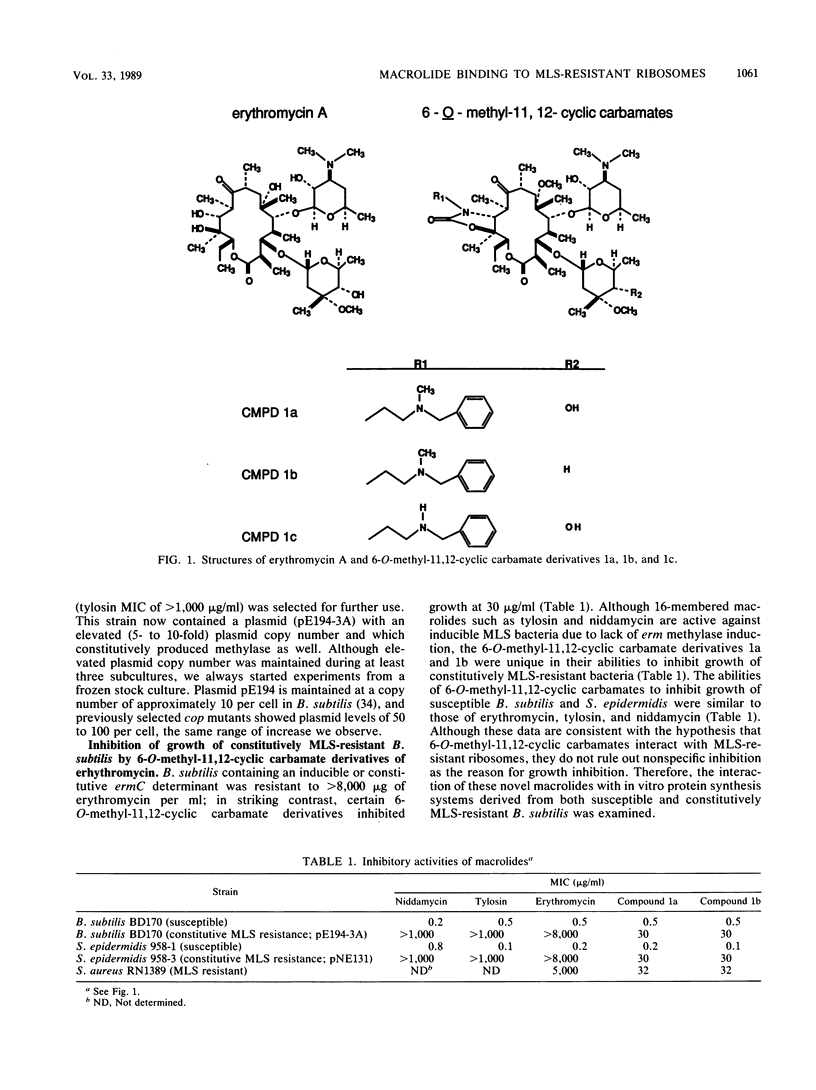
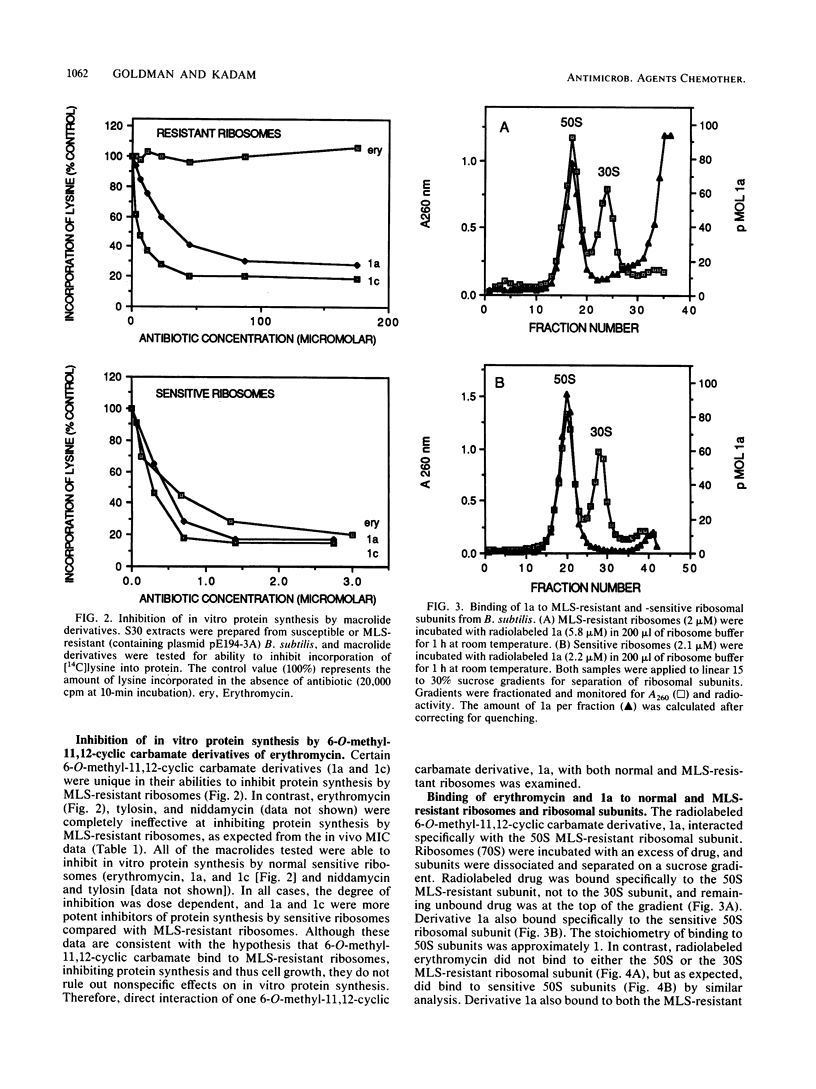
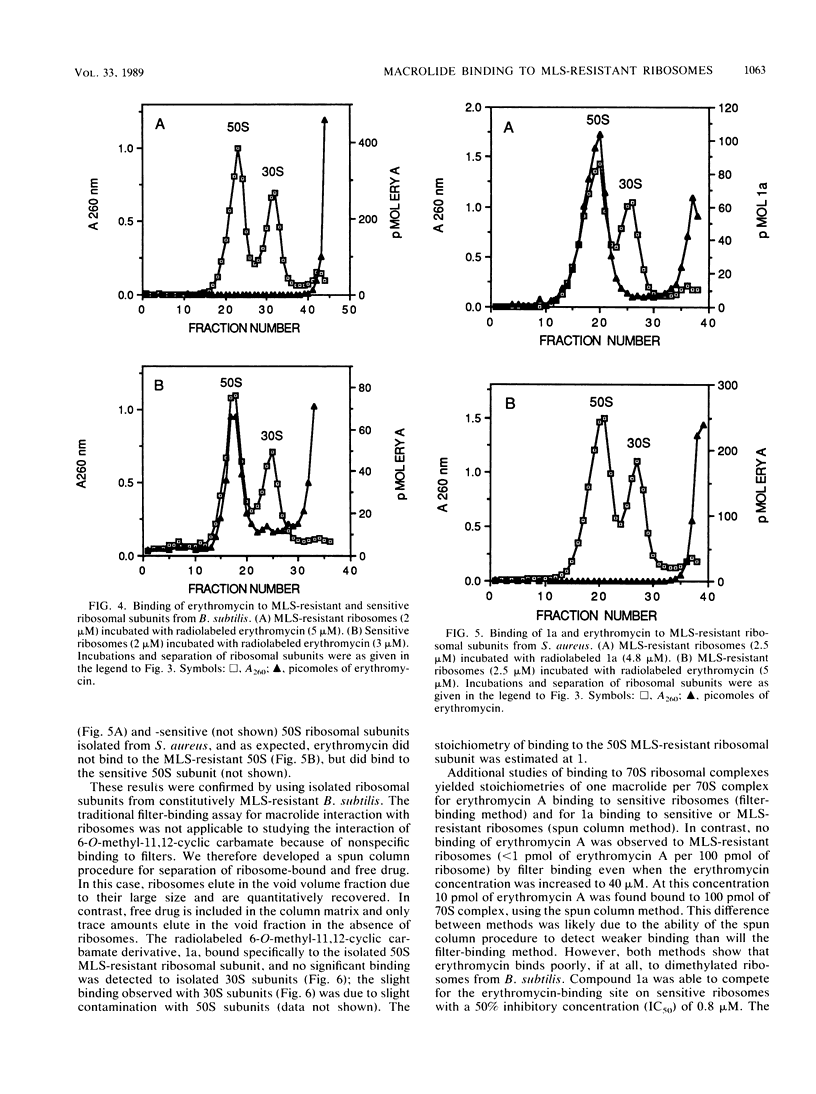
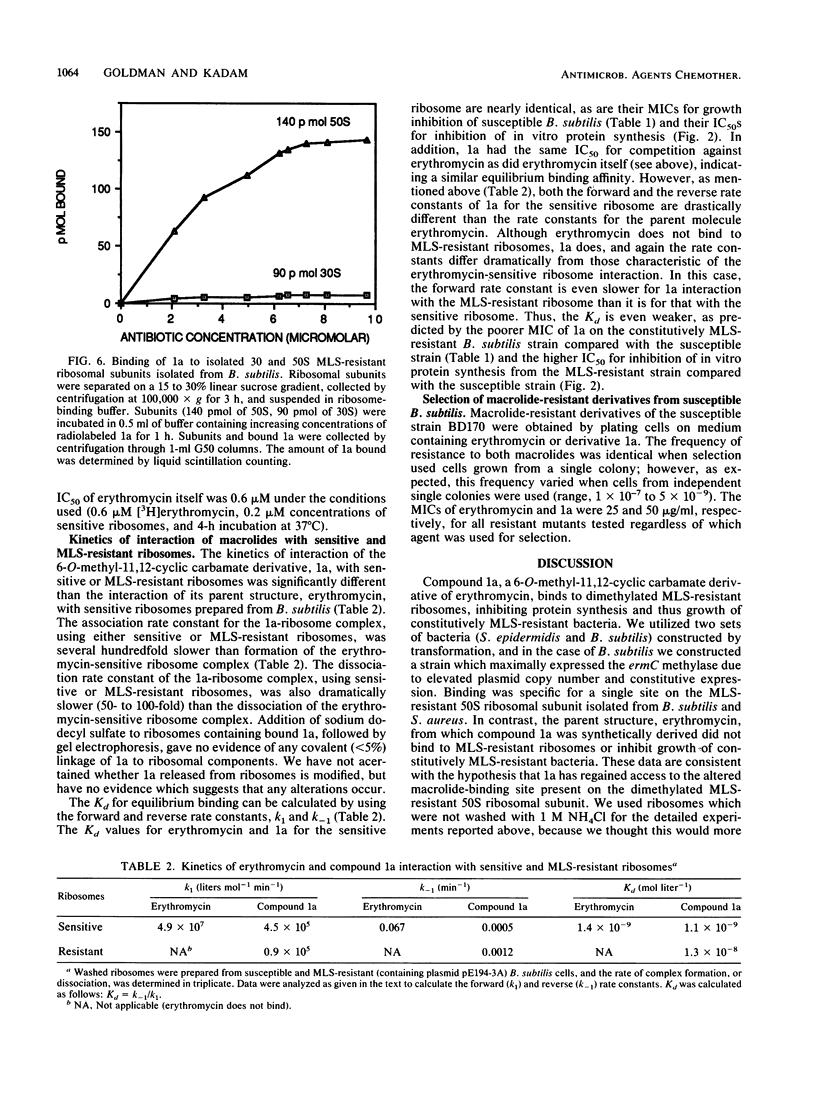
Abstract
Dimethylation of adenine 2058 in 23S rRNA renders bacteria resistant to macrolides, lincosamides, and streptogramin B (MLS resistance), because the antibiotic binding site on the altered 50S ribosomal subunit is no longer accessible. We now report that certain 6-O-methyl-11,12-cyclic carbamate derivatives of erythromycin are able to bind to dimethylated MLS-resistant 50S ribosomal subunits, thus inhibiting protein synthesis and cell growth. One of these novel structures, an 11-deoxy-11-(carboxyamino)-6-O-methylerythromycin A 11,12-(cyclic ester) derivative, structure 1a, was studied in detail. It inhibited in vitro protein synthesis in extracts prepared from both susceptible and MLS-resistant Bacillus subtilis with 50% inhibitory concentrations of 0.4 and 20 microM, respectively. The derivative bound specifically to a single site on the 50S subunit of MLS-resistant ribosomes prepared from B. subtilis and Staphylococcus aureus, and no binding to 30S subunits was observed. The association rate constant of derivative 1a with sensitive and resistant ribosomes was 100- and 500-fold slower, respectively, than that of the parent compound, erythromycin, with sensitive ribosomes. The dissociation rate constant of 1a from sensitive and resistant ribosomes was 50- to 100-fold slower than the rate of erythromycin dissociation from sensitive ribosomes. Furthermore, 1a binding to sensitive 50S subunits led to induction of ermC and ermD, while binding to resistant 50S subunits did not, showing that perturbation of sensitive and resistant 50S subunit function by 1a differs. These data demonstrated that 1a is unique in its interaction with MLS-resistant ribosomes and that this interaction causes a novel allosteric perturbation of ribosome function.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arthur M., Andremont A., Courvalin P. Distribution of erythromycin esterase and rRNA methylase genes in members of the family Enterobacteriaceae highly resistant to erythromycin. Antimicrob Agents Chemother. 1987 Mar;31(3):404–409. doi: 10.1128/aac.31.3.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Brisson-Noël A., Courvalin P. Origin and evolution of genes specifying resistance to macrolide, lincosamide and streptogramin antibiotics: data and hypotheses. J Antimicrob Chemother. 1987 Dec;20(6):783–802. doi: 10.1093/jac/20.6.783. [DOI] [PubMed] [Google Scholar]
- Contente S., Dubnau D. Characterization of plasmid transformation in Bacillus subtilis: kinetic properties and the effect of DNA conformation. Mol Gen Genet. 1979 Jan 2;167(3):251–258. doi: 10.1007/BF00267416. [DOI] [PubMed] [Google Scholar]
- Dette G. A., Knothe H., Kaula S. Modifying effects of pH and temperature on (14C)erythromycin uptake into Staphylococcus aureus--relation to antimicrobial activity. Zentralbl Bakteriol Mikrobiol Hyg A. 1987 Jul;265(3-4):393–403. [PubMed] [Google Scholar]
- Di Giambattista M., Engelborghs Y., Nyssen E., Cocito C. Kinetics of binding of macrolides, lincosamides, and synergimycins to ribosomes. J Biol Chem. 1987 Jun 25;262(18):8591–8597. [PubMed] [Google Scholar]
- Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16(2):103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- Duval J. Evolution and epidemiology of MLS resistance. J Antimicrob Chemother. 1985 Jul;16 (Suppl A):137–149. doi: 10.1093/jac/16.suppl_a.137. [DOI] [PubMed] [Google Scholar]
- Fernandes P. B., Baker W. R., Freiberg L. A., Hardy D. J., McDonald E. J. New macrolides active against Streptococcus pyogenes with inducible or constitutive type of macrolide-lincosamide-streptogramin B resistance. Antimicrob Agents Chemother. 1989 Jan;33(1):78–81. doi: 10.1128/aac.33.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro J. F., Hardisson C., Salas J. A. Involvement of cell impermeability in resistance to macrolides in some producer streptomycetes. J Antibiot (Tokyo) 1988 Jan;41(1):142–144. doi: 10.7164/antibiotics.41.142. [DOI] [PubMed] [Google Scholar]
- GARDNER R. S., WAHBA A. J., BASILIO C., MILLER R. S., LENGYEL P., SPEYER J. F. Synthetic polynucleotides and the amino acid code. VII. Proc Natl Acad Sci U S A. 1962 Dec 15;48:2087–2094. doi: 10.1073/pnas.48.12.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci U S A. 1971 Apr;68(4):856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson B. C., von David W., Parisi J. T. Novel mechanism for plasmid-mediated erythromycin resistance by pNE24 from Staphylococcus epidermidis. Antimicrob Agents Chemother. 1986 Nov;30(5):653–658. doi: 10.1128/aac.30.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois R., Cantor C. R., Vince R., Pestka S. Interaction between the erythromycin and chloramphenicol binding sites on the Escherichica coli ribosome. Biochemistry. 1977 May 31;16(11):2349–2356. doi: 10.1021/bi00630a007. [DOI] [PubMed] [Google Scholar]
- Langlois R., Lee C. C., Cantor C. R., Vince R., Pestka S. The distance between two functionally significant regions of the 50 S Escherichia coli ribosome: the erythromycin binding site and proteins L7/L12. J Mol Biol. 1976 Sep 15;106(2):297–313. doi: 10.1016/0022-2836(76)90087-5. [DOI] [PubMed] [Google Scholar]
- Legault-Demare L., Chambliss G. H. Natural messenger ribonucleic acid-directed cell-free protein-synthesizing system of Bacillus subtilis. J Bacteriol. 1974 Dec;120(3):1300–1307. doi: 10.1128/jb.120.3.1300-1307.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M., Perutz M. F. Aromatic rings act as hydrogen bond acceptors. J Mol Biol. 1988 Jun 20;201(4):751–754. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- Mao J. C., Putterman M. Accumulation in gram-postive and gram-negative bacteria as a mechanism of resistance to erythromycin. J Bacteriol. 1968 Mar;95(3):1111–1117. doi: 10.1128/jb.95.3.1111-1117.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. C., Putterman M. The intermolecular complex of erythromycin and ribosome. J Mol Biol. 1969 Sep 14;44(2):347–361. doi: 10.1016/0022-2836(69)90180-6. [DOI] [PubMed] [Google Scholar]
- Morell P., Smith I., Dubnau D., Marmur J. Isolation and characterization of low molecular weight ribonucleic acid species from Bacillus subtilis. Biochemistry. 1967 Jan;6(1):258–265. doi: 10.1021/bi00853a040. [DOI] [PubMed] [Google Scholar]
- Nakayama T., Munoz L., Doi R. H. A procedure to remove protease activities from Bacillus subtilis sporulating cells and their crude extracts. Anal Biochem. 1977 Mar;78(1):165–170. doi: 10.1016/0003-2697(77)90020-3. [DOI] [PubMed] [Google Scholar]
- O'Hara K., Kanda T., Kono M. Structure of a phosphorylated derivative of oleandomycin, obtained by reaction of oleandomycin with an extract of an erythromycin-resistant strain of Escherichia coli. J Antibiot (Tokyo) 1988 Jun;41(6):823–827. doi: 10.7164/antibiotics.41.823. [DOI] [PubMed] [Google Scholar]
- Pestka S. Binding of [14C]erythromycin to Escherichia coli ribosomes. Antimicrob Agents Chemother. 1974 Oct;6(4):474–478. doi: 10.1128/aac.6.4.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath L. D., Lorian V., Gerstein D., Loder P. B., Finland M. Enhancing effect on alkalinization of the medium on the activity of erythromycin against gram-negative bacteria. Appl Microbiol. 1968 Sep;16(9):1288–1292. doi: 10.1128/am.16.9.1288-1292.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock R. A., Leighton T., Wittmann H. G. Macrolide and aminoglycoside antibiotic resistance mutations in the bacillus subtilis ribosome resulting in temperature-sensitive sporulation. Mol Gen Genet. 1981;183(3):538–543. doi: 10.1007/BF00268778. [DOI] [PubMed] [Google Scholar]
- Skinner R., Cundliffe E., Schmidt F. J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem. 1983 Oct 25;258(20):12702–12706. [PubMed] [Google Scholar]
- Sparling P. F., Blackman E. Mutation to erythromycin dependence in Escherichia coli K-12. J Bacteriol. 1973 Oct;116(1):74–83. doi: 10.1128/jb.116.1.74-83.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince R., Weiss D., Pestka S. Binding of N-substituted erythromycyclamines to ribosomes. Antimicrob Agents Chemother. 1976 Jan;9(1):131–136. doi: 10.1128/aac.9.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B., Graham M. Y., Gryczan T., Dubnau D. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J Bacteriol. 1979 Jan;137(1):635–643. doi: 10.1128/jb.137.1.635-643.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild D. G. Reversion from erythromycin dependence in Escherichia coli: strains altered in ribosomal sub-unit association and ribosome assembly. J Gen Microbiol. 1988 May;134(5):1251–1263. doi: 10.1099/00221287-134-5-1251. [DOI] [PubMed] [Google Scholar]
- Wiley P. F., Baczynskyj L., Dolak L. A., Cialdella J. I., Marshall V. P. Enzymatic phosphorylation of macrolide antibiotics. J Antibiot (Tokyo) 1987 Feb;40(2):195–201. doi: 10.7164/antibiotics.40.195. [DOI] [PubMed] [Google Scholar]
- Yamagishi S., Nakajima Y., Inoue M., Oka Y. Decrease in accumulation of macrolide antibiotics as a mechanism of resistance in Staphylococcus aureus. Jpn J Microbiol. 1971 Jan;15(1):39–52. doi: 10.1111/j.1348-0421.1971.tb00549.x. [DOI] [PubMed] [Google Scholar]


