Abstract
The Zα domain of human double-stranded RNA adenosine deaminase 1 binds specifically to left-handed Z-DNA and stabilizes the Z-conformation. Here we report spectroscopic and analytical results that demonstrate that Zα can also stabilize the left-handed Z-conformation in double-stranded RNA. Zα induces a slow transition from the right-handed A-conformation to the Z-form in duplex r(CG)6, with an activation energy of 38 kcal mol−1. We conclude that Z-RNA as well as Z-DNA can be accommodated in the tailored binding site of Zα. The specific binding of Z-RNA by Zα may be involved in targeting double-stranded RNA adenosine deaminase 1 for a role in hypermutation of RNA viruses.
Double-stranded RNA (dsRNA) adenosine deaminase (ADAR1) is an enzyme that modifies the genetic message by deaminating adenosine in pre-mRNAs; the resulting inosine acts like guanosine in translation. ADAR1 requires a dsRNA substrate, which often is provided by the pairing of exons with introns (1); therefore, editing must take place before the removal of introns from pre-mRNAs. In addition to a deaminase domain and three dsRNA binding domains, ADAR1 contains a Z-DNA binding domain, Zab, at its N terminus (2, 3). The Zα subdomain of Zab has been isolated and binds left-handed Z-DNA with a low nanomolar dissociation constant (4, 5). Recently, Zα has been crystallized with a segment of left-handed Z-DNA, and the resulting structure reveals the tailored fitting of the Zα domain to a number of specific geometrical and electrostatic features of the left-handed nucleic acid conformation (6). Z-DNA is stabilized in vivo by negative supercoiling of DNA, which occurs upstream of a moving RNA polymerase (7). This stabilization by supercoiling has given rise to the suggestion that the Z-DNA binding domain of ADAR1 may target the enzyme to actively transcribing genes, thereby ensuring that editing can precede splicing (8).
When an RNA virus such as measles infects a cell, the antiviral interferon response leads to increased activity of interferon-inducible genes, including the ADAR1 gene, which is strongly up-regulated and produces the full-length protein, including the Zα domain (9). In addition, the distribution of ADAR1 changes from primarily nuclear localization to both nuclear and cytoplasmic localization. The measles virus replicates in the cytoplasm (as do most RNA viruses), and late in infection it has been observed that the viral RNA has been subjected to hypermutation in which a significant fraction of adenines have been changed to guanines, and uracil residues to cytosines (10). Such mutations are the expected result of the action of ADAR1 on the viral RNA replication system and may be an attempt on the part of the host cell to disable the virus. Hypermutation similar to that found in the measles virus has also been found in the RNA of vesicular stomatitis virus, respiratory syncytial virus, and para-influenza virus 3 (11, 12).
RNA viruses generally use a double-stranded intermediate during some period of their life cycle (13). Little is known about the conformation of this dsRNA or the forces acting upon it during replication. We address the question of whether the Zα domain, known to bind left-handed Z-DNA, can also bind left-handed Z-RNA, in which case it might participate in viral hypermutational activities of the ADAR1 enzyme.
Although the low-energy forms of right-handed duplexes of B-DNA and A-RNA are structurally very different, they adopt similar left-handed Z-conformations (14–16). The Z-DNA conformation, favored by alternating purines and pyrimidines, is stabilized in vitro by high concentrations of salt and other agents that screen repulsion between electronegative phosphate residues, which are closer together in the Z-conformation (17, 18). The transition from the right-handed A-form of duplex RNA to the left-handed Z-form is much less favorable than the B → Z-DNA transition; consequently, higher concentrations of chaotrophic salts or low dielectric solvents combined with elevated temperatures are required to induce the transition in vitro (19, 20). Like the B → Z transition in DNA, the A → Z-RNA conformational transition can be observed by characteristic changes in the CD spectrum, as well as shifts in the Raman spectrum. Both of these techniques are used to demonstrate that Zα can convert dsRNA from the A-conformation to the Z-form and generate a stable complex.
Materials and Methods
Sample Preparation and CD Spectroscopy.
RNA oligonucleotides were synthesized (Dharmacon, Boulder, CO) and were purified by denaturing PAGE. Zα was expressed and purified as described (3). Zα⋅RNA complexes were prepared by incubating samples containing 5 μM r(CG)6 duplex and 30 μM Zα (500-μl vol) at 45°C for 30 min and then cooling to the experimental temperature. CD spectra were collected in 1-nm steps from 350 to 200 nm by using an Aviv 202 spectrometer in 0.2- or 0.5-cm quartz cuvettes in 10 mM Na2HPO4 (pH 7), 20 mM NaCl, and 0.5 mM EDTA at 25°C, unless specified otherwise. Titrations were performed manually by stepwise addition of the titrant.
Raman Spectroscopy.
Laser Raman spectroscopy data were acquired at 25°C with samples of duplex r(CG)6 (5 mM), Zα (5 mM), or the Zα⋅Z-RNA complex [5 mM r(CG)6 duplex; 15 mM Zα] in 10 μl of 10 mM Na2HPO4 (pH 7), by using 1-mm quartz capillaries; the salt-induced Z-RNA sample contained 6.5 M NaBr. Spectra were collected in a 180° back-scattering geometry, by using 200 mW of holographically filtered (HLBF-647.1; Kaiser Optical Systems, Ann Arbor, MI) 647-nm krypton laser light (Innova 90C; Coherent Radiation, Palo Alto, CA). A modified spectrograph (model 1877; Spex Industries, Metuchen, NJ) with slits set to 4 cm−1 resolution and holographic Rayleigh rejection (HNPF-647–2; Kaiser Optical Systems) was used with a thermoelectrically cooled CCD for detection (TE/CCD 1110PB; Princeton Instruments, Trenton, NJ). Spectra were collected for 10 s, and 10–15 acquisitions were averaged and corrected for cosmic rays and spectral response. Wavelength was calibrated by using the spectral features of a 50% (vol/vol) acetonitrile/toluene standard. Calibration and baseline corrections were performed with the grams 5.1 program (Galactic Industries, Salem, NH).
A → Z-RNA Transition Kinetics.
CD kinetic profiles of the A → Z conformation change were collected at 285 nm for 4–16 h at 1-s intervals. Zα and r(CG)6 were separately preequilibrated for 20 min at each temperature, and then the Zα solution was rapidly injected into the RNA sample. After a 3-s dead time, data acquisition was initiated. Final sample conditions were 5 μM duplex r(CG)6 and 30 μM Zα in 500 μl. Averages of at least two experiments were fit to single exponential curves, and first-order rate constants were obtained by using origin 5.0 (Microcal Software, Northampton, MA). Arrhenius plots were constructed and analyzed to obtain activation energies.
Results
Z-RNA Can Be Stabilized by Zα.
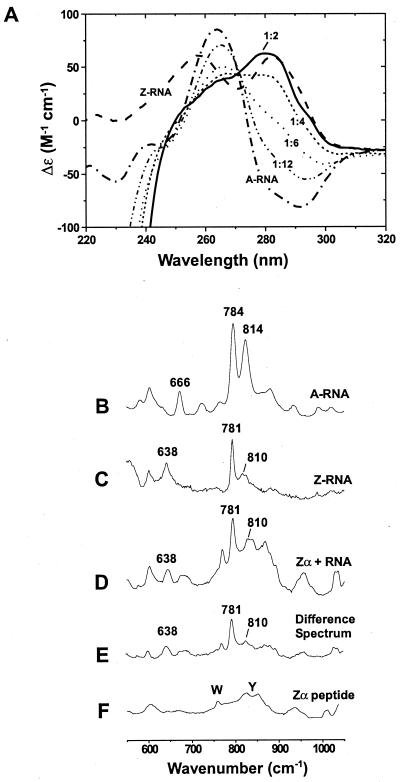
Previous studies have demonstrated that B → Z-DNA transitions can be induced in alternating purine–pyrimidine sequences by the addition of Zα (3–5, 21, 22). We asked whether Zα could also induce the Z-conformation in dsRNA. When Zα is added to the self-complementary r(CG)6 duplex and the temperature is raised to 37 or 45°C, the A → Z-RNA transition occurs readily, as seen in the strong rise of the CD spectrum near 285 nm and a decrease at 266 nm (Fig. 1A). This spectrum is similar to that of Z-RNA induced by high concentrations of salt (19, 23) (Fig. 1A). Titration of 5 μM r(CG)6 duplex with increasing amounts of Zα shows that the spectrum is fully converted at a Zα concentration of 30 μM (a molar ratio of 1 Zα protein per 2 RNA bp) (Fig. 1A). This end point is similar to the stoichiometry seen for Zα⋅Z-DNA complex formation (22) and is consistent with the Zα⋅Z-DNA crystal structure (6). Prior studies have shown that Zα binds d(CG)n oligonucleotides and polymers stabilized in the Z-form with a dissociation constant (Kd) in the low nanomolar range (4, 5). We expect that the affinity for Z-RNA is of the same order of magnitude as the affinity for Z-DNA. The exposed surface of Z-RNA is very similar to that of Z-DNA (14–16). Molecular modeling studies have shown that the 2′-hydroxyl groups could be added to the Zα⋅Z-DNA crystal structure without great perturbation to the model (B.A.B., unpublished results). It is likely that common structural features are involved in Zα recognition of both Z-RNA and Z-DNA.
Figure 1.
The Z-RNA conformation can be stabilized by Zα, as shown by CD and Raman spectroscopy. CD studies: (A) Spectra are shown for 5 μM duplex r(CG)6 in the A-form (–⋅–⋅–). All samples contained 10 mM Na2HPO4 (pH 7), 20 mM NaCl, and 0.5 mM EDTA. In 6.5 M NaClO4, the typical Z-RNA spectrum is seen (– – –). The A-RNA spectrum changes as Zα is added (Zα has no CD signal above 250 nm, but a strong negative ellipticity below 250 nm). Spectra are shown for the addition of 5 μM Zα (-⋅⋅-⋅⋅-), which is 1 Zα:12 bp; 10 μM Zα (⋅ ⋅ ⋅ ⋅), 1:6; 15 μM Zα (- - - -), 1:4; and 30 μM Zα (——), 1:2. Inversion of the CD bands around 285 nm and the decrease in signal at 266 nm are characteristic of the A → Z transition. Raman spectroscopy: (B) The A-form of r(CG)6 (5 mM duplex) has doublet peaks at 784 and 814 cm−1, which are characteristic of the A-conformation. (C) Z-conformation of r(CG)6 (5 mM duplex) induced by 6.5 M NaBr. The bands at 781 and 638 cm−1 and markedly reduced intensity at 810 cm−1 distinguish the Z-RNA conformation from the A-form (B). (D) Zα⋅r(CG)6 complex containing 15 mM Zα and 5 mM r(CG)6 duplex (1 Zα:4 bp RNA). This spectrum has several features identical to those of the salt-induced Z-RNA spectrum (C), notably intensities at 638, 781, and 810 cm−1. (E) Difference spectrum created by digitally subtracting the Zα spectrum (F) from that of the Zα⋅RNA complex (D). (F) Zα peptide spectrum (15 mM Zα). Raman bands for the aromatic amino acids tryptophan and tyrosine are labeled.
Raman Spectroscopy Confirms That RNA Is in the Z-Conformation.
Raman spectroscopy has previously been used to characterize the salt-induced A → Z-RNA transitions in duplex r(CG)3 (24) and poly[r(GC)] (19, 25). We were able to obtain similar spectra with r(CG)6, using Zα to induce the conformational change. The A-form of duplex r(CG)6 has distinct doublet Raman bands at 784 and 814 cm−1 (Fig. 1B); the intensity at 784 cm−1 arises from a cytosine ring breathing mode, and the 814 cm−1 band is a phosphodiester stretching vibration. A band at 666 cm−1 is characteristic of guanosine residues in the familiar anti-glycosyl orientation. In the presence of 6.5 M NaBr, the Raman signal at 814 cm−1 markedly decreases in intensity and shifts to 810 cm−1 (Fig. 1C), reflecting conformational changes in the phosphate groups. The peak at 784 cm−1 shifts to 781 cm−1, associated with changes in cytosine stacking. In addition, the band at 666 cm−1 is replaced by a band at 638 cm−1, indicating guanosines in the syn-glycosyl conformation. Similar frequency shifts and loss of intensity at 814 cm−1 were observed in the salt-induced A → Z-RNA conformational changes of r(CG)3 (24) and poly[r(GC)] (19, 25).
In the presence of Zα, r(CG)6 has a Raman spectrum similar in amplitude and frequency to that observed for salt-induced Z-RNA (Fig. 1D). A narrow, intense band appears at 781 cm−1, and a reduced intensity (superimposed on a tyrosine ring vibration) is present at 810 cm−1. The syn-guanosine band at 638 cm−1 is also detectable. On digital subtraction of the Zα Raman spectrum (Fig. 1F) from that of the Zα⋅Z-RNA complex (Fig. 1D), the difference spectrum (Fig. 1E) clearly shows similarity to the salt-induced Z-RNA spectrum (Fig. 1C). These results support the interpretation that the observed CD changes are the result of an A → Z-RNA transition.
Zα Is Required to Maintain RNA in the Z-Conformation.
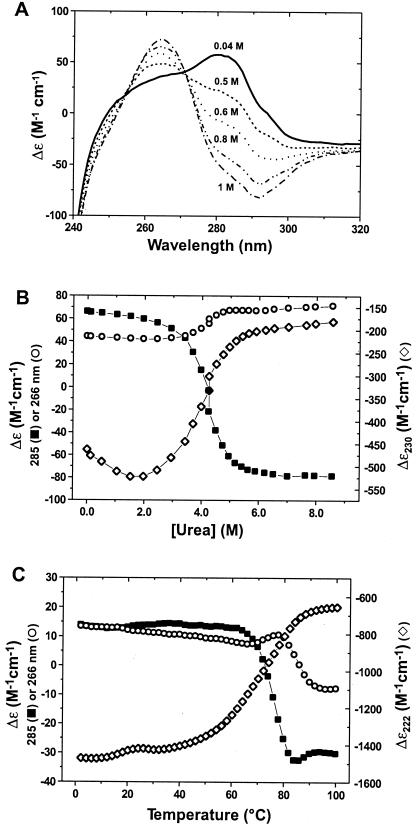
The addition of salt to the Zα⋅Z-RNA complex perturbs electrostatic interactions between protein and nucleic acid and causes disruption of the complex. A salt titration of the Zα⋅r(CG)6 complex is shown in Fig. 2A. In low salt, the RNA is in the Z-conformation, as indicated by the broad peak at 285 nm. The Zα⋅Z-RNA complex is stable at concentrations up to about 200 mM NaCl, somewhat greater than typical physiological ionic strength. As more NaCl is added, the ellipticity at 285 nm becomes negative, and the band at 266 nm, diagnostic of A-RNA, is observed. The loss of Z-RNA signal is proportional to the appearance of the A-RNA signal. Thus, as electrostatic interactions stabilizing the Zα⋅Z-RNA complex are perturbed, the Z-RNA reverts to the A-conformation.
Figure 2.
Perturbation of Zα⋅r(CG)6 complex results in reversion of Z-RNA to the A-conformation, as seen in CD spectra. (A) NaCl titration of Zα⋅r(CG)6 complex (30 μM Zα, 5 μM r(CG)6 duplex; 1 Zα:2 bp RNA). As NaCl is added, the broad Z-RNA peak around 285 nm gradually decreases, and then inverts, signifying reversion to the A-conformation. Representative points shown are 0.04 M NaCl (——), 0.5 M (- - - -), 0.6 M (⋅ ⋅ ⋅ ⋅), 0.8 M (–⋅⋅–⋅⋅–), and 1 M NaCl (–⋅–⋅–). The midpoint of the titration is ca. 0.7 M NaCl. (B) Urea denaturation of Zα⋅r(CG)6 complex. The urea titration was monitored at 285 (■), 266 (○), and 230 nm (⋄), corresponding to major ellipticity signals of Z-RNA, A-RNA, and Zα protein, respectively. The relative amount of Z-RNA decreases coincident with an increase in the A-RNA signal, as Zα is denatured by increasing urea. (C) Thermal denaturation of the Zα⋅r(CG)6 complex monitored at 285 (■), 266 (○), and 222 nm (⋄), corresponding to the major signal intensities for Z-RNA, A-RNA, and Zα, respectively. As the temperature is increased, the relative amount of Z-RNA decreases as Zα begins to denature (ca. 60°C). In addition, as the Z-RNA signal decreases, the A-RNA signal increases from 60°C up to approximately 80°C, above which the RNA denatures and the ellipticity drops at 266 nm, with a slight rise at 285 nm. Individual melting experiments yielded Tm values of 69, 86, and 77°C for Zα, r(CG)6, and Zα⋅r(CG)6, respectively.
Similar results are seen when urea is added to Zα⋅Z-RNA. The complex is relatively unaffected up to about 2 M urea (Fig. 2B). The ellipticity signal at 230 nm, a measure of Zα peptide secondary structure, becomes more negative linearly with urea concentration up to about 1.5 M, possibly as a result of increased ordering of loop regions. At urea concentrations above 2 M, the signal at 230 nm approaches zero proportionally to urea concentration, indicating denaturation of the Zα protein. At the same time, the Z-RNA band (285 nm) begins to decrease, paralleled by an increase of the A-RNA band (266 nm). These results indicate that as Zα is denatured, the stabilized Z-RNA reverts to the A-conformation.
These changes could also be followed by thermal denaturation of the Zα-RNA complex (Fig. 2C). As temperature is increased, the 266-nm signal characteristic of A-RNA decreases slightly up to ca. 65°C. As in the above experiments, once Zα begins to denature (signal at 222 nm, T ≥ 60°C), the Z-RNA signal (285 nm) drops precipitously, coincident with an increase of the A-RNA signal (266 nm). At temperatures above 85°C, the r(CG)6 duplex denatures and the CD bands at 285 and 266 nm reflect the single-stranded RNA.
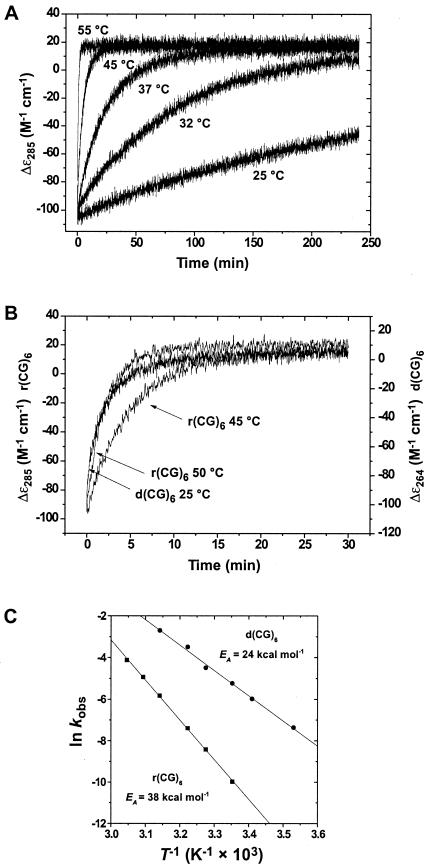
The A → Z-RNA Transition Occurs Slowly and Has a High Activation Energy.
The rate of the Zα-induced A → Z-RNA transition in duplex r(CG)6 occurs more rapidly at higher temperatures. To quantitate the transition rate, the change in ellipticity at 285 nm was monitored over time at temperatures between 25°C and 55°C (Fig. 3A). The observed rate constants (kobs) for the Zα-induced A → Z-RNA transition for duplex r(CG)6 are shown in Table 1. Parallel experiments were carried out for the B → Z-DNA transition of duplex d(CG)6 (Table 1). Both A-RNA and B-DNA convert to the left-handed Z-conformation with first-order kinetics (Fig. 3B). An Arrhenius plot was constructed from the temperature dependence of the observed rate constants (Fig. 3C) and used to obtain the activation energy (EA) for both transitions. The activation energy for the A → Z-RNA transition of r(CG)6 duplex is calculated to be 38.1 ± 0.5 kcal·mol−1 of duplex RNA (159 kJ·mol−1) or 3.2 kcal·mol−1/bp. The B → Z-DNA transition for the d(CG)6 duplex has an activation energy of 24.1 ± 0.8 kcal·mol−1 duplex (101 kJ·mol−1) or 2 kcal·mol−1/bp. Thus, the transition to left-handed Z-RNA requires more energy (≈1.2 kcal·mol−1/bp) than its DNA counterpart.
Figure 3.
Temperature dependence of the A → Z-RNA transition. (A) Kinetics of the A → Z transition as a function of temperature. CD traces at 285 nm of the Zα-induced conformational transitions are shown for 25, 32, 37, 45, and 55°C (the 50°C trace was omitted for clarity). The kinetics for the A → Z transition are first-order and were fit to single exponentials to obtain rate constants (Table 1). (B) The rates of the A → Z transition for the r(CG)6 duplex at 45 and 50°C (285 nm) are comparable to the rate of B → Z transition of d(CG)6 (264 nm) at 25°C, demonstrating the higher-energy requirements of the A → Z transition. (C) Temperature dependence for Z-conformational transition of r(CG)6 (■) or d(CG)6 (●). Arrhenius plots constructed from rate data as a function of temperature yielded activation energies of 38.1 ± 0.5 and 24.0 ± 0.8 kcal mol−1 for r(CG)6 and d(CG)6, respectively. Correlation coefficients for the Arrhenius plots of r(CG)6 and d(CG)6 were 0.999 and 0.998, respectively.
Table 1.
Temperature dependence of observed rate constants
| Molecule |
kobs,
s−1, ×105
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 10°C | 20°C | 25°C | 32°C | 37°C | 45°C | 50°C | 55°C | |
| r(CG)6 | — | — | 4.5 ± 0.0069 | 22 ± 0.044 | 60 ± 0.12 | 285 ± 0.90 | 706 ± 4.0 | 1586 ± 18.0 |
| d(CG)6 | 62 ± 1.0 | 246 ± 3.0 | 520 ± 4.0 | 1092 ± 20 | 2957 ± 110 | 7781 ± 609 | — | — |
Discussion
The results presented here describe the interaction between the Zα domain of human ADAR1 and r(CG)6. Zα induces an A → Z-RNA conformational change in duplex r(CG)6, and the transition occurs slowly and has a high activation energy. The Zα⋅r(CG)6 complex, stabilized by both ionic and conformation-specific interactions, can withstand a range of perturbations before dissociating. However, when the complex is dismantled, the dsRNA immediately reverts to the right-handed A-conformation, indicating that Zα actively maintained the RNA in the left-handed Z-form.
We have shown that the A → Z-RNA transition requires more energy than the B → Z-DNA transition for the same sequence. The major conformational differences between dsRNA and dsDNA are related to the pucker (pseudorotation) of the respective furanose rings. In dsRNA, ribose generally adopts the 3′-endo conformation, whereas in dsDNA, deoxyribose adopts the 2′-endo conformation (26). Ribose has a 2′-hydroxyl group that stabilizes its pucker, and, consequently, more energy is required to alter the pucker in dsRNA than in dsDNA. Z-DNA is exactly intermediate in these two conformations because the nucleotides along its chain alternate in the 2′-endo and 3′-endo conformations. Duplex A-RNA is generally stiff, compared with the more flexible B-DNA molecule. In large part this rigidity is attributable to the influence of the 2′-hydroxyl on ring pucker and is reflected in the larger energy of activation for the A → Z-RNA conformational transition compared with DNA. The rate at which the reaction occurs at 25°C is significantly slower than that at 37°C (Fig. 3A). This slower rate accounts for our failure to observe the interaction of ADAR1 with r(CG)12 in earlier experiments (2).
The cocrystal structure of Zα and Z-DNA showed that the Zα domain is specific for the left-handed Z-conformation but independent of the DNA sequence. The same is likely true for the interaction between Zα and Z-RNA. Further experiments must be performed so that we can investigate the interaction of Zα with Z-RNA containing A⋅U base pairs. Previous studies have shown that A⋅T or A⋅U base pairs make the salt-induced conversion to the Z-form more difficult in DNA or RNA (27, 28); it is likely that this will also be true for Z-RNA stabilized by Zα.
It is not surprising that proteins that bind to B-DNA do not bind to A-RNA, and vice versa, because the structures of these right-handed duplexes differ significantly; however, the left-handed Z-form duplexes are very similar. Zα may be the first nucleic acid binding domain discovered that specifically binds to both duplex DNA and RNA. The role of this domain in the hypermutation of RNA viruses has yet to be explored. A great deal is known about the negative superhelicity generated by the transcription of dsDNA, but little is known about negative torsional strain in replicating RNA molecules. This subject needs to be more fully explored to understand the possible participation of the Zα binding domain in hypermutational activities of ADAR1 during infections by RNA viruses.
Acknowledgments
We thank Alekos Athanasiadis, Wilfredo Blasini, Alan Herbert, Yang Gyun Kim, Stefan Maas, Thomas Schwartz, and P. Shing Ho for useful discussions, comments on the manuscript, and experimental assistance. This work was supported by grants to A.R. from the National Institutes of Health and the National Science Foundation. Raman measurements were conducted at the National Institutes of Health-funded Laser Biomedical Research Center at the G. R. Harrison Spectroscopy Laboratory. B.A.B. is an Aid for Cancer Research postdoctoral fellow.
Abbreviations
- ADAR1
double-stranded RNA adenosine deaminase 1
- ds
double-stranded
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240464097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240464097
References
- 1.Higuchi M, Single F N, Kohler M, Sommer B, Sprengel R, Seeburg P H. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 2.Herbert A, Lowenhaupt K, Spitzner J, Rich A. Proc Natl Acad Sci USA. 1995;92:7550–7554. doi: 10.1073/pnas.92.16.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz T, Lowenhaupt K, Kim Y G, Li L, Brown B A, II, Herbert A, Rich A. J Biol Chem. 1999;274:2899–2906. doi: 10.1074/jbc.274.5.2899. [DOI] [PubMed] [Google Scholar]
- 4.Herbert A, Alfken J, Kim Y G, Mian I S, Nishikura K, Rich A. Proc Natl Acad Sci USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schade M, Behlke J, Lowenhaupt K, Herbert A, Rich A, Oschkinat H. FEBS Lett. 1999;458:27–31. doi: 10.1016/s0014-5793(99)01119-9. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz T, Rould M A, Lowenhaupt K, Herbert A, Rich A. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 7.Liu L F, Wang J C. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herbert A, Rich A. J Biol Chem. 1996;271:11595–11598. doi: 10.1074/jbc.271.20.11595. [DOI] [PubMed] [Google Scholar]
- 9.Patterson J B, Samuel C E. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cattaneo R, Billeter M A. Curr Top Microbiol Immunol. 1992;176:63–74. doi: 10.1007/978-3-642-77011-1_5. [DOI] [PubMed] [Google Scholar]
- 11.Cattaneo R. Curr Opin Genet Dev. 1994;4:895–900. doi: 10.1016/0959-437x(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 12.Bass B L. Trends Biochem Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs B L, Langland J O. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura Y, Fujii S, Urata H, Uesugi S, Ikehara M, Tomita K. Nucleic Acids Symp Ser. 1985;16:29–32. [PubMed] [Google Scholar]
- 15.Teng M K, Liaw Y C, van der Marel G A, van Boom J H, Wang A H. Biochemistry. 1989;28:4923–4928. doi: 10.1021/bi00438a001. [DOI] [PubMed] [Google Scholar]
- 16.Davis P W, Adamiak R W, Tinoco I., Jr Biopolymers. 1990;29:109–122. doi: 10.1002/bip.360290116. [DOI] [PubMed] [Google Scholar]
- 17.Pohl F M, Jovin T M. J Mol Biol. 1972;67:375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- 18.Rich A, Nordheim A, Wang A H. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- 19.Tinoco I, Jr, Cruz P, Davis P W, Hall K, Hardin C C, Mathies R A, Puglisi J D, Trulson M O, Johnson W C, Jr, Neilson T. In: Structure and Dynamics of RNA. van Knippenberg P H, Hilbers C W, editors. New York: Plenum; 1986. pp. 55–68. [Google Scholar]
- 20.Klump H H, Jovin T M. Biochemistry. 1987;26:5186–5190. doi: 10.1021/bi00390a043. [DOI] [PubMed] [Google Scholar]
- 21.Berger I, Winston W, Manoharan R, Schwartz T, Alfken J, Kim Y G, Lowenhaupt K, Herbert A, Rich A. Biochemistry. 1998;37:13313–13321. doi: 10.1021/bi9813126. [DOI] [PubMed] [Google Scholar]
- 22.Herbert A, Schade M, Lowenhaupt K, Alfken J, Schwartz T, Shlyakhtenko L S, Lyubchenko Y L, Rich A. Nucleic Acids Res. 1998;26:3486–3493. doi: 10.1093/nar/26.15.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall K, Cruz P, Tinoco I, Jr, Jovin T M, van de Sande J H. Nature (London) 1984;311:584–586. doi: 10.1038/311584a0. [DOI] [PubMed] [Google Scholar]
- 24.Nishimura Y, Tsuboi M, Uesugi S, Ohkubo M, Ikehara M. Nucleic Acids Symp Ser. 1985;16:25–28. [PubMed] [Google Scholar]
- 25.Trulson M O, Cruz P, Puglisi J D, Tinoco I, Jr, Mathies R A. Biochemistry. 1987;26:8624–830. doi: 10.1021/bi00400a020. [DOI] [PubMed] [Google Scholar]
- 26.Sanger W. In: Principles of Nucleic Acid Structure. Cantor C E, editor. New York: Springer; 1984. pp. 55–78. [Google Scholar]
- 27.Taboury J A, Taillandier E. Nucleic Acids Res. 1985;13:4469–4483. doi: 10.1093/nar/13.12.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vorlickova M, Kypr J, Jovin T M, Planck M. Biopolymers. 1990;29:385–392. doi: 10.1002/bip.360290210. [DOI] [PubMed] [Google Scholar]