Abstract
The β3-adrenoceptor (AR) differs from the β1-AR and β2-ARs in having introns within and downstream of the coding block. This study demonstrates two splice variants of the mouse β3-AR which differ within the coding region.
Reverse transcription/polymerase chain reaction with intron-spanning primers was used to demonstrate the splice variant of the mouse β3-adrenoceptor. The novel β3b-AR has 17 amino acids encoded by exon 2 (SSLLREPRHLYTCLGYP) which differ from the 13 in the known β3a-AR (RFDGYEGARPFPT).
β3b-AR mRNA is differentially expressed in mouse tissues, with levels relative to β3a-AR mRNA highest in hypothalamus, cortex and white adipose tissue, and lower in ileum smooth muscle and brown adipose tissue.
Keywords: β3-adrenoceptor, splice variants, mouse, adipose tissue, ileum, brain
Introduction
The β3-adrenoceptor (AR) is a potential target for anti-obesity and anti-diabetic drugs, mediating stimulation of lipolysis in white adipose tissue (WAT) and lipolysis and thermogenesis in brown adipose tissue (BAT). Functional β3-ARs are also found in the stomach and in other regions of the gastrointestinal tract where they mediate relaxation of smooth muscle (Cohen et al., 1995; Manara et al., 1996; Roberts et al., 1997). The β3-AR agonist SR 58611A has antidepressant effects in mice (Simiand et al., 1992), and infusion of BRL 37344 into the third cerebral ventricle of rats suppresses food intake (Tsujii & Bray, 1992). mRNA encoding the β3-AR has been detected in a variety of mammalian tissues, including WAT and BAT, stomach, colon, ileum, gallbladder and brain (Krief et al., 1993; Summers et al., 1995; Evans et al., 1996). In mice fed a high-fat diet and in genetically obese ob/ob C57BL/6J mice, chronic treatment with β3-selective agonists promotes weight loss, improves insulin sensitivity and lowers plasma insulin levels (Arch et al., 1984; Arbeeny et al., 1995; Collins et al., 1997), possibly by a combination of actions on adipose tissue, gut smooth muscle and the central nervous system. The role of the β3-AR in mediating responses to endogenous neuronal or hormonal stimulation has been investigated using knockout and transgenic mice (Susulic et al., 1995; Grujic et al., 1997; Revelli et al., 1997). Mice with targeted disruption of the β3-AR gene have increased lipid stores and are more susceptible to weight gain when fed on a high-fat diet, indicating that the β3-AR participates in the regulation of energy balance. Acute responses to the β3-AR agonist CL 316243, including increased lipolysis and oxygen consumption, increased circulating insulin levels and reduced food intake, are completely absent in β3-AR knockout mice (Susulic et al., 1995).
There appears to be redundancy in the response mediated by β3-ARs and classical β1/β2-ARs in various cell types. For example, β1-, β2- and β3-ARs can all mediate lipolysis in adipocytes, their relative contributions depending on the type of fat depot, the species, and other physiological influences (Arch & Wilson, 1996). Stimulation of all three β-ARs increases adenosine 3′5′ cyclic monophosphate (cyclic AMP) production via activation of GS, and the effects of β-AR agonists can be mimicked by cyclic AMP analogues or forskolin (Deng et al., 1997). However in some cell types, although the predominant receptor present is the β3-AR, increases in cyclic AMP occur primarily through β1- and β2-ARs (Jockers et al., 1998). The β3-AR may mediate responses via other signalling pathways. For example we have shown that relaxation of rat ileum smooth muscle to β3-AR agonists is not accompanied by an increase in cyclic AMP (S.J. Roberts, unpublished data). In isolated adipocytes the stimulation of cyclic AMP by BRL 37344 is limited by coupling of the β3-AR to Gi (Chaudhry et al., 1994). Similarly, β2-ARs in heart couple strongly to Gi, preventing agonist stimulation of myocyte contractility (Xiao et al., 1999). In contrast β1-ARs do not couple to Gi under normal conditions in either heart or adipocytes (Chaudhry et al., 1994; Xiao et al., 1999). β3-ARs differ from classical β1- and β2-ARs in their regulatory properties. At the protein level, desensitization of β1- and β2-AR responses upon agonist stimulation involves phosphorylation of occupied receptors, uncoupling and internalization (Summers et al., 1997). Both β1- and β2-ARs have serine and threonine residues in the C-terminal tail which act as substrates for G protein-coupled receptor kinases, and consensus sequences for phosphorylation by cyclic AMP-dependent protein kinase (PKA). The β3-AR lacks a PKA phosphorylation site, and has fewer serine and threonine residues in the C-terminal tail. Experimentally, the β3-AR is resistant to short term agonist-promoted desensitization. Studies on chimeric β2/β3-ARs show that domains within the C-terminal tail and second and third intracellular loops of the β2-AR are the major determinants of desensitization (Liggett et al., 1993; Jockers et al., 1996).
The gene encoding the β3-AR differs from the β1- and β2-AR genes in that it contains three exons and two introns (van Spronsen et al., 1993; Granneman & Lahners, 1995). Previous studies in mice demonstrate that the use of alternate splice acceptor sites results in the generation of two β3-AR transcripts which differ in their 3′ untranslated regions (van Spronsen et al., 1993). Here we report that alternative splicing also generates two isoforms of the β3-AR which have interesting differences in the C-terminal tail and are differentially expressed in mouse tissues. In addition we have demonstrated using primers located in the 3′ untranslated region that at least five transcripts of the β3-AR gene could be identified.
Methods
Tissue dissection and RNA preparation
Male C57BL/6J mice (8 weeks of age) were anaesthetized with 80% CO2/20% O2 and decapitated. Total RNA was extracted from white adipose tissue (WAT) from epididymal fat pads, interscapular brown adipose tissue (BAT), hypothalamus, cortex, heart ventricle and ileal smooth muscle by homogenization in TRIZOL (Life Technologies). The yield and quality of the RNA were assessed by measuring absorbance at 260 and 280 nm, and by electrophoresis on 1.2% agarose gels. Total RNA was treated with DNase to remove any contaminating genomic DNA. The reaction mix contained 20 μg RNA, sodium acetate (pH 7.0) 100 mM, MgSO4 5 mM, dithiothreitol 5 mM, 36 U RNasin (Promega), and 10 U DNase I (Amersham Pharmacia Biotech) in a total volume of 40 μl. Following digestion at 37°C for 30 min, the solution was diluted to 400 μl with H2O and extracted with an equal volume of phenol : chloroform (1 : 1). The RNA was precipitated with 1.0 ml of ethanol and 40 μl of 2 M sodium acetate. The yield and quality of DNase-treated RNA were determined as above.
Reverse transcription/PCR
cDNAs were synthesized by reverse transcription of 1.0 μg of each total RNA using oligo (dT)15 as a primer. The RNA in a volume of 7.5 μl was heated to 70°C for 5 min then placed on ice for 2 min prior to the addition of reaction mix containing 1×reverse transcription (RT) buffer (supplied by Promega), dNTPs 1 mM, MgCl2 5 mM, 18 U RNasin (Promega), 20 U AMV reverse transcriptase (Promega), and 50 μg ml−1 oligo(dT)15 in a volume of 12.5 μl. Following brief centrifugation, the reactions were incubated at 42°C for 45 min, then at 95°C for 5 min. The completed reverse transcription reactions were stored at −20°C and used for polymerase chain reaction (PCR) without further treatment. A negative control for each RNA sample was produced by setting up the RT reaction as usual but omitting the reverse transcriptase. PCR amplification was carried out on cDNA equivalent to 100 ng of starting RNA, using intron-spanning primers. For amplification of β3-AR cDNA, the primers were forward, mb3.f1 or mb3.f2 and reverse, mb3.r1, mb3.r2, or mb3.r3 (Table 1). PCR mixes contained 2.5 pmol each of the forward and reverse primers, 1 U Taq polymerase (Life Technologies), Tris-HCl (pH 8.4) 20 mM, KCl 50 mM, dNTPs 200 μM, Mg-acetate 3.0 mM, and cDNA in a volume of 10 μl. Following heating at 95°C for 2 min, amplification cycles were 30 s at 95°C, 30 s at 64°C, 30 s extension at 72°C. Appropriate cycle numbers were determined as described in Figure 3 and by Evans et al. (1998). PCR products were separated by gel electrophoresis on 1.3% agarose and transferred onto Hybond N+ (Amersham) in 0.4 M NaOH/1 M NaCl. β3-AR genomic sequences were amplified for 30 cycles from C57BL/6J DNA.
Table 1.
Oligonucleotides used as PCR primers and hybridization probes, and expected PCR fragmentation sizes
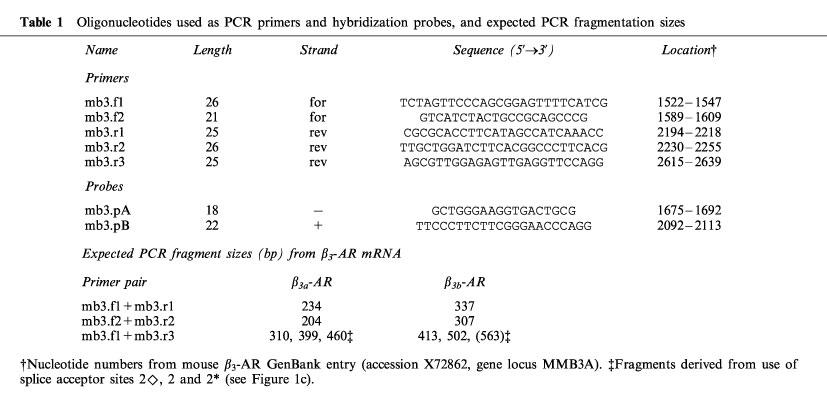
Figure 3.
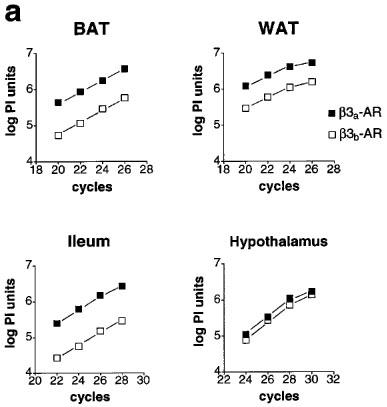
Relative levels of β3a-AR and β3b-AR mRNA. (a) Product/cycle relationship for β3-AR PCR performed on cDNA from BAT, WAT, ileum and hypothalamus. The cDNA was produced by reverse transcription of 1 μg total RNA, and one tenth used for PCR with β3-AR primers (mb3.f1 and mb3.r1). For each tissue, a single PCR mix was aliquotted into four separate capillaries which were removed sequentially from the cycler and placed on ice after the cycle numbers shown. Following gel electrophoresis and transfer to Hybond N+, PCR products were detected by hybridization with a [33P]-labelled β3-AR probe (mb3.pA) and exposure to phosphorimager plates for 16 h. The β3a-AR and β3b-AR products were quantitated using ImageQuaNT software (Molecular Dynamics), and the log of the volume (phosphorimager units, PI) plotted against cycle number. Note that the lines representing accumulation of the β3a-AR and β3b-AR PCR products are parallel, indicating proportional amplification. (b) β3b-AR splice variant mRNA as a percentage of total β3-AR mRNA in BAT, WAT, ileum, hypothalamus and cortex. Bars show the mean (s.e.mean). β3a-AR and β3b-AR fragments were quantitated as in (a) from multiple tissue samples (5 to 12).
Detection and measurement of PCR products
PCR products were fixed to nylon membranes (u.v. light 2 min) before prehybridization at 42°C for 4 h in a buffer containing 5×SSC, 0.5% SDS, 100 μg ml−1 salmon sperm DNA (pre-heated to 95°C for 5 min), 5×Denhardts solution (1 mg ml−1 ficoll type 400, 1 mg ml−1 polyvinylpyrrolidone, 1 mg ml−1 bovine serum albumin) and 0.1 mM ATP. Products were hybridized in the same buffer at 42°C for 16 h to a β3-AR probe, mb3.pA, or a probe selective for the splice variant sequence, mb3.pB (Table 1). Each probe (10 pmol) was end-labelled with 15 μCi [γ-33P]-ATP (2000 Ci mmol−1; Bresatec) and T4 polynucleotide kinase (Pharmacia). Filters were washed in 2×SSC/0.1% SDS, rinsed at room temperature then washed at 30°C for 20–30 min then at 37°C for 5 min. Radioactivity was detected using a Molecular Dynamics SI phosphorimager, and bands were quantitated using the ‘Volume Report' function of ImageQuaNT software (Molecular Dynamics).
Subcloning and sequencing
Following excision from low melting temperature agarose gels, PCR products were sublconed into the PCR-Script vector (Stratagene) according to the manufacturer's instructions. Positive colonies were detected by hybridization to the mb3.pA probe, and minipreps from five independent clones were sequenced (Micromon, Monash University, Australia).
Reagents
TRIZOL reagent, oligo(dT)15, oligonucleotide primers and probes, Taq polymerase, 10×PCR buffer and salmon sperm DNA were from Life Technologies (Gaithersburg, MD, U.S.A.); RNasin, 10×RT buffer and AMV reverse transcriptase were from Promega (Madison, WI, U.S.A.); dNTPs (100 mM each nucleotide), 100 base pair (bp) DNA ladder, One-Phor-All Plus buffer and T4 polynucleotide kinase were from Amersham Pharmacia Biotech (Uppsala, Sweden); and [γ-33P]-ATP (2000 Ci mmol−1) was from Bresatec (Adelaide, S. Australia).
Results
We have found consistently that with intron-spanning primers, RT–PCR on RNA from mouse tissues expressing β3-AR mRNA produces 2 bands. For example, primers mb3.f1 and mb3.r1 give the expected β3-AR product of 234 bp, but also an extra product of 337 bp (Figure 1a and b). As the proportion of this larger product was highest in the brain sample, RNA from mouse hypothalamus was subjected to RT–PCR then three additional rounds of PCR, and the products excised from an agarose gel. The upper 337 bp band corresponding to the novel transcript was subcloned into PCR-Script and five different subclones sequenced. This showed that the β3-AR mRNA corresponding to the 337 bp fragment is produced by alternative splicing at a novel acceptor site 100 bp upstream from the previously characterized start of exon 2 (van Spronsen et al., 1993). The β3b-AR encoded by the alternately spliced mRNA has a C-terminus which has 17 amino acids (SSLLREPRHLYTCLGYP) following the sequence encoded by the first exon region compared to 13 (RFDGYEGARPFPT) in the known receptor (β3a-AR, Figure 1c). We have shown subsequently both by hybridization and by sequencing that BAT expresses the same mRNA encoding the β3b-AR. Also, we see similar proportions of β3b-AR relative to total β3-AR mRNA in tissues from C57BL/6 and Swiss mice (data not shown).
Figure 1.
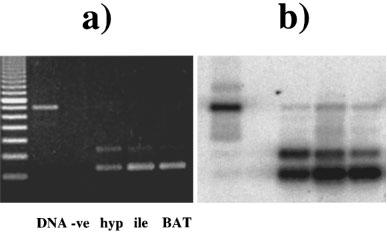
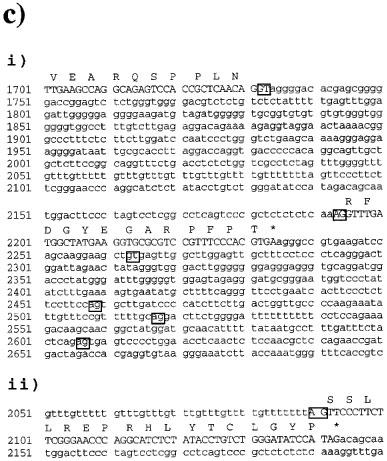
Detection of the β3-adrenoceptor splice variant mRNA in mouse tissues. (a) ethidium bromide-stained gel showing PCR products generated using primers mb3.f1 and mb3.r1. PCR was performed as described in Methods using C57BL/6J genomic DNA, and RT–PCR using RNA from hypothalamus, ileum smooth muscle, and brown adipose tissue. The negative control contained no added RNA in the reverse transcription reaction. Cycle numbers were 28 for hypothalamus and ileum, and 24 for BAT. Fragment sizes were determined by comparison with 100 bp DNA ladder (Pharmacia). (b) Following transfer to Hybond N+, PCR products were detected by hybridization with a [33P]-labelled β3-AR probe (mb3.pA) and exposure to phosphorimager plates for 16 h. (c) Sequence of exon 2 and adjacent sequences in the mouse β3-AR gene. (i) shows the splice donor-1 (nucleotide 1732) and acceptor-1 (nuc. 2193) sites utilized in β3a-AR mRNA. Also boxed are the intron B donor-2 site (nuc. 2263) and three potential acceptor sites within the 3′-untranslated region, acceptor-2⋄ (nuc. 2458), acceptor-2 (nuc. 2517) and acceptor-2* (nuc. 2606). Nucleotide numbers correspond to those in the original mouse β3-AR GenBank entry (accession number X72862, locus MMB3A). Amino acids are shown in one-letter code over the third nucleotide of each codon. (ii) shows the novel splice acceptor site utilized in β3b-AR mRNA. The results shown in Figure 4 demonstrate that exon 2 in this mRNA continues through the coding and 3′-untranslated regions found in β3a-AR mRNA.
Because the β3b-AR transcript was detected by RT–PCR, we have conducted several experiments to ensure that it is not an artefact (Figures 2 and 3). First, total RNA from BAT, WAT, ileum, hypothalamus, cortex and heart was treated with DNase to remove contaminating genomic DNA. Any genomic DNA containing the full intron A sequence would give a PCR product of 697 bp rather than the observed 337 bp. Also, two independent sets of PCR primers both gave the upper band corresponding to β3b-AR mRNA (Figure 2a and b). The PCR amplifications shown in Figure 2 were done on pairs of RT reactions with and without reverse transcriptase added. In the absence of enzyme, there was negligible PCR product corresponding to either the β3a-AR or β3b-AR transcript. Over a much larger number of RNA samples which have not been treated with DNase and thus have variable levels of genomic DNA contamination, we find no correlation (either positive or negative) between the amount of genomic DNA band (697 bp) and the amount of β3b-AR band (data not shown). These observations all tend to rule out the possibility of DNA artefacts due to template jumping or other occurrences during PCR. Also, artefactual products would be very unlikely to show splicing at the bona fide donor and acceptor sites identified in the β3b-AR transcript. A second consideration on RT–PCR is that for tissues that have a low abundance of β3-AR mRNA, the reaction might favour amplification of the minor β3b-AR species. Figure 3a shows, however, that the relative levels of β3a-AR and β3b-AR product in BAT, WAT, ileum and hypothalamus are maintained over a range of cycle numbers. mRNA coding for the β3b-AR represents 7.6±0.4% of the total β3-AR transcripts in BAT, 21±0.9% in WAT and 16±0.8% in ileum smooth muscle. The highest proportion of β3b-AR transcripts occurs in brain, with 38±1.9% in hypothalamus and 32±1.3% in cortex (Figure 3b).
Figure 2.
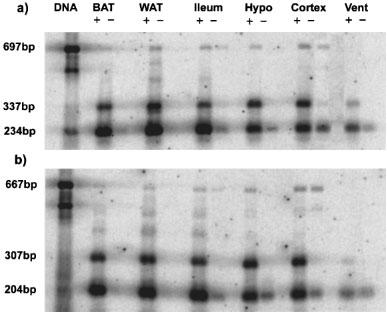
Confirmation of the presence of β3-adrenoceptor splice variant mRNA in mouse tissues. PCR was performed as described in Methods using C57BL/6J genomic DNA, and RT–PCR using RNA from BAT, WAT, ileum smooth muscle, hypothalamus, cortex and heart ventricle. For each tissue, PCR was carried out using RT reactions performed with all components except in the presence (left) or absence (right) of reverse transcriptase. PCR primers were either mb3.f1 and mb3.r1 (a) or mb3.f2 and mb3.r2 (b), and cycle numbers were 24 for WAT and BAT cDNA and 28 for ileum, hypothalamus and cortex (see Figure 3a). Fragment sizes were determined by comparison with 100 bp DNA ladder (Pharmacia). Following transfer to Hybond N+, PCR products were detected by hybridization with a [33P]-labelled β3-AR probe (mb3.pA) and exposure to phosphorimager plates for 16 h.
We were next interested in whether the alternate splice sites in the 3′-untranslated region previously described by van Spronsen and co-workers (1993) are used in conjunction with the novel splice site in intron A. Primers mb3.f1 and mb3.r3 were used for PCR amplification of all possible β3-AR mRNAs (Figure 1c), and the products examined by hybridization with a probe, specific for the splice variant sequence, mb3.pB (Figure 4a) or general β3-AR probe, mb3.pA (Figure 4b). If we consider the standard β3a-AR mRNA, the expected PCR products derived from the use of acceptor-2 or acceptor-2* (van Spronsen et al., 1993) are 399 and 310 bp respectively. As shown in Figure 4b, acceptor-2* predominates over acceptor-2. Quantitation of the two bands indicates that the shorter fragment from acceptor-2* represents 70–80% of the β3a-AR transcripts in all three tissues. PCR fragments of 502 and 413 bp derived from the use of acceptor-2 or acceptor-2* in the β3b-AR mRNA are seen in Figure 4a and b. Again the shorter 413 bp fragment represents about 70% of β3b-AR transcripts in hypothalamus, ileum and BAT. In addition to the four expected bands, there is another band of 460 bp which hybridizes only with the general β3-AR probe and which shows a tissue distribution consistent with β3a-AR mRNA, namely BAT>ileum>hypothalamus. This may represent β3a-AR transcripts derived from a third acceptor site in the 3′-untranslated region (acceptor 2⋄ in Figure 1c).
Figure 4.
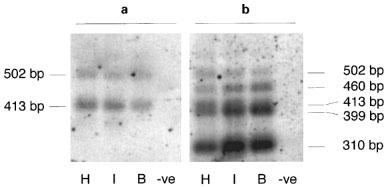
(a) Hybridization of a β3b-AR specific probe (splice variant) or (b) a general β3-AR probe to PCR fragments from hypothalamus (H), ileum smooth muscle (I) and brown adipose tissue (B), or a negative control RT–PCR containing no added RNA. PCR was done using primers mb3.f1 and mb3.r3 described in Methods, and fragment sizes were determined from the ethidium bromide-stained gel by comparison to 100 bp DNA ladder. The reverse primer mb3.r3 is located 3′ to the distal acceptor-2* site described by van Spronsen et al. (1993) and therefore detects all possible β3-AR splice variants.
Discussion
Several studies have demonstrated multiple transcripts of the β3-AR gene with differing degrees of polyadenylation or the use of alternate promoters (van Spronsen et al., 1993; Granneman & Lahners, 1994). The presence of introns in the β3-AR gene (Bensaid et al., 1993; Granneman et al., 1992) further stimulated the search for multiple isoforms of the receptor. A detailed study of the structure of the human and mouse β3-AR genes (van Spronsen et al., 1993) showed that in addition to the well recognized 2.2–2.4 kb mRNA species, there are larger size transcripts which could result from the use of alternate promoters. In the mouse, there was clear evidence for the production of transcripts which differed in their 3′ untranslated regions due to the use of alternate splice acceptor sites. These transcripts differed in their tissue expression with the shorter transcript predominant in WAT and the longer in BAT (van Spronsen et al., 1993). However in the study reported here we have also demonstrated alternative splicing of the mouse β3-AR transcript within the coding region, resulting in the production of a mRNA encoding a β3-AR variant (β3b-AR). mRNA coding for this receptor represents 7.6% of the β3-AR transcripts in BAT, 21% in WAT and 16% in ileum smooth muscle. The highest proportion of β3b-AR transcripts occurs in brain, with 38% in hypothalamus and 32% in cortex.
Despite a significant representation of β3b-AR mRNA in mouse WAT, this splice variant has not been detected by other groups (van Spronsen et al., 1993; Granneman & Lahners, 1995). We believe that the apparent absence of the β3b-AR transcript in these studies can be explained by the procedures used for detection of splice variants. The primers used for RT–PCR by van Spronsen and co-workers (1993) gave fragments of 743 and 832 bp corresponding to transcripts derived from splice acceptor sites 2* and 2 respectively. However, the major β3b-AR transcript incorporating the acceptor-2* site would give a PCR fragment of 846 bp, which would not have separated clearly from the 832 bp fragment following gel electrophoresis. The PCR product derived from the β3b-AR/acceptor-2 transcript would be expected to have lower abundance, and therefore would be undetectable on an ethidium bromide-stained gel. In the study of Granneman & Lahners (1995), a ribonuclease protection assay was used to detect and quantitate β3-AR splice variant mRNAs. In both BAT and WAT the probe was found to protect a 171 bp fragment in β3-AR mRNA derived from acceptor-2, or a 129 bp fragment in mRNA derived from acceptor-2*. However β3b-AR/acceptor-2 transcripts would give protected fragments of 68, 61 and 42 bp, while β3b-AR/acceptor-2* transcripts would produce only the 68 and 61 bp fragments. As these fragments are much shorter than those expected, they may have been disregarded as artefacts of the ribonuclease protection assay. We believe that our PCR data using two different sets of primers and a variety of mouse tissues correctly indicate the presence of the β3b-AR splice variant.
The mouse β3b-AR has a unique C-terminus with 17 amino acids (SSLLREPRHLYTCLGYP) quite distinct from the 13 (RFDGYEGARPFPT) present in the known β3a-AR. In particular, the β3b-AR C-terminal region has a cysteine residue additional to the palmitoylation site cysteine (Cys)358 which is the sole site available in the mouse β3a-AR (Figure 5). It is tempting to speculate that Cys400 may represent an alternative or competing palmitoylation site in the β3b-AR, a possibility which also exists in the human β3-AR which has two cysteine residues in the C-terminal region after the putative palmitoylation site (Strosberg, 1997). This does not occur in the human β2-AR where the primary site for palmitoylation is clearly Cys341 despite the presence of Cys378 and Cys406 (O'Dowd et al., 1989). Precedents do exist however for palmitoylation of cysteines located more distally in other receptors such as the mGLUR4 receptor (Morello & Bouvier, 1996).
Figure 5.
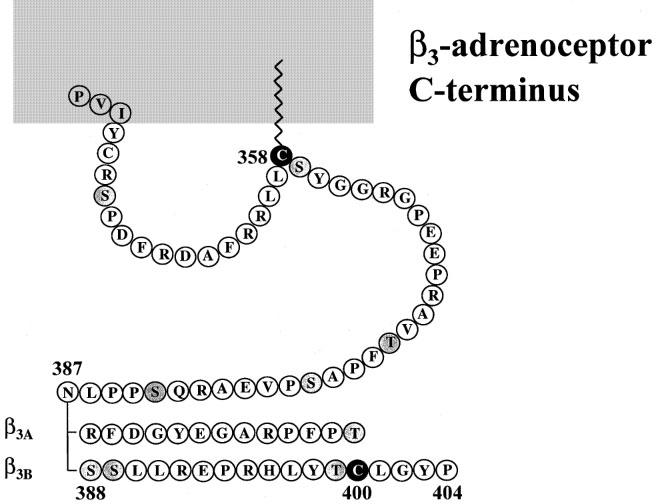
C-terminal tail of the β3a-AR and β3b-AR. Asn387 is the final common amino acid between the β3a-AR and β3b-AR. Cys358 is conserved amongst mouse, human and rat β3-AR, and also between the β3-AR and β2-AR in each species. In the β2-AR the corresponding Cys341 is known to be palmitoylated (O'Dowd et al., 1989). Serine and threonine residues which are potential phosphorylation sites are shaded.
A second characteristic of the β3b-AR compared to the β3a-AR C-terminal tail is the presence of two additional serine residues (Figure 5). These are potentially important targets for phosphorylation and may thereby mediate desensitization of the β3b-AR. The distal serine, Ser389 may be a target for protein kinase C as there is a basic residue arginine (Arg) in close proximity towards the C terminus (Bouvier et al., 1991). Although superficially similar, the human β3-AR has the basic residue (Arg) located more distally to its corresponding Ser390. Phosphorylation by G protein-coupled receptor kinases is less likely, as the human β3-AR has seven serine residues in the C-terminal tail versus eight serine and threonine residues in the β3b-AR, and the human receptor is resistant to short-term desensitization by catecholamines (Nantel et al., 1993). It will be of interest to determine whether the mouse β3b-AR demonstrates a greater tendency towards phosphorylation and associated desensitization.
A number of G-protein coupled receptors including the 5-HT4 (Gerald et al., 1995) and 5-HT7 (Heidmann et al., 1997) receptors, α1A/C adrenoceptor (Hirasawa et al., 1995) and prostaglandin EP3 receptor (Namba et al., 1993) are synthesized as separate isoforms with different C-terminal tails due to alternate splicing within the coding region. In the case of the prostaglandin EP3 receptor, the four isoforms have been shown to couple to different G proteins with resulting differences in intracellular signalling. We suggest that generation of variant G protein-coupled receptors by alternative splicing may be relatively common when the receptors are encoded by genes containing introns, further increasing the diversity of responses to particular hormones. However, alternative splicing of a particular receptor mRNA will not necessarily occur across all mammalian species. We have demonstrated a splice variant of the mouse β3-AR, whereas in the rat there is no evidence for alternative splicing within the coding region (Granneman & Lahners, 1995; Evans et al., 1996). Although there is an AG sequence at the equivalent position in the rat gene, the sequences upstream and downstream are substantially different. We have also analysed human tissues which express β3-AR mRNA, including gallbladder, colon and SK-N-MC neuroblastoma cells, for the presence of splice variants (Hamilton & Evans, unpublished data). RT–PCR using several sets of forward and reverse primers indicates no alternative acceptor sites either upstream or downstream of that previously described (van Spronsen et al., 1993). The presence of the β3b-AR variant in mouse but not rat or human may indicate somewhat divergent roles for the β3-AR, perhaps reflecting differences in energy homeostasis in small mammals which have high thermogenic requirements.
In conclusion, the present study shows that there is an additional novel β3-AR that is differentially expressed in mouse tissues with the highest proportion of the splice variant present in hypothalamus and cortex. We have expressed the β3a-AR and β3b-AR in CHO-K1 cells and find that both are functional. We are currently determining whether the novel receptor differs from the known receptor in signalling and regulatory properties.
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council of Australia. B.A. Evans is a Monash University Biomedical Research Fellow.
Abbreviations
- AR
adrenoceptor
- Arg
arginine
- BAT
brown adipose tissue
- bp
base pairs
- cyclic AMP
adenosine 3′5′ cyclic monophosphate
- Cys
cysteine
- PCR
polymerase chain reaction
- PKA
cyclic AMP-dependent protein kinase
- RT
reverse transcription
- Ser
serine
- WAT
white adipose tissue
References
- ARBEENY C.M., MEYERS D.S., HILLYER D.E., BERGQUIST K.E. Metabolic alterations associated with the antidiabetic effect of β3-adrenergic receptor agonists in obese mice. Am. J. Physiology. 1995;31:E678–E684. doi: 10.1152/ajpendo.1995.268.4.E678. [DOI] [PubMed] [Google Scholar]
- ARCH J.R., AINSWORTH A.T., CAWTHORNE M.A., PIERCY V., SENNITT M.V., THODY V.E. Atypical β-adrenoceptor on brown adipocytes as target for anti-obesity drugs. Nature. 1984;309:163–165. doi: 10.1038/309163a0. [DOI] [PubMed] [Google Scholar]
- ARCH J.R.S., WILSON S. Prospects for β3-adrenoceptor agonists in the treatment of obesity and diabetes. Int. J. Obesity. 1996;20:191–199. [PubMed] [Google Scholar]
- BENSAID M., KAGHAD M., RODRIGUEZ M., LE FUR F.G., CAPUT D. The rat β3-adrenergic receptor gene contains an intron. FEBS Lett. 1993;318:223–226. doi: 10.1016/0014-5793(93)80516-w. [DOI] [PubMed] [Google Scholar]
- BOUVIER M., GUILBAULT N., BONIN H. Phorbol-ester-induced phosphorylation of the β2-adrenergic receptor decreases its coupling to GS. FEBS Lett. 1991;279:243–248. doi: 10.1016/0014-5793(91)80159-z. [DOI] [PubMed] [Google Scholar]
- CHAUDHRY A., MACKENZIE R.G., GEORGIC L.M., GRANNEMAN J.G. Differential interaction of β1- and β3-adrenergic receptors with Gi in rat adipocytes. Cellular Signalling. 1994;6:457–465. doi: 10.1016/0898-6568(94)90093-0. [DOI] [PubMed] [Google Scholar]
- COHEN M.L., GRANNEMAN J.G., CHAUDHRY A., SCHENCK K.W., CUSHING D.J., PALKOWITZ A.D. Is the atypical β-receptor in the rat stomach fundus the rat β3 receptor. J. Pharmacol. Exp. Ther. 1995;272:446–451. [PubMed] [Google Scholar]
- COLLINS S., DANIEL K.W., PETRO A.E., SURWIT R.S. Strain-specific response to β3-adrenergic receptor agonist treatment of diet-induced obesity in mice. Endocrinology. 1997;138:405–413. doi: 10.1210/endo.138.1.4829. [DOI] [PubMed] [Google Scholar]
- DENG C., MOINAT M., CURTIS L., NADAKAL A., PREITNER F., BOSS O., ASSIMACOPOULOS-JEANNET F., SEYDOUX J., GIACOBINO J. Effects of β-adrenoceptor subtype stimulation on obese gene messenger ribonucleic acid and on leptin secretion in mouse brown adipocytes differentiated in culture. Endocrinology. 1997;138:548–552. doi: 10.1210/endo.138.2.4922. [DOI] [PubMed] [Google Scholar]
- EVANS B.A., PAPAIOANNOU M., ANASTASOPOULOS F., SUMMERS R.J. Differential regulation of β3-adrenoceptors in gut and adipose tissue of genetically obese (ob/ob) C57BL/6J mice. Br. J. Pharmacol. 1998;124:763–771. doi: 10.1038/sj.bjp.0701867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EVANS B.A., PAPAIOANNOU M., BONAZZI V.R., SUMMERS R.J. Expression of β3-adrenoceptor mRNA in rat tissues. Br. J. Pharmacol. 1996;117:210–216. doi: 10.1111/j.1476-5381.1996.tb15176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GERALD C., ADHAM N., KAO H.T., OLSEN M.A., LAZ T.M., SCHECHTER L.E., BARD J.A., VAYSSE P.J., HARTIG P.R., BRANCHEK T.A., WEINSHANK R.L. The 5-HT4 receptor: molecular cloning and pharmacological characterization of two splice variants. EMBO J. 1995;14:2806–2815. doi: 10.1002/j.1460-2075.1995.tb07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRANNEMAN J.G., LAHNERS K.N. Analysis of human and rodent β3-adrenergic receptor messenger ribonucleic acids. Endocrinology. 1994;135:1025–1031. doi: 10.1210/endo.135.3.8070345. [DOI] [PubMed] [Google Scholar]
- GRANNEMAN J.G., LAHNERS K.N. Regulation of mouse β3-adrenergic receptor gene expression and mRNA splice variants in adipocytes. Am. J. Physiol. – Cell Physiology. 1995;37:C1040–C1044. doi: 10.1152/ajpcell.1995.268.4.C1040. [DOI] [PubMed] [Google Scholar]
- GRANNEMAN J.G., LAHNERS K.N., RAO D.D. Rodent and human β3-adrenergic receptor genes contain an intron within the protein-coding block. Mol. Pharmacol. 1992;42:964–970. [PubMed] [Google Scholar]
- GRUJIC D., SUSULIC V.S., HARPER M.E., HIMMS-HAGEN J., CUNNINGHAM B.A., CORKEY B.E., LOWELL B.B. β3-adrenergic receptors on white and brown adipocytes mediate β3-selective agonist-induced effects on energy expenditure, insulin secretion, and food intake. A study using transgenic and gene knockout mice. J. Biol. Chem. 1997;272:17686–17693. doi: 10.1074/jbc.272.28.17686. [DOI] [PubMed] [Google Scholar]
- HEIDMANN D.E., METCALF M.A., KOHEN R., HAMBLIN M.W. Four 5-hydroxytryptamine7 (5-HT7) receptor isoforms in human and rat produced by alternative splicing: species differences due to altered intron-exon organization. J. Neurochem. 1997;68:1372–1381. doi: 10.1046/j.1471-4159.1997.68041372.x. [DOI] [PubMed] [Google Scholar]
- HIRASAWA A., SHIBATA K., HORIE K., TAKEI Y., OBIKA K., TANAKA T., MURAMOTO N., TAKAGAKI K., YANO J., TSUJIMOTO G. Cloning, functional expression and tissue distribution of human α1C-adrenoceptor splice variants. FEBS Lett. 1995;363:256–260. doi: 10.1016/0014-5793(95)00330-c. [DOI] [PubMed] [Google Scholar]
- JOCKERS R., DA SILVA A., STROSBERG A.D., BOUVIER M., MARULLO S. New molecular and structural determinants involved in β2-adrenergic receptor desensitization and sequestration–delineation using chimeric β3/β2-adrenergic receptors. J. Biol. Chem. 1996;271:9355–9362. doi: 10.1074/jbc.271.16.9355. [DOI] [PubMed] [Google Scholar]
- JOCKERS R., ISSAD T., ZILBERFARB V., DE COPPET P., MARULLO S., STROSBERG A.D. Desensitization of the β-adrenergic response in human brown adipocytes. Endocrinology. 1998;139:2676–2684. doi: 10.1210/endo.139.6.6050. [DOI] [PubMed] [Google Scholar]
- KRIEF S., LONNQVIST F., RAIMBAULT S., BAUDE B., VAN SPRONSEN A., ARNER P., STROSBERG A.D., RICQUIER D., EMORINE L.J. Tissue distribution of β3-adrenergic receptor mRNA in man. J. Clin. Invest. 1993;91:344–349. doi: 10.1172/JCI116191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIGGETT S.B., FREEDMAN N.J., SCHWINN D.A., LEFKOWITZ R.J. Structural basis for receptor subtype-specific regulation revealed by a chimeric β3/β2-adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3665–3669. doi: 10.1073/pnas.90.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANARA L., BADONE D., BARONI M., BOCCARDI G., CECCHI R., CROCI T., GIUDICE A., GUZZI U., LANDI M., LEFUR G. Functional identification of rat atypical β-adrenoceptors by the first β3-selective antagonists, aryloxypropanolaminotetralins. Br. J. Pharmacol. 1996;117:435–442. doi: 10.1111/j.1476-5381.1996.tb15209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORELLO J.P., BOUVIER M. Palmitoylation: a post-translational modification that regulates signalling from G-protein coupled receptors. Biochem. Cell Biol. 1996;74:449–457. doi: 10.1139/o96-049. [DOI] [PubMed] [Google Scholar]
- NAMBA T., SUGIMOTO Y., NEGISHI M., IRIE A., USHIKUBI F., KAKIZUKA A., ITO S., ICHIKAWA A., NARUMIYA S. Alternative splicing of C-terminal tail of prostaglandin E receptor subtype EP3 determines G-protein specificity. Nature. 1993;365:166–170. doi: 10.1038/365166a0. [DOI] [PubMed] [Google Scholar]
- NANTEL F., BONIN H., EMORINE L.J., ZILBERFARB V., STROSBERG A.D., BOUVIER M., MARULLO S. The human β3-adrenergic receptor is resistant to short term agonist-promoted desensitization. Mol. Pharmacol. 1993;43:548–555. [PubMed] [Google Scholar]
- O'DOWD B.F., HNATOWICH M., CARON M.G., LEFKOWITZ R.J., BOUVIER M. Palmitoylation of the human β2-adrenergic receptor. Mutation of Cys341 in the carboxyl tail leads to an uncoupled nonpalmitoylated form of the receptor. J. Biol. Chem. 1989;264:7564–7569. [PubMed] [Google Scholar]
- REVELLI J.P., PREITNER F., SAMEC S., MUNIESA P., KUEHNE F., BOSS O., VASSALLI J.D., DULLOO A., SEYDOUX J., GIACOBINO J.P., HUARTE J., ODY C. Targeted gene disruption reveals a leptin-independent role for the mouse β3-adrenoceptor in the regulation of body composition. J. Clin. Invest. 1997;100:1098–1106. doi: 10.1172/JCI119620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBERTS S.J., PAPAIOANNOU M., EVANS B.A., SUMMERS R.J. Functional and molecular evidence for β1-, β2- and β3-adrenoceptors in human colon. Br. J. Pharmacol. 1997;120:1527–1535. doi: 10.1038/sj.bjp.0701056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMIAND J., KEANE P.E., GUITARD J., LANGLOIS X., GONALONS N., MARTIN P., BIANCHETTI A., LE FUR G., SOUBRIE P. Antidepressant profile in rodents of SR 58611A, a new selective agonist for atypical β-adrenoceptors. Eur. J. Pharmacol. 1992;219:193–201. doi: 10.1016/0014-2999(92)90296-g. [DOI] [PubMed] [Google Scholar]
- STROSBERG A.D. Structure and function of the β3-adrenergic receptor. Ann. Rev. Pharmacol. Toxicol. 1997;37:421–450. doi: 10.1146/annurev.pharmtox.37.1.421. [DOI] [PubMed] [Google Scholar]
- SUMMERS R.J., KOMPA A., ROBERTS S.J. β-Adrenoceptor subtypes and their desensitization mechanisms. J. Autonomic Pharmacol. 1997;17:331–343. doi: 10.1046/j.1365-2680.1997.00055.x. [DOI] [PubMed] [Google Scholar]
- SUMMERS R.J., PAPAIOANNOU M., HARRIS S., EVANS B.A. Expression of β3-adrenoceptor mRNA in rat brain. Br. J. Pharmacol. 1995;116:2547–2548. doi: 10.1111/j.1476-5381.1995.tb17205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUSULIC V.S., FREDERICH R.C., LAWITTS J., TOZZO E., KAHN B.B., HARPER M.E., HIMMSHAGEN J., FLIER J.S., LOWELL B.B. Targeted disruption of the β3-adrenergic receptor gene. J. Biol. Chem. 1995;270:29483–29492. doi: 10.1074/jbc.270.49.29483. [DOI] [PubMed] [Google Scholar]
- TSUJII S., BRAY G.A. Food intake of lean and obese Zucker rats following ventricular infusions of adrenergic agonists. Brain Research. 1992;587:226–237. doi: 10.1016/0006-8993(92)91001-u. [DOI] [PubMed] [Google Scholar]
- VAN SPRONSEN A., NAHMIAS C., KRIEF S., BRIEND S.M., STROSBERG A.D., EMORINE L.J. The promoter and intron/exon structure of the human and mouse β3-adrenergic-receptor genes. Eur. J. Biochem. 1993;213:1117–1124. doi: 10.1111/j.1432-1033.1993.tb17861.x. [DOI] [PubMed] [Google Scholar]
- XIAO R.-P., AVDONIN P., ZHOU Y.-Y., CHENG H., AKHTER S.A., ESCHENHAGEN T., LEFKOWITZ R.J., KOCH W.J., LAKATTA E.G. Coupling of β2-adrenoceptor to Gi proteins and its physiological relevance in murine cardiac myocytes. Circulation Res. 1999;84:43–52. doi: 10.1161/01.res.84.1.43. [DOI] [PubMed] [Google Scholar]


