Abstract
The causes and consequences of chromosome loss in bacteria with multiple chromosomes are unknown. Vibrio cholerae, the causative agent of the severe diarrheal disease cholera, has two circular chromosomes. Like many other bacterial chromosomes, both V. cholerae chromosomes contain homologues of plasmid partitioning (par) genes. In plasmids, par genes act to segregate plasmid molecules to daughter cells and thereby ensure plasmid maintenance; however, the contribution of par genes to chromosome segregation is not clear. Here, we show that the chromosome II parAB2 genes are essential for the segregation of chromosome II but not chromosome I. In a parAB2 deletion mutant, chromosome II is mislocalized and frequently fails to segregate, yielding cells with only chromosome I. These cells divide once; their progeny are not viable. Instead, chromosome II-deficient cells undergo dramatic cell enlargement, nucleoid condensation and degradation, and loss of membrane integrity. The highly consistent nature of these cytologic changes suggests that prokaryotes, like eukaryotes, may possess characteristic death pathways.
Keywords: chromosome segregation, parA, parB, apoptosis
The molecular mechanisms that ensure the accurate segregation of bacterial genomes are incompletely understood. Partitioning (par) genes have been known for many years to play an essential role in the stable inheritance of certain plasmids. Plasmid partitioning systems consist of two proteins encoded by genes, often known as parA and parB, and a cis-acting centromere-like site, parS. Plasmids deleted for their par loci no longer localize to particular regions of the cell and are readily lost from host cells (1–4). ParB proteins bind to their cognate parS site(s) and form a nucleoprotein complex. ParA proteins possess Walker-type ATPase motifs and show weak ATPase activity. ParA proteins interact with the ParB–parS complex, and their ATPase activity is essential for plasmid partitioning (5, 6). Several ParA homologues have been shown to form ATP-dependent polymers in vitro and to oscillate in vivo (7–10). Still, the molecular mechanisms by which the interactions of ParA with the ParB–parS complex mediate plasmid localization and partitioning are not well understood, although several models have been proposed to account for Par-mediated plasmid segregation (9–13). Homologues of plasmid Par proteins are also encoded in most bacterial chromosomes (11, 14); however, less is known about the function of chromosomal parABS loci.
It has become clear in recent years that some bacteria, including several pathogens, have complex genomes that consist of more than one circular chromosome (15). The genome of Vibrio cholerae, the curved Gram-negative rod that causes the severe diarrheal disease cholera, is divided between two circular chromosomes (16). Chromosome I [chrI; 2.96 megabases (Mb)] is larger than chromosome II (chrII; 1.07 Mb) and contains the majority of the genes considered essential for cell growth. ChrII contains some essential genes, including rpmI, rplT, infC, and thrS, which encode ribosome proteins L22 and L35, a translation initiation factor, and an aminoacyl tRNA synthetase, respectively (16). Interestingly, chrII also encodes 13 toxin-antitoxin (TA) loci; similar TA loci are often found in plasmids, where they ensure maintenance by preventing the survival of plasmid-free cells (17–19). The presence of many TA loci, as well as other features of chrII, led to the suggestion that this chromosome originated as a megaplasmid acquired by an ancestral Vibrio species (16). However, the presence of essential genes on chrII qualifies this replicon as a bona fide chromosome (15, 16).
Although the two V. cholerae chromosomes initiate replication in coordinated fashion and have some shared replication requirements (20, 21), different mechanisms appear to control their segregation, and their origin regions have distinct subcellular distributions and dynamics. In newborn cells, the origin of replication of chrI (oriCIvc) is found near one of the cell poles, whereas the origin of replication of chrII (oriCIIvc) is found near the cell center (22, 23). The origin region of chrI undergoes an asymmetric segregation process: one copy of the duplicated oriCIvc remains at the pole while the other copy traverses the cell to the opposite pole (22–24). In contrast, the duplicated copies of oriCIIvc segregate in a symmetric fashion from the cell center to the quarter positions of the cell, the sites of the new cell centers (22, 23). The pattern of oriCIvc segregation is very similar to that reported for the Caulobacter crescentus chromosome (25), whereas the pattern of oriCIIvc segregation is similar to that reported for P1 and the F plasmid (3, 26).
Each of the V. cholerae chromosomes has parAB genes near its replication origin (16). Phylogenetic analyses suggest that the Par proteins encoded on chrI (ParA1 and ParB1) cluster with other chromosome-encoded Par proteins, whereas the Par proteins encoded on chrII (ParA2 and ParB2) tend to cluster with plasmid and phage Par proteins (11, 14, 16). Given the importance of par loci in plasmid segregation, we have initiated studies to evaluate the role of parAB1 and parAB2 in V. cholerae chromosome segregation. Recently, we found that parA1 is required for the polar localization and asymmetric segregation pattern of chrI but does not influence the segregation of chrII. Furthermore, parA1 mutants did not have an appreciable cell growth defect and did not lose chrI (24, 27). Here, we investigated the role of parAB2 in the maintenance and segregation of the V. cholerae chromosomes. We found that ParAB2 functions in a chromosome-specific manner to promote the accurate subcellular localization and maintenance of chrII but not chrI. A parAB2 deletion mutant yields a high frequency of cells that lack chrII. Cells containing only chrI divide once and then become “CHUB” cells (an acronym we created based on some of the cells' properties: condensed nucleoid, hypertrophic, undividing bacteria). Also, some of the characteristic cytologic changes that occur in cells lacking chrII appear to be attributable to the activity of some of the toxins encoded in the TA loci on chrII.
Results and Discussion
parABS2 Is Functional in a Heterologous Host.
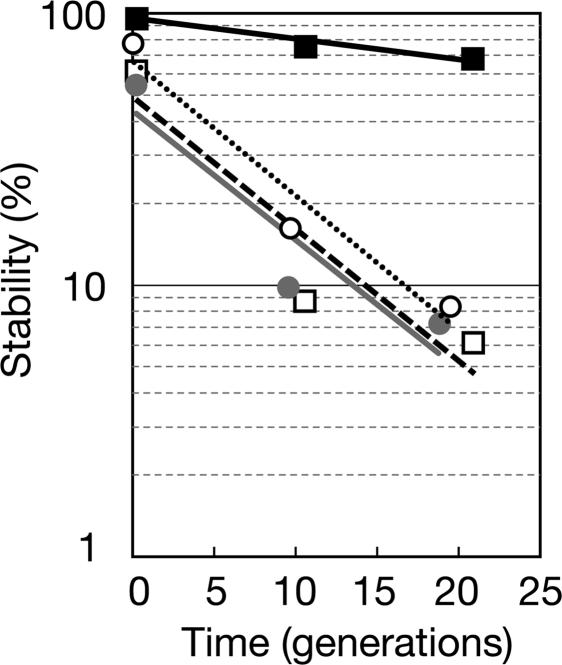
Several previous studies have introduced chromosomal parABS loci into an unstable derivative of the F plasmid to test their functionality in a heterologous host, Escherichia coli (11, 28–31). To perform parallel experiments, we defined ParB2-binding sites (parS2). We found that six of nine parS2 sites on chrII were fairly close to oriCIIvc, but none were found within the parAB2 genes (our unpublished work). Then we explored whether parAB2, along with parS2, can mediate the partitioning of an unstable plasmid in E. coli. Although the unstable mini-F plasmid was readily lost from E. coli in the absence of selection, insertion of parAB2 and parS2 dramatically stabilized the plasmid (Fig. 1). The ability of V. cholerae parABS2 to stabilize this plasmid in E. coli indicates that the chrII par loci encode a functional partitioning system and suggests that they may mediate DNA partitioning by the same general mechanism used by related Par proteins of some plasmids. Furthermore, this observation suggests that parABS2 does not require any additional V. cholerae-specific factors to mediate partitioning. Insertion of either parS2 or parAB2 alone did not stabilize the mini-F plasmid (Fig. 1), suggesting that all three components of the chrII par system are required for the activity of this partitioning system.
Fig. 1.
V. cholerae parABS2 stabilizes a mini-F plasmid in E. coli. Stability is measured as the percentage of cells harboring mini-F plasmid derivatives over time. The mini-F derivatives are pYB145 (parABS2+; filled squares, solid line), pYB095 (parAB2+; filled circles, gray line), pYB141 (parS2+; open squares, broken line), and control vector pXX705 (open circles, dotted line).
parAB2 Is Required for Wild-Type (WT) Cell Growth.
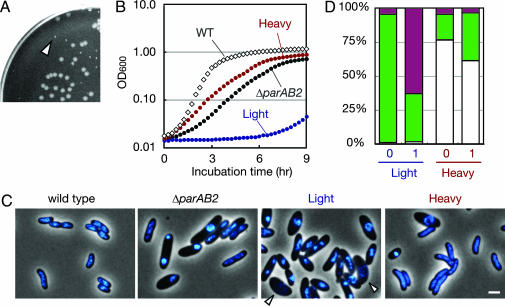
Using a standard allele exchange method (32), we found that we were unable to delete parAB2 unless a WT copy of these genes was provided on a plasmid (pYB085). However, pYB085 could be lost from the host deletion mutant. Mutant cells that lost pYB085 were easily identified because they generated tiny colonies [Fig. 2A and supporting information (SI) Fig. 6]. These cells also had a severe growth defect in liquid culture (Fig. 2B) that could be complemented by providing a plasmid-borne copy of parAB2 (SI Fig. 6 and data not shown). The severe growth defect of the parAB2 mutant is in marked contrast to the lack of an appreciable growth defect in parA1 mutants (24, 27).
Fig. 2.
Growth defect and heterogeneity of cell size, shape, and nucleoid morphology in a ΔparAB2 V. cholerae strain. (A) Heterogeneous colony size of a ΔparAB2 strain harboring a parAB2 complementing plasmid grown on nonselective LB plates. All of the normal-sized colonies were found to carry the parAB2-bearing plasmid, whereas all of the tiny colonies (arrowhead) lost the parAB2-bearing plasmid (data not shown). (B) Growth in LB at 37°C of WT (diamonds), unfractionated ΔparAB2 cells (black circles), light fraction (blue circles), and heavy fraction (red circles). (C) Merged phase-contrast and fluorescence images of DAPI-stained fixed cells. The arrowheads point to cells with fragmented nucleoids. (Scale bar, 2 μm.) (D) BacLight live/dead assay of fractionated cells. Cells were assayed immediately after fractionation (time 0) or after growth in LB medium at 37°C for 1 h. Normal and CHUB cells were distinguished by morphology. Depicted are normal-live cells (white), normal-dead cells (black), CHUB-live cells (green), and CHUB-dead cells (magenta).
The ΔparAB2 strain consists of two phenotypically different types of cells. About one-third of the ΔparAB2 cells were normal in size, shape, and nucleoid morphology, and about two-thirds of the cells were significantly larger, irregularly shaped, and had condensed nucleoids (CHUB cells; Fig. 2C and Table 1). The CHUB phenotype is directly associated with a ParAB2 deficiency, inasmuch as CHUB cells were not seen in the WT population or in ΔparAB2 cells that contain a parAB2 complementing plasmid. CHUB cells also have a lower density than WT cells, allowing them to be isolated by density gradient centrifugation (33). The “heavy” fraction consisted of ≈80% normal-appearing cells; DAPI staining revealed diffuse nucleoids similar to those of WT cells (Fig. 2C and Table 1). In contrast, the vast majority of the cells in the “light” fraction were large and had the condensed nucleoid(s) characteristic of CHUB cells. In addition, CHUB cells appear to be inviable; there was virtually no increase in the optical density (OD) of cultures of cells isolated from the light fraction for up to 6 h after inoculation in LB (Fig. 2B). The plating efficiency (per OD unit) of cells from the light fraction was nearly 1/300 that of cells from the heavy fraction (Table 1). Furthermore, although isolated CHUB cells were initially scored as “live” according to BacLight (Molecular Probes, Eugene, OR) live/dead analysis, after 1 h of incubation at 37°C in LB, these cells apparently lost membrane integrity, allowing propidium iodide staining and their classification as “dead” (Fig. 2D). Overall, the growth defect of the parAB2 mutant appears to be largely attributable to the emergence of inviable CHUB cells in this background.
Table 1.
Characteristics of ΔparAB2 V. cholerae before and after fractionation on a density gradient
| Fraction | Morphology* |
cfu (/ml/OD600)* | qPCR† |
||
|---|---|---|---|---|---|
| Normal | CHUB | oriCIIvc/oriCIvc | TerIIvc/TerIvc | ||
| Unfractionated | 33.6% | 66.4% | 5.6 × 107 | 0.30 ± 0.02 | 0.34 ± 0.09 |
| Light | 0.7% | 99.3% | 5.7 × 105 | 0.020 ± 0.006 | 0.020 ± 0.006 |
| Heavy | 83.0% | 17.0% | 1.7 × 108 | 0.62 ± 0.12 | 0.63 ± 0.12 |
*Average of three independent experiments.
†Mean ± SD of three independent experiments.
parAB2 Is Required for Maintenance of ChrII.
The ability of parAB2 to stabilize an unstable plasmid, and the severe growth defect of the parAB2 mutant, raised the possibility that these genes were important for V. cholerae chromosome segregation. To examine the chromosome content of the parAB2 strain, we used quantitative PCR (qPCR) to measure chromosome-specific DNA at multiple sites on both chromosomes. In contrast to WT cells (for which we set the ratio chrII/chrI equal to 1), in parAB2 cells the ratio chrII/chrI was only ≈0.3 (Table 1). Thus, in the absence of parAB2, chrII appears to be frequently lost. Furthermore, CHUB cells appear to completely lack chrII (Table 1). The small percentage of purified CHUB cells containing chrII DNA (≈2%) is probably contaminating non-CHUB cells. Taken together, these observations suggest that the loss of chrII due to the absence of parAB2 results in the development of CHUB cells.
parAB2 Is Required for Proper Subcellular Localization and Segregation of ChrII.
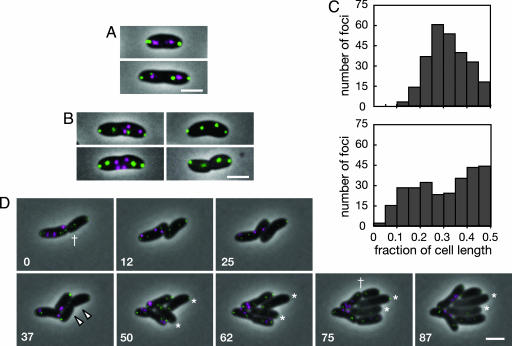
A fluorescent protein fused to ParB2 (e.g., CFP-ParB2) can be used to determine the subcellular location of the parS2 sites, which on chrII are mostly clustered near the origin of replication (our unpublished work). Similarly, the ParB1 protein binds to parS1 sites near the origin of chrI, and a fluorescent fusion protein (e.g., YFP-ParB1) can be used to visualize the subcellular localization of the chrI origin region (24). Most WT V. cholerae had a YFP-ParB1 focus close to each pole, and some cells had additional (presumably segregating) foci at intermediate positions between the poles (Fig. 3A). The majority of WT cells also had two CFP-ParB2 foci, one near each quarter position (Fig. 3 A and C). The subcellular locations of YFP-ParB1 and CFP-ParB2 foci in WT cells are consistent with previous studies of chrI and chrII origin localization (22, 23). Thus, YFP-ParBI and CFP-ParB2 mark the subcellular locations of the origin region of each chromosome.
Fig. 3.
Number and subcellular location of YFP-ParB1 and CFP-ParB2 foci in WT and ΔparAB2 V. cholerae. (A and B) Static images of representative WT (A) and ΔparAB2 (B) cells expressing YFP-ParB1 and CFP-ParB2. (C) Histograms showing the localization of CFP-ParB2 foci in WT (Upper) and ΔparAB2 (Lower) cells. The distance from each focus to the nearest pole was measured and divided by cell length. A total of 261 foci from 118 WT cells and 274 foci from 103 ΔparAB2 cells were analyzed. (D) Time-lapse images of ΔparAB2 cells taken at 12-min intervals at 35°C. The number in the lower left corner of each image indicates the time (min). YFP is shown in green, and CFP is shown in magenta. The dagger shows a cell division yielding a cell lacking chrII. The arrowheads show cells that will become CHUBs. Asterisks show enlarging CHUB cells. (Scale bar, 2 μm.)
We compared the localization and segregation of both chromosomes in WT and parAB2 mutant cells by using YFP-ParB1 and CFP-ParB2 fusion proteins. Nearly 100% of WT cells with any detectable foci had both YFP-ParB1 and CFP-ParB2 foci (Fig. 3A and Table 2), suggesting that both chrI and chrII were present. This was strikingly not the case for the ΔparAB2 strain. Only ≈24% of ΔparAB2 cells with detectable foci had both YFP-ParB1 and CFP-ParB2 foci (Table 2), whereas ≈76% had only YFP-ParB1 foci, consistent with the loss of chrII in parAB2 cells as measured by qPCR (see Table 1).
Table 2.
Percentages of cells with both YFP-ParB1 and CFP-ParB2, or with YFP-ParB1 only or CFP-ParB2 only, in WT and ΔparAB2 cells
| Strain | YFP-ParB1 and CFP-ParB2 | YFP-ParB1 only | CFP-ParB2 only |
|---|---|---|---|
| WT | 99.4% | 0.3% | 0.3% |
| ΔparAB2 | 23.5% | 75.9% | 0.6% |
In the ΔparAB2 cells, the location of the YFP-ParB1 foci was similar to that observed in WT cells (Fig. 3B). However, the subcellular distribution of CFP-ParB2 foci in ΔparAB2 cells was markedly different from that observed in the WT cells. In the ΔparAB2 cells, the CFP-ParB2 foci were not found predominantly near the quarter positions; instead, these foci appeared to be distributed fairly randomly throughout the cell (Fig. 3 B and C). Thus, parAB2 is required for subcellular positioning of chrII. In the absence of parAB2, the random subcellular localization of chrII apparently prevents accurate division of replicated chrII copies among daughter cells. Because parAB2 genes do not appear to influence chrI localization, our observations suggest that these genes are key components of a chrII-specific partitioning system.
Time-lapse studies revealed that loss of chrII was often detectable in ΔparAB2 cells (Fig. 3D) but never in WT cells. Cells lacking chrII underwent one more round of cell division (Fig. 3D, white arrowheads), yielding daughter cells in which the YFP-ParB1 foci were still apparent. These cells then progressively enlarged to become CHUB cells (Fig. 3D, asterisks). Thus, newborn cells lacking chrII are precursors of CHUB cells (pre-CHUBs), and pre-CHUBs are capable of one round of cell division, suggesting that there is no mechanism to block cell division in cells with an incomplete set of chromosomes. CHUB cells appear to lack the capacity to divide, inasmuch as no CHUB cell divisions were observed in numerous time-lapse experiments.
Degradation of ChrI in CHUB Cells.
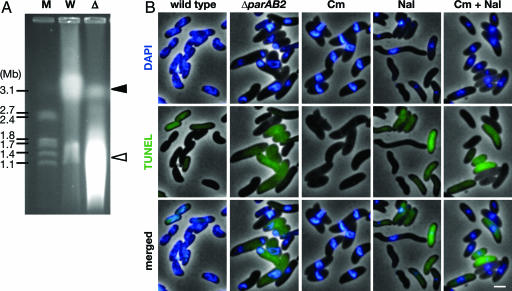
The nucleoids of some of the CHUB cells appeared as broken rings (Fig. 2C, arrowheads), suggestive of nucleoid fragmentation. Pulsed-field gel electrophoresis of uncut DNA isolated from ΔparAB2 cells also suggested that there was significant degradation of the chromosomal DNA in this background (Fig. 4A). The presence of DNA fragmentation in ΔparAB2 cells was confirmed by TUNEL assays. TUNEL is commonly used to assess nuclear DNA fragmentation in eukaryotes associated with apoptosis (34), and this technique has also been shown to be useful for detecting DNA damage in prokaryotes (35). Approximately 70% of CHUB cells had intense and diffuse fluorescence (Fig. 4B), suggesting that there is massive degradation of chrI in these cells.
Fig. 4.
ChrI degradation in CHUB cells. (A) Pulsed-field gel electrophoresis of intact (undigested) genomic DNA prepared from WT (W) and ΔparAB2 (Δ) cells. Molecular weight standards (M) are shown at left. The positions of chrI and chrII are indicated by the black and white arrowheads, respectively. In principal, the more rapidly migrating DNA in the ΔparAB2 cells could be derived from increased initiations of replication of chrI; however, overinitiation of chrI does not appear to occur in ΔparAB2 cells, inasmuch as the ratio oriCIvc/TerIvc in these cells was 0.88, compared with 1.0 in WT, as determined by qPCR. (B) TUNEL assay of ΔparAB2 cells and WT cells with no treatment or with treatment with chloramphenicol (Cm), nalidixic acid (Nal), or both. (Scale bar, 2 μm.)
It is possible that cells that have lost chrII are subject to the activity of some or all of the 13 toxins encoded in chrII TA loci, because toxins are more stable than their cognate antitoxins. Ten of the toxins are thought to inhibit translation, and the other three are thought to inhibit DNA gyrase (18, 19, 36). We used antibiotics that inhibit translation (chloramphenicol, Cm) or DNA gyrase (nalidixic acid, Nal) to begin to assess whether the action of one or more of the chrII toxins could account for the CHUB phenotype. Cm-treated cells were enlarged and had condensed nucleoids similar to CHUB cells, but TUNEL fluorescence in these cells was not as frequent or intense as in CHUB cells (Fig. 4B and SI Fig. 7). Nal-treated cells had similar TUNEL fluorescence as CHUB cells but were smaller and had more punctate nucleoids than CHUB cells (Fig. 4B and SI Fig. 7). Cells treated with both antibiotics also had similar TUNEL fluorescence as CHUB cells but did not entirely recapitulate the CHUB phenotype; compared with CHUB cells, these cells were usually smaller in length and width and had more punctate nucleoids (Fig. 4B). Thus, the combined action of chrII-encoded toxins likely contributes to some aspects of the CHUB phenotype.
Conclusions.
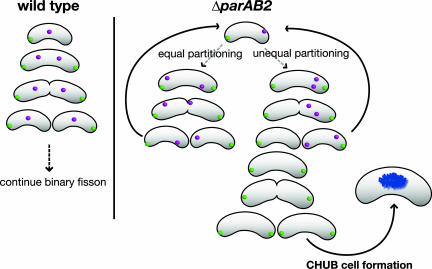
There appears to be no redundancy in the mechanisms that account for chrII segregation in V. cholerae. In the parAB2 mutant, chrII appears to have a random distribution (Fig. 3C) that results in its frequent missegregation and the generation of cells lacking this chromosome (Fig. 5). The high frequency of chrII loss in the parAB2 mutant appears to explain the growth defect of this strain. In contrast, redundant mechanisms likely mediate chrI segregation; deletion of parA1 abrogates the polar localization of oriCIvc, but chrI is faithful partitioned in this background and cell growth is not altered (24, 27). Redundant mechanisms also seem to operate in the segregation of the Bacillus subtilis and E. coli chromosomes (37, 38). In contrast, low-copy-number plasmids usually depend on a single par-based partitioning mechanism. Thus, the lack of redundant mechanisms governing chrII partitioning supports the hypothesis that this chromosome arose from a plasmid or a phage. Perhaps consistent with this hypothesis is our observation that the Par systems of the two chromosomes appear to function independently of one another; i.e., there was no detectable mislocalization of oriCIvc in the parAB2 background and no oriCIIvc mislocalization in the parA1 background (24, 27).
Fig. 5.
Schematic representation of chromosome segregation and the generation of CHUB cells in ΔparAB2 V. cholerae. (Left) Schematic representation showing the pattern of segregation of the origin regions of chrI (green) and chrII (magenta) in WT cells. (Right) The chrII origin region is randomly localized in the ΔparAB2 mutant. Cell division can therefore result in daughter cells that either contain or lack this chromosome. Cells lacking chrII divide only once, and then their progeny develop into CHUB cells.
The missegregation of chrII in ΔparAB2 V. cholerae routinely yields cells that contain only chrI, and these aneuploid cells undergo a highly consistent set of cytologic changes terminating in cell death. The cells divide once; the resulting daughter cells, which are no longer capable of division, then enlarge, undergo nucleoid condensation and degradation, and lose membrane integrity. At least some of these cytologic changes may be attributable to the activity of the toxins encoded on the chrII superintegron. It has been proposed that TA loci can promote the genetic stability of superintegrons (39); our findings raise the possibility that TA loci may also contribute to the overall integrity of the V. cholerae genome. Future studies should address whether the cytologic changes observed in cells that contain only chrI can be entirely explained by the action of chrII toxins, or whether the absence of ≈25% of the V. cholerae genetic information also contributes. The cytologic changes in cells containing only chrI bear some similarity to programmed cell death in eukaryotes, but it is not clear whether they reflect a particular V. cholerae genetic program. To date, the pathology of prokaryotic cell death has largely been ignored. Our findings suggest that characterizing such death pathways will be a fruitful area for study. Finally, it was proposed that V. cholerae cells containing a single chromosome could act as “drones”: metabolically active, nondividing cells that could contribute to the population (16). Our findings demonstrate that generation of V. cholerae cells containing only chrI is possible, but the contribution of CHUB cells to the cell population remains to be explored.
Materials and Methods
Strains and Plasmids.
The strains and plasmids used this study are listed in SI Table 3 and SI Table 4, respectively. All V. cholerae strains used in this study were derived from the sequenced El Tor clinical isolate N16961 (16). E. coli strains DH5α, DH5a λpir, and TOP10 were used for DNA cloning. SM10 λpir was used to mobilize DNA into V. cholerae, and MC1061 was the host strain for the plasmid stability assay. The growth curves shown in Fig. 2B were obtained by using a SynergyHT microplate reader and KC4 software (BioTek, Winooski, VT).
Isolation of CHUB Cells.
Growing cells (11 ml; OD600 ≈ 0.3) and 3 ml of 40% (wt/vol) Ludox (Sigma, St. Louis, MO), filtered and pH-adjusted to ≈7.5 before use, were mixed in 15-ml cortex tubes and then centrifuged at 9,000 × g for 30 min. The top band was then carefully transferred to a new tube by pipette and washed three times with PBS.
Analyses of DNA Content.
Real-time qPCR to compare the content of chrI and chrII was performed as described previously (40), except that primers for oriCIIvc were 5′-CCAGTGATGAATGGTGTGT-3′ and 5′-ATGATCGAGACTTACGGTATG-3′. Standard curves were obtained with genomic DNA prepared from growing WT cells. Pulsed-field gel electrophoresis was carried out as described previously (41). DNA degradation was detected by TUNEL assay, using the In Situ Cell Death Detection Kit, Fluorescein (Roche Applied Science, Indianapolis, IN) in accordance with the manufacture's instructions. Samples were analyzed with either microscopy or a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ).
Microscopy.
For fluorescence microscopy experiments, strains were grown in LB medium at 37°C to the midexponential phase. Fluorescent-tagged proteins under the control of the PBAD promoter were induced with 0.08% l-arabinose for 1 h before cells were transferred to an agarose pad on a microscope slide. Vectashield mounting medium with DAPI (Vector Laboratories, Burlingame, CA) was used to observe the nucleoid of cells fixed with 3% paraformaldehyde. The BacLight live/dead bacterial viability kit for microscopy (Molecular Probes) was used for the membrane integrity assay. All images were acquired and processed as described previously (22, 37).
Supplementary Material
Acknowledgments
We thank H. Niki (National Institute of Genetics, Mishima, Japan) for plasmids and M.K.W. laboratory members for useful advice and comments on the manuscript. M.K.W. was supported by National Institutes of Health Grant AI42347 and Howard Hughes Medical Institute.
Abbreviations
- chrI
chromosome I
- chrII
chromosome II
- CHUB
condensed nucleoid hypertrophic undividing bacteria
- TA
toxin-antitoxin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608341104/DC1.
References
- 1.Ogura T, Hiraga S. Cell. 1983;32:351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- 2.Austin S, Abeles A. J Mol Biol. 1983;169:373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- 3.Niki H, Hiraga S. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Dabrazhynetskaya A, Youngren B, Austin S. Mol Microbiol. 2004;53:93–102. doi: 10.1111/j.1365-2958.2004.04111.x. [DOI] [PubMed] [Google Scholar]
- 5.Ebersbach G, Gerdes K. Annu Rev Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 6.Hayes F, Barillà D. Nat Rev Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- 7.Ebersbach G, Gerdes K. Proc Natl Acad Sci USA. 2001;98:15078–15083. doi: 10.1073/pnas.261569598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebersbach G, Gerdes K. Mol Microbiol. 2004;52:385–398. doi: 10.1111/j.1365-2958.2004.04002.x. [DOI] [PubMed] [Google Scholar]
- 9.Lim GE, Derman AI, Pogliano J. Proc Natl Acad Sci USA. 2005;102:17658–17663. doi: 10.1073/pnas.0507222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebersbach G, Ringgaard S, Moller-Jensen J, Wang Q, Sherratt DJ, Gerdes K. Mol Microbiol. 2006;61:1428–1442. doi: 10.1111/j.1365-2958.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- 11.Yamaichi Y, Niki H. Proc Natl Acad Sci USA. 2000;97:14656–14661. doi: 10.1073/pnas.97.26.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Austin S. Mol Microbiol. 2002;46:63–74. doi: 10.1046/j.1365-2958.2002.03156.x. [DOI] [PubMed] [Google Scholar]
- 13.Adachi S, Hori K, Hiraga S. J Mol Biol. 2006;356:850–863. doi: 10.1016/j.jmb.2005.11.088. [DOI] [PubMed] [Google Scholar]
- 14.Gerdes K, Moller-Jensen J, Bugge Jensen R. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 15.Egan ES, Fogel MA, Waldor MK. Mol Microbiol. 2005;56:1129–1138. doi: 10.1111/j.1365-2958.2005.04622.x. [DOI] [PubMed] [Google Scholar]
- 16.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, et al. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes F. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 18.Gerdes K, Christensen SK, Lobner-Olesen A. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 19.Pandey DP, Gerdes K. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egan ES, Waldor MK. Cell. 2003;114:521–530. doi: 10.1016/s0092-8674(03)00611-1. [DOI] [PubMed] [Google Scholar]
- 21.Egan ES, Lobner-Olesen A, Waldor MK. Curr Biol. 2004;14:R501–R502. doi: 10.1016/j.cub.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 22.Fogel MA, Waldor MK. Mol Microbiol. 2005;55:125–136. doi: 10.1111/j.1365-2958.2004.04379.x. [DOI] [PubMed] [Google Scholar]
- 23.Fiebig A, Keren K, Theriot JA. Mol Microbiol. 2006;60:1164–1178. doi: 10.1111/j.1365-2958.2006.05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fogel MA, Waldor MK. Genes Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon GS, Sitnikov D, Webb CD, Teleman A, Straight A, Losick R, Murray AW, Wright A. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 27.Saint-Dic D, Frushour BP, Kehrl JH, Kahng LS. J Bacteriol. 2006;188:5626–5631. doi: 10.1128/JB.00250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartosik AA, Lasocki K, Mierzejewska J, Thomas CM, Jagura-Burdzy G. J Bacteriol. 2004;186:6983–6998. doi: 10.1128/JB.186.20.6983-6998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubarry N, Pasta F, Lane D. J Bacteriol. 2006;188:1489–1496. doi: 10.1128/JB.188.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godfrin-Estevenon AM, Pasta F, Lane D. Mol Microbiol. 2002;43:39–49. doi: 10.1046/j.1365-2958.2002.02735.x. [DOI] [PubMed] [Google Scholar]
- 31.Jakimowicz D, Chater K, Zakrzewska-Czerwínska J. Mol Microbiol. 2002;45:1365–1377. doi: 10.1046/j.1365-2958.2002.03102.x. [DOI] [PubMed] [Google Scholar]
- 32.Donnenberg MS, Kaper JB. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evinger M, Agabian N. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavrieli Y, Sherman Y, Ben-Sasson SA. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohwer F, Azam F. Appl Environ Microbiol. 2000;66:1001–1006. doi: 10.1128/aem.66.3.1001-1006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen-Dalsgaard M, Gerdes K. Mol Microbiol. 2006;62:397–411. doi: 10.1111/j.1365-2958.2006.05385.x. [DOI] [PubMed] [Google Scholar]
- 37.Yamaichi Y, Niki H. EMBO J. 2004;23:221–233. doi: 10.1038/sj.emboj.7600028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee PS, Grossman AD. Mol Microbiol. 2006;60:853–869. doi: 10.1111/j.1365-2958.2006.05140.x. [DOI] [PubMed] [Google Scholar]
- 39.Rowe-Magnus DA, Guerout AM, Biskri L, Bouige P, Mazel D. Genome Res. 2003;13:428–442. doi: 10.1101/gr.617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duigou S, Knudsen KG, Skovgaard O, Egan ES, Lobner-Olesen A, Waldor MK. J Bacteriol. 2006;188:6419–6424. doi: 10.1128/JB.00565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaichi Y, Iida T, Park KS, Yamamoto K, Honda T. Mol Microbiol. 1999;31:1513–1521. doi: 10.1046/j.1365-2958.1999.01296.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.