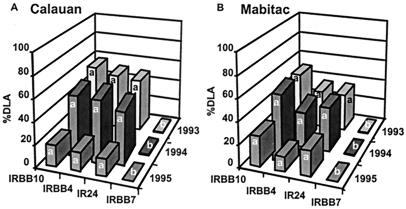
Severity of disease was measured over 3 years (six cropping seasons) at both sites (Mabitac and Calauan) to compare the effectiveness of the R genes in rice lines IRBB4 (Xa4), IRBB10 (Xa10), and IRBB7 (Xa7) and IR24 (no R gene). Bacterial blight was severe on both IR24 and IRBB4, which are known to be susceptible to the indigenous Xoo population (30), in both locations throughout the study (Fig. 1). Despite the fact that Xa10 had not been deployed in the Philippines, bacterial blight was also severe on the rice line with this gene (IRBB10). In contrast, little disease (less than 1% DLA) was observed on IRBB7. Consistent with the disease severity profiles, all leaf samples (196 per site per year) from IRBB10 and IR24 yielded Xoo, whereas only 62 of a total of 392 (15.8%) samples from IRBB7 yielded Xoo. Because epidemics occurred on IRBB10 but not on IRBB7, we suggest that evolution to virulence on Xa7 but not Xa10 was associated with a cost in pathogenic fitness to Xoo.
Figure 1.
Bacterial blight disease expressed as % DLA on near-isogenic rice lines with different R genes at the two experimental sites, Calauan (A) and Mabitac (B) in the Philippines. Little disease was observed on IRBB7. Common letters across lines are not significantly different (P = 0.01).
Pathogen Population Structure and Evolution of Virulence on Xa7.
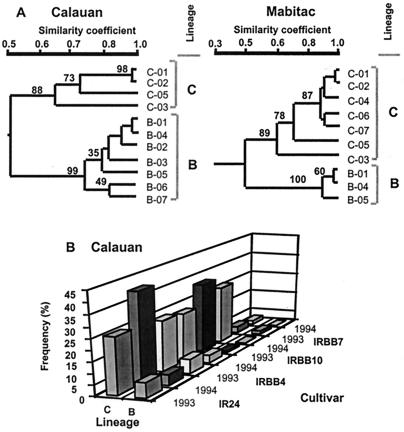
To monitor changes in the population structure that resulted in the evolution to virulence on Xa7, we compared genetic differences in Xoo populations in 1993 and 1994 by analyzing PCR-amplified profiles of 1,417 strains of Xoo from Calauan and Mabitac. Fourteen haplotypes that grouped into two lineages (B and C, ref. 30) were distinguished in the two locations (Fig. 2A). Lineage C dominated the population of Xoo and there was no significant difference in the frequency of lineage C on all rice lines at both locations and in both years (Calauan, Fig. 2B; Mabitac, data not shown). Although both lineages were isolated from IR24 and IRBB4, only lineage C strains were isolated from IRBB10 and IRBB7.
Figure 2.
(A) Genetic structure of Xoo populations in Calauan and Mabitac assessed in 1993 and 1994. Using UPGMA, Xoo lineages B and C were identified from genomic profiles generated by PCR assays. Lineages B and C were separated by cluster analysis at approximately 48% and were robust based on bootstrap analysis (values of 88–100%, ref. 35). Numbers on branches of dendrograms indicate the stability of branch arrangement based on 2,000 iterations of resampling. (B) Frequency of lineages B and C in Calauan on four rice lines in 1993 and 1994. Lineage C dominated the pathogen population in all rice lines.
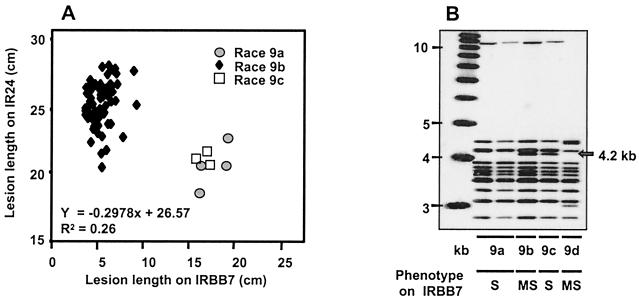
All seven lineage C haplotypes were virulent to IRBB10, whereas only two (C-01 and C-05) were virulent to IRBB7. All 62 strains isolated from IRBB7 and a subset of 124 C-01 and C-05 strains isolated from IR24 were inoculated to rice differential lines to determine virulence spectrum. The 62 strains isolated from IRBB7 and 47% (58/124) of the strains from IR24 were race 9 (the remaining 53%, 66/124, were race 3). The strains of race 9 haplotypes C-01 and C-05 fell into four groups based on (i) the lengths of lesions they caused on IRBB7 (Fig. 3A), (ii) the presence or absence of the XorI restriction modification system (data not shown), and (iii) the presence or absence of a 4.2-kb BamHI fragment that corresponds to avrXa7 (Fig. 3B). In DNA blot analysis, at least 11 other members of the avrBs3 gene family and the 4.2-kb BamHI fragment that corresponds to avrXa7 hybridized to a probe from the avrXa10 gene (16). The data combining all analyses are summarized in Fig. 4. C-01 strains causing lesions of 15–20 cm on IRBB7 were grouped as 9a and 9c (7% of the total C-01 strains), and those causing intermediate and variable lesions (4.5–10 cm) were grouped as race 9b (46%). All C-05 race 9 strains caused lesions of between 10 and 15 cm on IRBB7 and were grouped as race 9d.
Figure 3.
(A) Relationships between lesion lengths on rice lines IRBB7 (Xa7) and IR24 induced by strains of Xoo haplotype C-01, races 9a, 9b and 9c, that were recovered from IRBB7 and IR24 over 3 years. Xoo strains that were more adapted to Xa7 (race 9a and 9c) were less aggressive (caused shorter lesions) on IR24 than the less adapted strains (race 9b). The lesions induced on IRBB7 by 9a and 9c strains were shorter than those induced on IR24 by the 9b strains. (B) DNA blot analysis of Xoo strains recovered from IRBB7 that represent the different haplotypes and virulence groups (haplotype C-01 contains race 9a, 9b, and 9c, and haplotype C-05 contains race 9d). A 4.2-kb BamHI fragment that hybridizes to the avrXa10 gene corresponds to all but 0.17 kb of the avrXa7 gene.
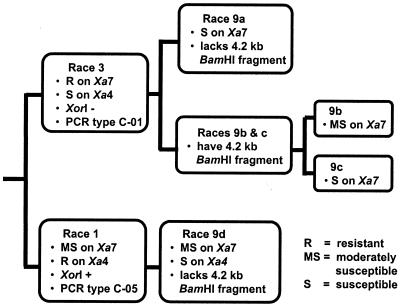
Figure 4.
Evolutionary pathways of Xoo to race 9a, 9b, 9c, and 9d are proposed based on the reaction phenotype (R, resistant; S, susceptible; MS, moderately susceptible) on rice lines with R genes Xa7 or Xa4, presence or absence of the XorI restriction modification system (32), genotype (haplotype) as determined by PCR assays (30, 31), and the presence or absence of a 4.2-kb BamHI fragment that corresponds to most of the coding region for avrXa7.
Although both C-05 (9d) and C-01 (9a, 9b, 9c) strains are race 9, the likely origin of the two haplotypes was different (Fig. 4). Based on genetic haplotype and the presence or absence of the XorI restriction modification system, C-05 strains virulent to Xa7 likely originated from Xoo race 1 whereas C-01 strains that were race 9a, 9b, and 9c virulent to Xa7 likely originated from race 3, the prevalent race at both sites (Fig. 4).
Aggressiveness of Race 9 Strains.
Laboratory mutants in avrXa7 were overall less aggressive to rice (21). To determine whether adaptation to virulence on Xa7 in field strains of race 9 was associated with a significant loss of pathogenic aggressiveness, we focused on race 9a, 9b, and 9c (C-01) strains isolated from either IR24 or IRBB7 because these were genetically most similar to the race 3 strains that predominated in the population (Fig. 4). Race 9a and 9c strains produced longer lesions on IRBB7 than did race 9b strains, but the race 9b strains produced longer lesions on IR24 than did the 9a and 9c strains (Fig. 3A). Thus, adaptation to virulence on IRBB7 (loss of the avirulence function of avrXa7) was associated with reduced aggressiveness to rice (15–25 cm lesions on IR24 and IRBB7 for races 9a and 9c vs. 20–30 cm for race 9b on IR24).
Molecular Changes Associated with Loss of avrXa7 Function in Race 9b and 9c.
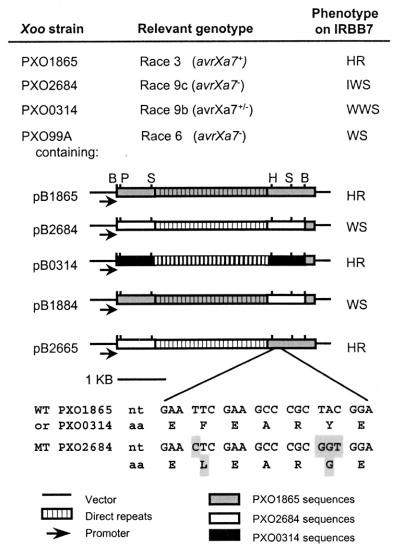
To characterize the molecular changes leading to virulence on IRBB7, the 4.2-kb BamHI fragments from a race 9b strain (PXO0314), a race 9c strain (PXO2684), and a wild-type race 3 strain (PXO1865) were cloned and sequenced. The 4.2-kb BamHI fragment that hybridizes to avrBs3 family members contains all but 0.17 kb of the avrXa7 gene. Experiments exchanging specific regions between the mutant and wild-type genes were performed to determine where the relevant mutations leading to loss of avrXa7 functions were located (Fig. 5). These experiments indicated that the mutations in the genes from both PXO0314 and PXO2684 were located in the 3′ terminus of the mutant avrXa7 genes. Because the 4.2-kb BamHI fragment from PXO0314 (race 9b) fused with the 0.17-kb terminus of the wild-type gene did not confer an altered avirulence phenotype, i.e., strains containing this construct conferred an HR (Figs. 5 and 6A), and because the sequences of the mutant and wild-type 4.2-kb BamHI fragments were identical (data not shown), we predicted that the mutation was contained within the 0.17-kb 3′ fragment of mutant allele from PXO0314.
Figure 5.
Location of mutated regions of avrXa7 alleles from field isolates of Xoo. Phenotypes indicated were determined by infiltration of rice line IRBB7 (Xa7) with Xoo strain PXO1865 (wild type), the mutants PXO0314, PXO2684, and PXO99A containing plasmids with the cloned 4.2-kb BamHI fragments from PXO1865, PXO2684, and PXO0314 or their derivatives with exchanged regions. The nucleotide and amino acid sequence alignment shows the four nucleotide changes detected in pB2684 as compared with the wild-type pB1865. These mutations, which result in two amino acid changes, alter avrXa7 function in pB2684. Phenotypes are intermediate (IWS), weak (WWS), and strong (WS) water-soaking of infiltration sites.
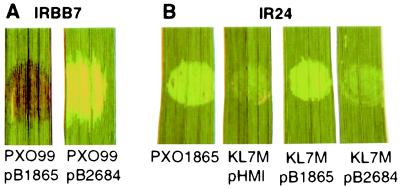
Figure 6.
The 4.2-kb BamHI fragment from Xoo field isolate PXO2684 does not complement avirulence or aggressiveness functions. (A) The avrXa7 allele from wild-type strain PXO1865, when introduced into Xoo strain PXO99A, confers a HR on IRBB7 whereas the fragment from strain PXO2684 does not. Plasmids containing the avrXa7 alleles (pB1865 and pB2684) were transformed into PXO99A and the transformants were inoculated by infiltration to rice line IRBB7 (Xa7). (B) The 4.2-kb BamHI fragment from PXO1865 restores the ability to elicit strong water-soaked lesions to KL7M, which harbors a mutation in avrXa7, whereas the fragment from PXO2684 does not.
Two recombinant plasmids containing fragments from PXO2684 (pB2684 and pB1884) conferred the mutant phenotype, i.e., water-soaked lesions on IRBB7 (Fig. 5), indicating the mutation in PXO2684 was contained within the HincII–BamHI fragment in the 3′ terminus. Sequence analysis of the entire mutant avrXa7 allele from PXO2684 (including the 0.17-kb 3′ terminus) showed only four nucleotide changes from the wild-type avrXa7 gene. These changes corresponded to two amino acid changes (Fig. 5).
To confirm that the avrXa7 allele in PXO2684 had lost both the avirulence and aggressiveness functions, plasmids with the wild-type (pB1865) and mutant (pB2684) avrXa7 fragments were used in complementation studies with Xoo strain KL7M, which is mutated at the avrXa7 locus (21). Transformants and parent strains were inoculated to rice lines IRBB7 and IR24 by infiltration and clip-inoculations. After infiltration, strains KL7M(pB1865) and PXO1865 elicited an HR on IRBB7, whereas PXO2684 and KL7M(pHM1) conferred intermediate and weak water-soaked phenotypes, respectively (data not shown). On rice line IR24, KL7M(pHM1) and KL7M(pB2684) produced weak water-soaked responses (i.e., a weak susceptible response), whereas KL7M(pB1865) and PXO1865 produced a strong water-soaked lesion, i.e., a strong susceptible response (Fig. 6B). At 15 days after clip-inoculation, KL7M(pB1865) and PXO1865 exhibited 13.2 ± 1.3-cm and 15.8 ± 1.3-cm lesions, respectively, on IR24, whereas the mutant KL7M(pHM1) and KL7M(pB2684) exhibited 4.7 ± 0.7-cm and 5.3 ± 0.9-cm lesions, respectively. These data show that the mutations in the avrXa7 allele in pB2684 were responsible for the loss of both avirulence and aggressiveness functions in the field strains.
Persistence of Race 9 Strains.
The ability of race 9 strains virulent to IRBB7 to persist and increase in the pathogen population over 3 years was assessed as a measurement of pathogenic fitness (Table 1). Based on previous studies (39), we assumed that the predominant source of inoculum for each crop was the pathogen population surviving in stubble from the previous crop. Race 9b strains, which are intermediate in virulence to Xa7, were detected in all 3 years at both sites, and at a higher frequency on IR24 than on IRBB7. Strains more aggressive to IRBB7, however, did not increase in the population during this study, and were not persistent over time. Race 9a was detected once in 1994 on IRBB7 in Calauan, race 9c was detected once on IRBB7 in Mabitac, and race 9d was detected once in 1993 on both IR24 and IRBB7 at both sites. Thus, strains that were more aggressive to Xa7 did not increase in number or persist over a long time in the pathogen population on any of the four rice lines in farmers' fields, whereas less aggressive strains persisted and increased over time.
Table 1.
Persistence of Xa7-adapted strains in two field sites over 3 years
| Year | No. strains of each race isolated from:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| IR24
|
IRBB7
|
|||||||
| 9a | 9b | 9c | 9d | 9a | 9b | 9c | 9d | |
| 1993 | 0 | 32 | 0 | 5 | 0 | 16 | 0 | 6 |
| 1994 | 0 | 26 | 0 | 3 | 4 | 13 | 3 | 0 |
| 1995 | 0 | 23 | 0 | 0 | 0 | 20 | 0 | 0 |
| % Total | 0 | 54 | 0 | 5 | 3 | 32 | 2 | 4 |
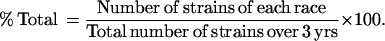 |
Discussion
Despite the presence of virulent Xoo strains in rice fields, the fitness penalty associated with loss of function of avrXa7 was apparently sufficient to prevent bacterial blight epidemics on rice lines with the Xa7 gene. This conclusion is based on field experiments showing that (i) no epidemic was observed on rice line IRBB7, which carries Xa7, over six cropping seasons, (ii) strains virulent to IRBB7 showed reduced aggressiveness to rice in general, and (iii) the more aggressive strains that were virulent to IRBB7 (race 9a, 9c, 9d) did not increase or persist in the field populations. We note that IRBB7 has been planted and monitored at one site (Calauan) for an additional 3 years beyond the end of this experiment and still exhibits very little disease (C.M.V.C., unpublished work). In addition, molecular characterization of mutant avrXa7 alleles from one group of field-adapted strains (race 9c) confirmed that the changes in avrXa7 were responsible for both the loss of avirulence function and reduced aggressiveness to rice. In contrast to avrXa7, there was no apparent fitness penalty associated with the loss of avrXa10 function (21) and in the field, IRBB10, the line containing the corresponding R gene Xa10, exhibited high severity throughout the study, despite the fact that Xa10 had not been deployed previously.
As observed for other bacterial avr genes (reviewed in refs. 14 and 15), some, but not all, Xoo avr genes function in pathogenic aggressiveness, and the contributions of these genes may be quantitatively different (21). Two Xoo avr genes (avrxa5 and avrXa7) and at least four related family members were shown to exhibit aggressiveness functions, and the contributions of these genes to aggressiveness were found to be gene-specific. In greenhouse tests, mutations of avrxa5, avrXa7, and other gene family members resulted in the loss of pathogen fitness whereas mutation of avrXa10 did not. Thus, we would predict that Xa7 would be relatively durable whereas Xa10 would not. The field studies presented here confirmed that loss of avrXa7 functions reduced fitness in Xoo, whereas loss of avrXa10 functions did not. Although there are likely many factors determining pathogen fitness, our complementary field and laboratory studies show that the contribution of avr genes to fitness is an important factor affecting durability. Our long-term goal is to prolong the durability of R genes through careful management practices, including the use of gene combinations and diversification of resistance sources. Because it can take many years to introduce the genes of interest into agronomically useful rice lines, and then several years of multilocation testing to assess or demonstrate durability, any means to predict the potential for durability in an R gene in advance would be extremely useful. The results of our studies suggest that durability of an R gene can be predicted based on an understanding of the avirulence and aggressiveness functions of the corresponding avr gene.
For many years, pathologists and breeders have attempted to identify unique characteristics of R genes that would be associated with durable resistance. This approach would seem promising with the successful cloning of many R genes from diverse plant species. Although detailed structural analyses of some R genes (e.g., the Rp1 locus in maize, ref. 40) revealed possible mechanisms for creating multiple specificities to counteract pathogen changes, they provide limited insight into the inherent properties of a durable R gene. Our results together with recent studies by others (reviewed in ref. 15) suggest that the answer may come from pathogen analysis. Indeed, based on careful field observations of multiple pathosystems, Van der Plank (41) proposed that the inherent quality of resistance is a function of the amount of fitness penalty imposed on the pathogen. This led to the hypothesis of “stabilizing selection” wherein the pathogen would suffer from a genetic load of “unnecessary virulence.” Because of the lack of genetic tools at that time, this important insight could not be experimentally tested. Extensive pathogen population analyses have provided clues to the effectiveness of R genes. We and others (reviewed in ref. 23) have shown that planting lines with single R genes, such as Xa4, imposes changes on the Xoo population structure. Although strains virulent to Xa4 (lineage B) were detected within 4–5 years after deployment (22), epidemics were not seen on Xa4 rice until the evolution of a group of lineage C strains virulent to Xa4 (30, 42, 43). Because avrXa4 has not yet been identified, the contribution of this gene, if any, to pathogen fitness has not been determined.
Based on our studies, we propose that the relative predicted durability of available R genes can be ranked by assessing the fitness attributed to avr gene mutations. The data suggest that combining xa5 and Xa7 into the same line would be more effective than using single R genes alone because of the greater fitness cost imposed by two different genes. A series of lines containing various combinations of xa5, Xa7, Xa4, and Xa21 have been produced to test this hypothesis (C.M.V.C., unpublished work). However, there are a number of caveats to this prediction scheme that need additional research. First, there could be secondary mutations that gradually would restore the fitness caused by a mutation in an avr gene (12). The race 9b strains observed in this study deserve additional monitoring and evaluation because these strains, although less aggressive than the race 3 strains, have persisted in the field. Second, fitness of a gene is a function of the genetic background; thus any fitness loss ascribed to an avr gene must be considered in the context of the genetic background of the strain. Although we have shown that Xa7 imposes a fitness penalty on the strains analyzed in the Philippines, it remains to be tested whether such effect also occurs in strains in other geographical regions. Preliminary data indicates that inactivation of avrXa7 in Korean Xoo strains also results in a large reduction in aggressiveness (S. H. Choi, personal communication). Third, aggressiveness is not the only factor contributing to pathogenic fitness; avr gene family members or other genes may contribute in other ways to overall pathogenic fitness. Finally, because we used near-isogenic rice lines, our experiments do not allow any comparison of the impact of various host genetic backgrounds on pathogen fitness. Such studies are important to extend the findings to breeding programs in different countries with different pathogen populations.
Acknowledgments
This work was supported by the Rockefeller Foundation (J.E.L., R.J.N., and H.L.), US Aid for International Development (J.E.L. and H.L.), and the Kansas State University Agricultural Experiment Station (#00–467-J).
Abbreviations
- Xoo
Xanthomonas oryzae pv. oryzae
- avr gene, avirulence gene
R gene, resistance gene
- HR
hypersensitive response
- DLA
diseased leaf area
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF262933 for the avrXa7 allele from Xoo PXO1865 and AF275267 for the allele from PXO2684).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250271997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250271997
References
- 1.Johnson R. Annu Rev Phytopathol. 1984;22:309–330. [Google Scholar]
- 2.Flor H H. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 3.Bent A F. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammond-Kosack K E, Jones J D G. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 5.Kearney B, Staskawicz B J. J Bacteriol. 1990;172:143–148. doi: 10.1128/jb.172.1.143-148.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swarup S, De Feyter R, Brlansky R H, Gabriel D W. Phytopathology. 1991;81:802–809. [Google Scholar]
- 7.Lorang J M, Shen H, Kobayashi D, Cooksey D, Keen N T. Mol Plant-Microbe Interact. 1994;7:508–515. [Google Scholar]
- 8.Ashfield T, Keen N, Buzzell R I, Innes R W. Genetics. 1995;141:1597–1604. doi: 10.1093/genetics/141.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritter C, Dangl J L. Mol Plant-Microbe Interact. 1995;8:444–453. doi: 10.1094/mpmi-8-0444. [DOI] [PubMed] [Google Scholar]
- 10.Chang J H, Rathjen J P, Bernal A J, Staskawicz B J, Michelmore R W. Mol Plant-Microbe Interact. 2000;13:568–571. doi: 10.1094/MPMI.2000.13.5.568. [DOI] [PubMed] [Google Scholar]
- 11.Rohe M, Gierlich A, Hermann H, Hahn M, Schmidt B, Rosah I S, Knogge W. EMBO J. 1995;14:4168–4177. doi: 10.1002/j.1460-2075.1995.tb00090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearney B, Staskawicz B J. Nature (London) 1990;346:385–386. doi: 10.1038/346385a0. [DOI] [PubMed] [Google Scholar]
- 13.Swords K M M, Dahlbeck D, Kearney B, Roy M, Staskawicz B J. J Bacteriol. 1996;178:4661–4669. doi: 10.1128/jb.178.15.4661-4669.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leach J E, White F F. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 15.Gabriel D W. Physiol Mol Plant Pathol. 1999;55:205–214. [Google Scholar]
- 16.Hopkins C M, White F F, Choi S-H, Guo A, Leach J E. Mol Plant-Microbe Interact. 1992;5:451–459. doi: 10.1094/mpmi-5-451. [DOI] [PubMed] [Google Scholar]
- 17.Bonas U, Stall R E, Staskawicz B J. Mol Gen Genet. 1989;218:127–136. doi: 10.1007/BF00330575. [DOI] [PubMed] [Google Scholar]
- 18.Herbers K, Conrads-Strauch J, Bonas U. Nature (London) 1992;356:172–174. [Google Scholar]
- 19.Yang Y, De Feyter R, Gabriel D W. Mol Plant-Microbe Interact. 1994;7:345–355. doi: 10.1094/mpmi-6-225. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Gabriel D W. J Bacteriol. 1995;177:4963–4968. doi: 10.1128/jb.177.17.4963-4968.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai J, Choi S H, Ponciano G, Leung H, Leach J E. Mol Plant-Microbe Interact. 2000;13:1322–1329. doi: 10.1094/MPMI.2000.13.12.1322. [DOI] [PubMed] [Google Scholar]
- 22.Mew T W, Vera Cruz C M, Medalla E S. Plant Dis. 1992;76:1029–1032. [Google Scholar]
- 23.Leach J E, Leung H, Nelson R J, Mew T W. Curr Opin Biotechnol. 1995;6:298–304. [Google Scholar]
- 24.Leung H, Nelson R J, Leach J E. In: Advances in Plant Pathology. Andrews J H, Tommerup I C, editors. New York: Academic; 1993. pp. 157–205. [Google Scholar]
- 25.Ogawa T, Yamamoto K, Khush G S, Mew T W. Jpn J Breed. 1991;41:523–529. [Google Scholar]
- 26.Oña I, Vera Cruz C M, Nelson R J, Leach J E, Mew T W. Plant Dis. 1998;82:1337–1340. [Google Scholar]
- 27.Karganilla A, Paris-Natural M, Ou S H. Philipp Agric. 1973;57:141–152. [Google Scholar]
- 28.Vera Cruz C M, Halda-Alija L, Louws F J, Skinner D Z, George M L, Nelson R J, DeBruijn F J, Rice C W, Leach J E. Int Rice Res Notes. 1995;20:23–24. [Google Scholar]
- 29.Louws F J, Fulbright D W, Stephens C T, De Bruijn F J. Appl Environ Microbiol. 1994;60:2286–2295. doi: 10.1128/aem.60.7.2286-2295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vera Cruz C M, Ardales E Y, Skinner D Z, Talag J, Nelson R J, Louws F J, Leung H, Mew T W, Leach J E. Phytopathology. 1996;86:1352–1359. [Google Scholar]
- 31.Adhikari T B, Mew T W, Leach J E. Phythopathology. 1999;89:687–694. doi: 10.1094/PHYTO.1999.89.8.687. [DOI] [PubMed] [Google Scholar]
- 32.Choi S H, Vera Cruz C M, Leach J E. Appl Environ Microbiol. 1998;64:1663–1668. doi: 10.1128/aem.64.5.1663-1668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kauffman H E, Reddy A P K, Hsiek S P V, Marca S D. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- 34.Mew T W. Annu Rev Phytopathol. 1987;25:359–382. [Google Scholar]
- 35.Yap, I. V. & Nelson, R. J. (1996) Int. Rice Res. Inst. Discuss. Pap. Ser.14, pp. 22.
- 36.Worley K C, Culpepper P, Wiese B A, Smith R F. Bioinformatics. 1998;14:890–891. doi: 10.1093/bioinformatics/14.10.890. [DOI] [PubMed] [Google Scholar]
- 37.Choi S H, Leach J E. Int Rice Res Notes. 1994;19:31–32. [Google Scholar]
- 38.Reimers P J, Leach J E. Physiol Mol Plant Pathol. 1991;38:39–55. [Google Scholar]
- 39.Ou S H. Rice Diseases. Kew: Commonwealth Mycological Institute; 1985. [Google Scholar]
- 40.Hulbert S H. Annu Rev Phytopathol. 1997;35:293–310. doi: 10.1146/annurev.phyto.35.1.293. [DOI] [PubMed] [Google Scholar]
- 41.Van der Plank J E. Genetic and Molecular Basis of Plant Pathogenesis. Berlin: Springer; 1978. [Google Scholar]
- 42.Nelson R J, Baraoidan M R, Vera Cruz C M, Yap I V, Leach J E, Mew T W, Leung H. Appl Environ Microbiol. 1994;60:3275–3283. doi: 10.1128/aem.60.9.3275-3283.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ardales E Y, Leung H, Vera Cruz C M, Mew T W, Leach J E, Nelson R J. Phytopathology. 1996;86:241–252. [Google Scholar]