Abstract
Septins are a family of conserved proteins that form hetero-oligomeric complexes that assemble into filaments. The filaments can be organized into linear arrays, coils, rings and gauzes. They serve as membrane-associated scaffolds and as barriers to demarcate local compartments, especially for the establishment of the septation site for cytokinesis. Studies in budding and fission yeast have revealed many of the protein–protein interactions that govern the formation of multi-septin complexes. GTP binding and phosphorylation direct the polymerization of filaments that is required for septin-collar assembly in budding yeast, whereas a homolog of anillin instructs timely formation of the ring of septin filaments at the medial cortex in fission yeast. These insights should aid understanding of the organization and function of the diverse septin structures in animal cells.
Introduction
Genes that encode septins were identified first more than 30 years ago in budding yeast (Saccharomyces cerevisiae) as temperature-sensitive mutations that prevent cytokinesis at a restrictive temperature, resulting in formation of chains of multinucleated and multibudded cells in which the buds are elongated [1]. S. cerevisiae cells express seven septins, five of which are involved in mitosis (described below). Homologous genes have been identified and characterized in many other eukaryotic species [2,3]. The genome of Drosophila melanogaster encodes five septins, Caenorhabditis elegans two and Homo sapiens thirteen (Table 1) [4] (for a comprehensive phylogeny, see [5]). In mammals, differential splicing and alternative translation-initiation sites generate an even greater variety of septin isoforms in specific cell types [5,6]. Interestingly, the genomes of aquatic green algae (Chlamydomonas reinhardtii) and marine phytoplankton (Nannochloris spp.) encode bona fide septin orthologs, but higher plants do not. Likewise, the genomes of protozoa, other eukaryotic protists (Giardia) and the cellular slime mold (Dictyostelium discoideum) seem to lack septins. Similarly, the genome of the planctomycete, Gemmata obscuriglobus (a budding microbe that harbors a membrane-bounded nucleoid, but is classified with the bacterial phyla based on the majority of its recognizable genes) lacks identifiable septins.
Table 1.
Classification of septins in several speciesa
| Budding yeast | Fission yeast | Worm | Fly | Human |
|---|---|---|---|---|
| Cdc3 | Spn1 | Unc-61 | Sep2, Sep5 | Sept6, Sept8, Sept10, Sept11 |
| Cdc10 | Spn2 | – | – | Sept3, Sept9, Sept12 |
| Cdc11 | Spn3, Spn5 | – | Sep1, Sep4 | Sept1, Sept2, Sept4, Sept5 |
| Cdc12, Spr3 | Spn4, Spn6 | Unc-59 | Pnut | Sept7, Sept13 |
| Shs1/Sep7, Spr28 | Spn7 | – | – | – |
Septins in a given species that belong to the same class are included in the same box. Septins shaded in the same color belong to the same phylogenetic branch, according to the analysis of Kinoshita [5]. As indicated by the change in hue, our classification of Cdc11- and Cdc12-like septins is based primarily on functional criteria rather than on close sequence similarity, as described in more detail in the text.
Septins are GTP-binding proteins that possess a characteristic primary structure (Figure 1a). Septin monomers assemble into hetero-oligomeric (multi-septin) complexes (models based on the available evidence are shown in Figure 1b). The complexes polymerize into filaments in vitro (Figure 2a) [7–10] and in vivo (Figure 2b) [11–13]. A model of filaments in S. cerevisiae is shown in Figure 2c. To date, no crystal structure has been reported for an individual septin, septin complex or septin filament. However, significant advances in analyzing the composition and organization of mitotic septin complexes in yeast indicate that general rules govern the assembly of septin complexes. In addition, new insights have been obtained in yeast about the mechanisms that control septin-filament formation spatially and temporally. These mechanisms are also likely to be conserved.
Figure 1. The primary structure of a typical septin and organization of septin complexes.
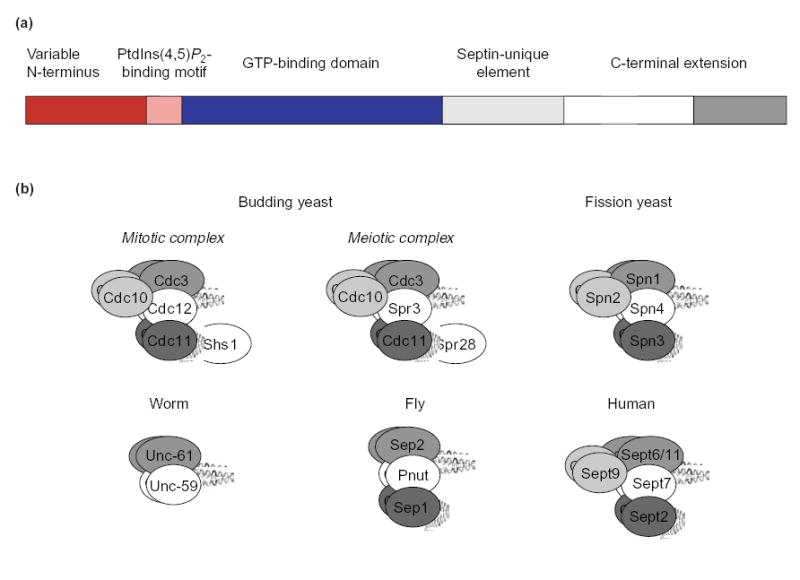
(a) The recognizable motifs and domains in the primary structure of a septin. The GTP-binding domain (solid blue) contains all five of the signature motifs (G-boxes) that are found in other members of the super-family of GTPases [81,82]. The predicted coiled-coil sequence in the C-terminal extension (dark gray box) is preceded by a sequence that is predicted to form an α-helix (open box). (b) Proposed organization of multi-septin hetero-oligomers in eukaryotes. Models of the mitotic complexes in budding and fission yeast are adapted from Versele et al. [10] and An et al. [33], respectively. The other models are inferred from the yeast archetypes, based on sequence relatedness, similarities in the primary structure and phylogenetic analysis [5]. Some models are supported by data on individual septin-septin interactions [7,9,45]. In the human complex, Sept9 is shown as the counterpart of Cdc10 (Table 1), based on sequence similarity and phylogenetic-tree analysis [5], the lack of a CTE, and the association between Sept9 and both Sept11 (a Cdc3 ortholog) and Sept7 (a Cdc12 ortholog) [83]. The N-terminal domain of Sept9 is required for its interaction with Sept7 and Sept11, which is similar to the interaction of Cdc10 with Cdc3 and Cdc12. However, there is evidence that the C-terminal portions of Sept7 and of Sept11 are required for their interaction with Sept9 [83], unlike the interactions of Cdc3 and Cdc12 with Cdc10 [10].
Figure 2. Septin filaments from budding yeast.
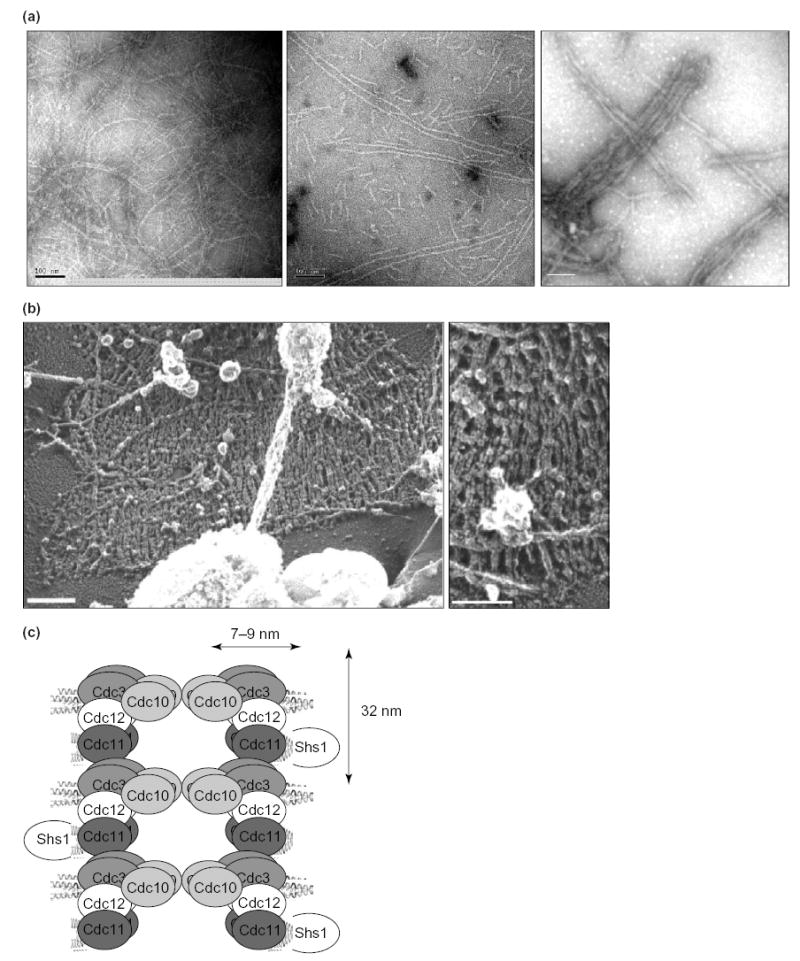
(a) EM images of negatively-stained preparations of filaments reconstituted from purified, recombinant septin complexes. A multimeric complex of budding yeast septins (Cdc3–Cdc12–Cdc11–Cdc10), purified from E. coli cells in which all four proteins are co-expressed, was dialysed into low-salt buffer. Previously unpublished images (left and middle panel) are courtesy of Sang-Shin Park (this laboratory) and Patricia Grob (laboratory of Eva Nogales, Univ. of California, Berkeley, USA); right panel, reproduced with permission from [10]. Scale bars, 100 nm. (b) EM images in negative-contrast of septin structures at the cell cortex of yeast spheroplasts, prepared by a rapid-freeze, deep-etch technique. Scale bars, 200 nm. Reproduced, with permission, from [13]. (c) Model of the polymerization of septin complexes into paired, linear filaments. End-to-end association of Cdc3 in one Cdc3–Cdc12–Cdc11 complex with Cdc11 in another complex yields filaments with a defined polarity; Cdc10 serves as a bridge to stabilize and pair the strands. A parallel arrangement of filaments is depicted, but an anti-parallel arrangement has not been ruled out by direct experimental evidence. Dimensions are from [8,10].
Septin functions
In budding yeast, septins (Cdc3, Cdc10, Cdc11, Cdc12 and Shs1/Sep7) form complexes that assemble into a tubular ‘collar’ of highly ordered filaments at the cortex of the mother-bud neck throughout the cell cycle, except for disassembly and reassembly during G1 (Figure 3). In mitotic cells, these septins are implicated in bud-site selection, the establishment and maintenance of polarized bud growth, the switch from polarized to isotropic bud growth, and spindle positioning [14–16]. In meiotic cells, two sporulation-specific septins (Spr3 and Spr28) replace Cdc12 and Shs1, respectively, and form an alternative complex with Cdc3, Cdc10 and Cdc11 that localizes to the prospore envelope as it forms [17,18]. In fission yeast, septins Spn1, Spn2, Spn3 and Spn4, which are homologous to Cdc3, Cdc10, Cdc11 and Cdc12, respectively (Table 1), colocalize at the medial cortex (See Glossary) as a double-ring structure at the onset of cytokinesis. However, unlike budding yeast mitotic septins, the mitotic septins in fission yeast are neither essential for cell viability nor are they required to define the location of the cell-cleavage plane because absence of all four septins only delays cytokinesis and results in a chain-of-cells phenotype [19,20]. These observations indicate that septins might contribute to the process of cell division through a more general role in either directing or enhancing the efficiency and specificity of membrane remodeling [21]. In agreement with a role in membrane synthesis, septins in animal cells associate with components of the secretory apparatus, such as the exocyst complex and a SNARE protein (syntaxin) [22,23]. Moreover, although in many higher cell types septins localize to the site of cytokinesis [2,3] and the metaphase plate [24], a subset of septins is expressed highly in post-mitotic tissues such as brain [25], which indicates that the contribution of septins to cell function is not confined to either cytokinesis or chromosome dynamics.
Figure 3.
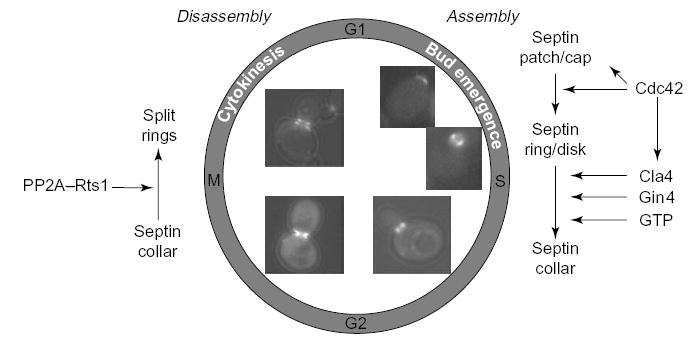
Schematic depiction of septin recruitment and septin-collar formation during the cell cycle of budding yeast. Recruitment of either a patch or cap of septins to the incipient bud site and its rapid conversion into a rimmed disk require the small GTPase, Cdc42. Stabilization of the disk and its transformation into a filamentous collar requires phosphorylation by two proteins kinases, Cla4 and Gin4 (and, perhaps, others), plus binding of GTP to Cdc10 and Cdc12 in the septin complex. At cytokinesis, the collar splits into two separate rings, which are disassembled after cell separation. Confocal fluorescence images (GFP–Cdc12) were kindly provided by Jeroen Dobbelaere (ETH, Zürich, Switzerland).
At the molecular level, two roles for septins have been defined in yeast. First, the filaments formed by septin complexes serve as a scaffold that recruits other proteins and, perhaps, activates them [26]. In S. cerevisiae, the localization of at least 21 proteins at the bud neck requires proper assembly of septin filaments. These proteins include many protein kinases (e.g. Cdc5, Cdc15, Dbf2, Elm1, Gin4, Hsl1, Kcc4 and Ssk2), other enzymes (e.g. chitin synthases) and components of the bud-site-selection machinery (for an exhaustive catalog, see [14,27]). In many instances, the binding is direct [28]. A well-characterized example of this scaffold role occurs in the so-called morphogenesis checkpoint (Box 1). The second function of septin filaments is to establish discrete cellular compartments. For example, the membrane-associated septins at the isthmus between a mother cell and its bud form a physical barrier that is necessary to prevent diffusion of bud-specific proteins into the mother cell [29,30]. During cytokinesis, the tubular collar of septin filaments is split into two separate ‘rings’ that seem to serve as gaskets that confine and concentrate between them factors that are involved directly in cytokinesis, such as the exocyst component Sec3, an adaptor protein Spa2 and an isoform of chitin synthase Chs2 [31]. This compartmentalization function appears to be conserved in fission yeast, where a double ring of septin filaments encompasses the contractile apparatus and provides for efficient dissolution of the primary septum [32,33]. It appears that septins might have a similar role in mammalian cells because septins are prominent components of the cleavage furrow and a diffusion barrier within the cleavage furrow has been demonstrated [34].
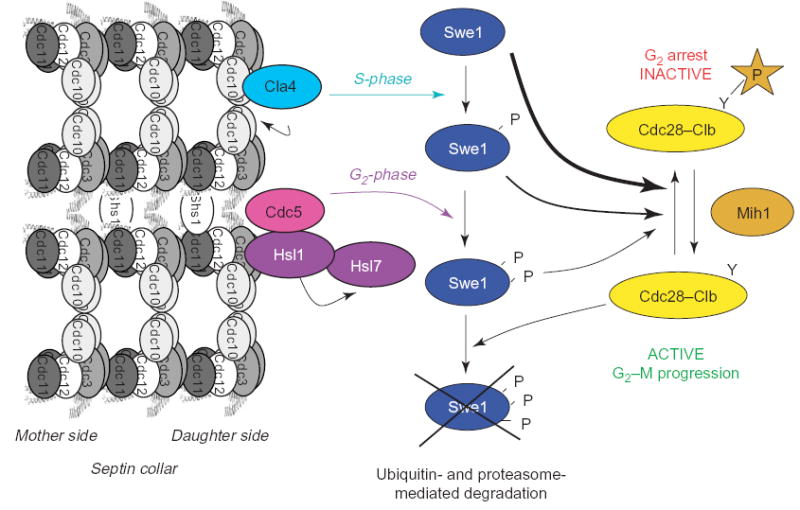
Box 1. The morphogenesis checkpoint
One set of proteins that is recruited to the septin collar in budding yeast includes the protein kinases Hsl1 and Swe1 (the S. cerevisiae homolog of Wee1 [87]) and a protein-arginine methyltransferase called Hsl7 [84–86] (Figure I). In response to perturbation of septin filament (or actin filament) organization, Swe1 is stabilized and, hence, its ability to phosphorylate a conserved tyrosine residue in the cyclin B-bound form of the Cdc28 cyclin-dependent kinase persists. This modification inhibits the activity of cyclin-B-bound Cdc28, thereby causing a significant G2 delay and thus blocking timely entry into mitosis (reviewed in [88]).
Swe1 is stabilized when morphogenesis at the bud neck is compromised because the mechanism for Swe1 inactivation and ubiquitin-mediated degradation requires its recruitment to the bud neck. Swe1 is marked for destruction by multi-site phosphorylation, which only occurs when it is located at the bud neck. Septins are required for the recruitment of Hsl1 and Hsl7 to the septin collar, and Hsl1 and Hsl7 are required, in turn, for efficient tethering of Swe1 at the bud neck. It is reported that direct binding of Cdc11 and Cdc12 to the C-terminal regulatory domain of Hsl1 relieves autoinhibition and stimulates Hsl1 phosphotransferase activity in vitro [89]. Hsl1 phosphorylates Hsl7 directly, but it does not phosphorylate Swe1 [90]. Instead, two other protein kinases, Cla4 (a PAK) and Cdc5 (the yeast Polo/PLK ortholog), which are targeted sequentially to the bud neck in a septin-dependent manner, are responsible for the stepwise phosphorylation and downregulation of Swe1 [91]. This scheme provides a mechanism that links the proper assembly of septin filaments with efficient passage into mitosis. Unanswered questions include the identity of the crucial substrates of Hsl1, and why Hsl7 is a protein-arginine methyltransferase, given that this activity is not involved in degradation of Swe1 [92,93].
Assembly of hetero-oligomeric multi-septin complexes in budding yeast
The GTP-binding domain of septins is distinct from, but homologous to, that of small Ras-like GTPases. N-terminal to the GTP-binding domain is a phosphoinositide-binding motif (Figure 1a) [35,36], and plasma membrane phosphatidylinositol (4,5)-bisphosphate is important for the maintenance of proper septin architecture in vivo [37]. To the C-terminal side, there is a highly conserved sequence of unknown function that is unique to and a hallmark of septins. Finally, a region at or near the C-terminus of most, but not all, septins is predicted to form a coiled coil because of the presence of an extended string of hydrophobic residues in the characteristic (but occasionally interrupted) 4–3 repeating pattern. Synthetic peptides that correspond to the predicted coiled-coils alone do not adopt a stable helical structure, either by themselves orwhenmixed withputative partner coiled-coil peptides from other septins [10]. A predicted α-helical region that precedes the coiled-coil segment appears to promote and stabilize coiled-coil formation, because a fragment of the C-terminus of Cdc12 that contains both the predicted α-helix and the coiled-coil sequence heterodimerizes with the C-terminus of Cdc3 [10]. Hereafter, we refer to the region that contains the predicted α-helix and the coiled-coil element as the C-terminal extension (CTE) (Figure 1a).
The contacts that mediate the interactions between the mitotic septins of S. cerevisiae have been mapped in detail [10]. Cdc3 and Cdc12 associate via their CTEs, and the CTEs are sufficient for this interaction. Cdc10, which lacks a CTE, binds to the globular ‘head’ domains of both Cdc3 and Cdc12, and binds more stably to the Cdc3–Cdc12 complex than to either septin alone. Cdc11 binds strongly to Cdc12 only, and independently of the CTE of either protein. Thus, Cdc12 serves as the linchpin of the complex because it binds Cdc3, Cdc10 and Cdc11 simultaneously via three distinct interfaces.
Shs1 (a non-essential septin) binds only to Cdc11, an interaction that is mediated largely by their CTEs. In addition, each septin (Cdc3, Cdc10, Cdc11 and Cdc12) can self-associate. In this regard, the apparent molecular weight of recombinant complexes isolated at a moderate salt concentration (0.25 M) is consistent with two copies of each septin in a 1:1:1:1 stoichiometry [10], whereas the amount of Cdc11 in native complexes that are isolated at higher salt concentrations (0.5–1.0 M) is substoichiometric [8,38]. Likewise, in high-salt conditions, the amount of Shs1 recovered in native complexes is substoichiometric [39]. Together, these data indicate a model for the complex that is illustrated in Figure 1b. The Cdc3–Cdc12 heterotetramer forms the core of the complex: without this pair of septins, no stable complexes are assembled and no polymerization occurs [10]. Indeed, CDC3 and CDC12 are essential genes in S. cerevisiae and truncation of the CTEs that mediate their interaction is lethal, which indicates that formation of this minimal complex is required for septin function in vivo. Consistent with this view, the only septins in C. elegans are orthologs of Cdc3 and Cdc12 [40,41]. Only ARTS, an atypical septin-like protein in mitochondria [42], is suggested to function individually.
Hetero-oligomeric, multi-septin complexes in fission yeast
For the most part, there is a one-to-one correspondence between septins in fission and budding yeast, based on primary sequence similarity (Table 1). Although Spn5, Spn6 and Spn7 from S. pombe have not been studied in detail, they probably represent sporulation-specific septins [43]. The structure of the septin complex in mitotic cells in fission yeast has been analyzed in detail [33], and is similar to that of budding yeast (Figure 1b). Spn1 (Cdc3) binds to Spn4 (Cdc12), and this complex is, in turn, stabilized by Spn2 (Cdc10). Spn3 (Cdc11) binds only to Spn4 (Cdc12). The amounts of the various subunits present in and the sedimentation coefficient of the complex are, again, consistent with two copies of each septin in a 1:1:1:1 stoichiometry. Moreover in agreement with the view that the Spn1 (Cdc3)–Spn4 (Cdc12) heterotetramer forms the core of the complex, deletion of either of these subunits causes a much more severe phenotype (cell-separation defect) than absence of either Spn2 (Cdc10) or Spn3 (Cdc11) [33].
Hetero-oligomeric, multi-septin complexes in animal cells
Phylogenetic analysis reveals that some mammalian septins (Sept3, Sept9 and Sept12) are the counterparts of Cdc10 (and Spn2) because they lack a CTE. Sept6, Sept8, Sept10 and Sept11 are mammalian counterparts of Cdc3 (Spn1), based on sequence similarity [5]. However, other animal septins cannot be classified readily as obvious orthologs of particular classes of septin in yeast, based on their primary structure alone [6]. However, all mammalian septin complexes characterized to date contain both Sept6 (or its close paralogs, Sept8, Sept10 and Sept11) and Sept7; in the fly, the orthologs of Sept6 and Sept7 are Sep2 and Pnut, respectively (Table 1) [44]. The association of Sept6 and Sept7 requires their CTEs [45]. Based on these considerations, we propose that the mammalian Sept6–Sept7 and fly Sep2–Pnut complexes are the counterparts of the yeast Cdc3–Cdc12 and Spn1–Spn4 complexes (Figure 1b). All septin complexes isolated from mammalian cells also contain either one or two additional septins: one from the Sept2 group (Sept1, Sept2, Sept4 and Sept5), the ortholog of which in the fly is Sep1; and/or, one from the Sept3 group (Sept3, Sept9 and Sept12), which, as mentioned previously, lack a CTE, like yeast Cdc10 (Spn2). Moreover, the apparent stoichiometry and molecular weight of all septin complexes from animal cells are most consistent with the presence of two copies of each subunit [7,9,45]. Based on sequence similarities (if clear-cut), the analogies in composition of the complexes, and the interrelationships discussed above, we propose that septins can be classified into conserved groups (Table 1) and that similar models for the corresponding complexes can be deduced (Figure 1b).
The exception that proves the rule?
In the nematode, C. elegans, only two septin genes have been found. Both proteins (Unc-59 and Unc-61) colocalize at the midbody, and each requires the other for proper localization, and the phenotype of single mutants is identical to that of a double mutant [40,41]. Also, although there is no direct biochemical evidence for their physical association, Unc-59 and Unc-61 interact in two-hybrid studies [46]. Based on these considerations and sequence similarities, it seems likely that Unc-59 and Unc-61 function as an obligate complex that is the counterpart of the yeast Cdc3–Cdc12 complex. These findings also support our conclusion that a Cdc3–Cdc12-like hetero-oligomer is the non-reducible core that is required in all eukaryotes for septin function in vivo. However, if Unc-59 and Unc-61 are the only septins in C. elegans and if, when purified, they form filaments in vitro, they represent an exception to our suggestion, based on the evidence from other organisms (described below), that three different types of septin (Cdc3-, Cdc11- and Cdc12-like) are the minimum necessary for the assembly of septin complexes into functional filaments.
Regulation of septin polymerization
Septins form filaments in all cell types that have been examined. In S. cerevisiae in G1, an apparent cap, or patch, of septins in which the subunits are highly mobile as judged by fluorescence recovery after photobleaching (FRAP) experiments [47,48] accumulates in the juxta-membrane region immediately subtending the incipient bud site. Septins seem to be more concentrated at the edge of this patch than at its center. As the cell cycle proceeds, concomitant with the emergence of the bud, the septin-containing structure transforms into a tube-like, hourglass-shaped collar that persists at the bud neck (Figure 3). At this stage, the subunits are basically immobile in FRAP studies; and, using electron microscopy (EM), filaments of 10 nm diameter are visualized readily at the neck [8,11,12]. During cytokinesis, this cylinder of filaments splits into two separate rings. Similarly, in fission yeast, the band of septin filaments that assembles at the medial cortex splits into two rings as the septum forms [19,20,32]. In mammalian cells, actin fibers act as templates for septin filaments, and when actin is perturbed in vivo, coils and rings of septins result [3,9,49]. In addition, SNARE proteins and microtubules are reported to contribute to the localization and/or assembly of septin filaments in animal cells [23,50–52]. Aberrant deposition of filamentous septin structures correlates with age-related neurodegenerative disorders such as Alzheimer’s and Parkinson’s disease [53,54], but whether these aggregates are a cause or a consequence of these maladies is not known.
Three conditions must be fulfilled for robust polymerization of septin complexes into filaments in vitro. First, a minimum complement of septin subunits must be present. With the notable exception of Xenopus laevis Sept2 [55], single septins and complexes that are composed of two different septins are not reported to polymerize in vitro. In all other cases examined, the capacity to polymerize into filaments requires the presence of complexes that contain at least three types of septin, for example, Cdc3–Cdc12–Cdc11 in budding yeast [10], Spn1–Spn4–Spn3 and Spn1–Spn4–Spn2 in fission yeast [33], Sept6–Sept7–Sept2 in mammalian cells [9,45] and Sep2–Pnut–Sep1 in flies [7]. Although the Cdc3–Cdc12–Cdc11-containing complex polymerizes into filaments in vitro, Cdc10 makes crucial contributions to septin-filament organization and stability in budding yeast (Box 2) [10], and Cdc10 counterparts in other organisms presumably serve similar functions.
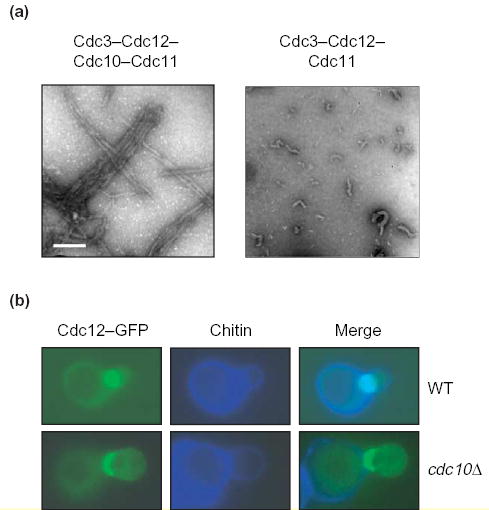
Box 2. Roles of Cdc10 in the organization of septin filaments
Although a purified recombinant Cdc3–Cdc12–Cdc11 complex polymerizes in vitro, the resulting filaments are unstable, short and display marked curvature (Figure Ia, right). By contrast, filaments generated from a purified Cdc3-Cdc12-Cdc11-Cdc10 complex are more stable, much longer and straighter (Figure Ia, left). Moreover, in the absence of Cdc10, filaments are not paired, whereas when Cdc10 is present, filaments are almost invariably paired and in register (Figure Ia; bar, 100 nm). In vivo, a septin collar forms in cells (cdc10Δ) that lack Cdc10, but the septins that are present (Cdc12–GFP in the example shown) are distributed asymmetrically at the bud neck, displaying a marked bias towards the bud (Figure Ib, bottom) compared with the distribution in normal cells (Figure Ib, top) [10,94]. Moreover, unlike wild-type cells, no filaments can be discerned by EM in negatively-stained grazing-sections of the bud necks of cdc10Δ cells [8]. The most abundant septin complex isolated from mammalian cells is composed of Sept6, Sept7 and Sept2. This is the presumed equivalent of Cdc3–Cdc12–Cdc11 (see Table 1 and Figure 1b). A corresponding hexameric complex with 2:2:2 stoichiometry can be reconstituted, via recombinant DNA-expression methods, and polymerizes into filaments [9,45]. However, in cell extracts, Sept9 (which, like Cdc10, lacks a CTE) co-purifies with the Sept6–Sept7–Sept2 complex. Thus, it seems reasonable to speculate that Sept9 might fulfill a similar role to Cdc10 and act as a linker between individual filaments. If so, including Sept9 in the complexes should result in reconstituted filaments that are more stable, longer and more cross-braced. In any event, the fact that heterodimers of Sept9–Sept7 and Sept9–Sept11 (a close paralog of Sept6) form readily in vitro [83] shows that the human Cdc10-like septin associates, as does yeast Cdc10, with Cdc3-like and Cdc12-like partners.
The other parameters that greatly influence septin polymerization in vitro are ionic strength and, to a lesser extent, pH. Filament formation increases in low-salt conditions [7–9]. In budding yeast [10], this effect has been attributed to the salt-sensitive nature of the interaction between Cdc3 and Cdc11, an interface that is pivotal to polymer formation (Figure 2). The physiological relevance of this property is unclear.
The last and, perhaps, most controversial, requirement for polymerization of septin complexes is GTP binding. The signature motifs (G-boxes) of the GTPase superfamily are present in all septin family members except ARTS, a splice variant of Sept4 that lacks the G4 box [56]. Many septins bind and hydrolyze GTP [7,28,45,55]. However, the rates of nucleotide exchange and hydrolysis observed in vitro are slow. This intrinsic property seems unaffected by cellular factors because the turnover of GTP in septin complexes in vivo, judged by mass spectrometry, is also very slow, at least in budding yeast [38]. Using radioactively labeled nucleotides (or fluorescent or photo-activatable analogs), it has been shown that some septin subunits, for example yeast Cdc10 and Cdc12, and fly Pnut and Sep1, bind GTP whereas others, for example yeast Cdc3 and Cdc11, and fly Sep2, do not [7,28]. Nonetheless, mutating residues that should contribute to GTP binding in many septins, based on analogy to other GTPases, alters formation, appearance, localization and/or function of septin filaments [3,36,57–59]. In budding yeast, mutations in the P-loop of Cdc10 and Cdc12 that prevent GTP binding in vitro, permit assembly of hetero-oligomeric septin complexes of normal composition, but prevent polymerization of these complexes into filaments and confer temperature-sensitive growth defects in vivo [28]. Preventing GTP binding to both Cdc10 and Cdc12 has a strongly synergistic effect and confers temperature-sensitive lethality. At the restrictive temperature, such a double mutant forms the initial septin patch at the incipient bud site, but does not assemble the filamentous septin collar at the mother-bud neck and, eventually, dies. Thus, it seems that guanine-nucleotide binding to specific septins is necessary to place the complex in a conformational state that is competent for polymerization.
The GDP:GTP ratio in septin complexes (~2) seems to be conserved [7,9,38]. Given the number of different septins in these hetero-oligomers, the effects of nucleotide are potentially even more complicated than in polymerization of αβ tubulin heterodimers into microtubules [60]. In α-tubulin, GTP is located at a non-exchangeable (N) site, whereas GTP binds to β-tubulin at an exchangeable (E) site; only αβ heterodimers in which the E site is occupied by GTP can polymerize. However, after polymerization, this nucleotide hydrolyzes and becomes non-exchangeable. Initially, a group who reported that addition of either GTP or GTPγS (but not GDP) promotes the polymerization of frog recombinant Sept2 suggested that this sort of model might apply to the assembly of septins into filaments [55]. However, no other published study shows that a single septin can polymerize; moreover, even when hetero-oligomeric septin complexes are used, regardless of species, no effect of exogenously added guanine nucleotide on the rate or extent of polymerization has been seen [28,45]. Because guanine nucleotides are present in purified complexes, but exogenously added guanine nucleotides have no ostensible effect on either the rate or the extent of assembly of these complexes into filaments in vitro, the tightly bound (non-exchangeable) guanine nucleotide that survives purification of the complex must be sufficient to promote polymerization.
Might GTP hydrolysis control the timing of the assembly (and/or the disassembly) of septin filaments? Two observations do not support such a regulatory role for formation and breakdown of the septin filaments at the bud neck in S. cerevisiae, at least. First, using isotopic labeling of cells and analysis of the nucleotide content of isolated septin hetero-oligomers by liquid chromatography and tandem mass spectrometry, it appears that the bound guanine nucleotides show either little or no turnover during one cell-cycle period [38]. Thus, GTP binding and hydrolysis within septin hetero-oligomers is too slow to account for the rate of assembly and disassembly of the filaments that is observed in vivo at the collar. However, the measurements made are an ensemble average and do not exclude the possibility that a subset of septins (that, perhaps, act as either nucleators or caps) might bind to and hydrolyze GTP much faster than the mean value determined for the mixtures of complexes isolated from cells. Second, and perhaps more tellingly, a cdc10 cdc12 double mutant in which both septins are deficient in GTP hydrolysis (but not in GTP binding), displays no detectable defect in assembly of the filamentous septin collar [28]. Thus, although GTP binding renders septin complexes competent for polymerization, factors other than GTP hydrolysis probably regulate the dynamics of septin filament formation and breakdown in the cell. Moreover, no GDP–GTP exchange factors (GEFs) and GTPase-activating proteins (GAPs) for septins have been identified.
Phosphorylation exerts temporal control on septin-collar assembly
The dynamics of septin-containing structures during the cell cycle of budding yeast is illustrated in Figure 3 (Reviewed in [61]). After completion of cytokinesis and separation of mother and daughter cells, the septin rings disassemble for a short period in G1. Whether disassembly involves the degradation of septin subunits or whether septins are recycled has not been determined, but a role for a specific post-translational modification (SUMOylation) by small ubiquitin-like modifier (SUMO) has been proposed [62]. In fact, septins are, by far, the most abundant substrates for SUMOylation in budding yeast. Preventing SUMO attachment to septins Cdc3 and Cdc11 by mutagenesis of the relevant Lys residues to Ala causes ‘old’ septin rings to persist. By contrast, preventing SUMO attachment to the same septins by inactivation of the major SUMO ligase (by a siz1Δ mutation) does not cause the same phenotype [63]. These newer findings cast doubt on the suggestion that SUMOylation is required for disassembly of septin rings after completion of cytokinesis. In the next cell cycle, the initial recruitment of the septin patch to the incipient bud site and its transition into a rimmed disk depends on function of the small, Rho-family GTPase, Cdc42 (Reviewed in [61]). It has been reported that the effects of Cdc42 require the conversion of Cdc42–GTP to Cdc42–GDP [64], via the action of either the cognate Cdc42–GAPs (Rga1, Rga2 and Bem3) [47] or, as yet, unknown effectors [65].
Phosphorylation by two protein kinases, Cla4 [28,66–68] and Gin4 [39,48,69], plays a prominent role in initiating and/or stabilizing filament assembly during collar formation during emergence of the bud. Cla4 is a clear-cut ortholog of mammalian p21-activated protein kinases (PAKs), and the closest mammalian relative of Gin4 is the neuron-specific, AMPK-related, kinase-family member, BRSK1. Evidence supports the view that both kinases are recruited to the bud site early in the cell cycle, at least in part, by direct binding to septins; and, Gin4 phosphorylates Shs1 exclusively, whereas Cla4 phosphorylates Cdc10, Cdc3 and Cdc11. Mutation of at least one of the Cla4 sites in Cdc10 (Ser256 to Ala) has readily discernible effects on cell morphology and septin architecture [28]. It is noteworthy that, as with other PAKs, Cla4 is activated through binding of Cdc42–GTP to an N-terminal Cdc42 and/or Rac-interactive-binding (CRIB) domain. Considering that Cdc42 function is implicated in bud emergence, stimulation of Cla4 activity by Cdc42 provides a mechanism to couple the timing of this event to assembly of the septin collar. Intriguingly, Cdc42 is also an important regulator of septin assembly in mammalian cells. This occurs, at least in part, through its binding to the Borg proteins, which are also septin-binding proteins [70], although there are no clear Borg orthologs in yeast [71]. To our knowledge, the role of PAK in the formation and function of septin filaments has not yet been investigated in mammals.
At the end of the cell cycle in budding yeast, during cytokinesis, septin-collar scission into separate rings correlates with dephosphorylation of Shs1 by phosphoprotein phosphatase 2A (PP2A), specifically an isoform that contains Rts1, a homolog of the mammalian B′-type regulatory subunit [48]. Also, direct phosphorylation by Cdc28, an ortholog of mammalian cyclin-dependent kinase 1 (Cdk1/Cdc2), of the extreme C-terminus of Cdc3 has been implicated in septin disassembly [72]. Conversely, two G1-cyclins (Pcl1 and Pcl2) that associate with a different cyclin-dependent kinase, Pho85 (ortholog of mammalian Cdk5) [73], localize to the bud neck at the time of septin-collar assembly [74]. In collaborative studies, we find that Pcl1–, Pcl2– or Pcl9–Pho85 complexes are able to phosphorylate Cdc3, Cdc10 and Cdc12 efficiently in vitro (R. Sopko, J. Moffat, M. Versele, J. Thorner and B.J. Andrews, unpublished), but the in vivo significance of these phosphorylation events is still under study. Furthermore, as mentioned earlier, there are additional protein kinases (and phosphoprotein phosphatases) that localize to the bud neck [14,27]. Clearly, we are just beginning to understand how phosphorylation of septins (and septin-associated proteins) influences the different facets of septin organization, dynamics and function.
Role of anillin in septin-ring formation
Anillin is an animal protein that interacts with septins and mediates the septin-filament assembly that is templated by actin [9,44,75,76]. Anillin also interacts with type II myosin and contributes to spatial regulation of the contractile activity of the actomyosin that is involved in cytokinesis [77]. Apparent anillin homologs in fission yeast, Mid1 and Mid2, which recruit type II myosins (Myo2 and Myp2), are important for organization of the septin rings at the medial cortex at the end of anaphase, and Mid2 binds septins directly [19,20,32]. In contrast to animal anillins, Mid2 does not contain an F-actin-binding domain. FRAP experiments show that the septin rings are destabilized if Mid2 is absent, which indicates that Mid2 is important for stability of septin filaments. Indeed, ectopic expression of a stable, truncated derivative of Mid2 (endogenous expression of Mid2 is restricted to anaphase) leads to persistent septin rings, presumably because disassembly is impeded. Although septins are recruited to the medial cortex of fission yeast cells in the absence of Mid2 (possibly by association with membrane phosphoinositides), they do not coalesce into a proper ring structure that, presumably, corresponds to authentic filament formation [33]. The closest counterpart of Mid2 in budding yeast appears to be Bud4, which localizes to the bud neck and associates with other markers of the bud neck, such as IQGAP (Iqg1) [78], which, in fission yeast, are also recruited early to the medial cytokinetic ring [32]. Together, these data make a strong case for an important role of anillin in septin-filament polymerization and stabilization to promote cytokinesis.
Possible role for chaperones in septin-filament dynamics
Septins, if purified individually and then mixed in vitro, do not assemble into hetero-oligomeric complexes. In contrast, stoichiometric hetero-oligomeric complexes can be purified readily if the different septin subunits are co-expressed in E. coli or insect cells [9,10,45]. It has been observed recently that overexpression of a dominant-negative mutant of the yeast chaperone Hsp104 (G217S T499I), which is also nucleotide-binding-defective, results in the formation of aberrant septin structures and altered bud morphology in otherwise wild-type cells, and the mutant also colocalizes with the misplaced septins [79]. Strikingly, the effect of the Hsp104 mutant is suppressed by point mutations in the CTE of Cdc12. These findings raise the possibility that assembly and/or disassembly of septin complexes and/or polymers involves Hsp104, but it remains to be shown whether the activity of wild-type Hsp104 is crucial for septin dynamics and whether other chaperones contribute when Hsp104 is absent (because hsp104Δ cells are viable). Nonetheless, a potential role for this universal family of chaperones (Hsp104 in eukaryotes and ClpB in prokaryotes) [80] in either assembly or preventing the non-specific aggregation of septin complexes might explain why hetero-oligomeric septin complexes cannot be reconstituted from purified individual subunits.
Concluding remarks
It is clear that septin filaments are obligate heteropolymers in all cell types examined, and that filament formation is required for the physiological function of septin-containing structures as scaffolds and compartment barriers. Moreover, the building block that polymerizes to form these filaments is itself a preformed, multimeric complex of at least three classes of septins (with the exception of C. elegans in which two different septins appear to suffice). Therefore, it seems more than appropriate to classify septin filaments in the same category as other, better-studied cytoskeletal polymers, such as actin microfilaments, microtubules and intermediate filaments. However, in contrast to these other elements of the cytoskeleton, septin polymerization is less dynamic and does not appear to be associated with performance of mechano–chemical work. These features are reflected in what is known about the mechanisms that control filament assembly. In budding yeast, at least, septin phosphorylation and dephosphorylation occur at the stately pace of the cell cycle, and GTP binding and hydrolysis occur exceedingly slowly. Although GTP binding is crucial to the appearance and function of septin structures in vivo, and to the ability of septin complexes to form filaments in vitro [28], how guanine nucleotides promote septin polymerization at the structural level is enigmatic. To address this issue, one major challenge in septin biochemistry is to find ways to manipulate the guanine nucleotide content of purified, pre-assembled septin complexes. Now that hetero-oligomeric septin complexes can be prepared and purified readily, a realistic, crucial goal is to determine high-resolution, three-dimensional structures of septin complexes (including mutants that assemble into complexes, but do not polymerize into filaments) and, eventually, septin filaments. Equally unexplained and relatively unexplored, but no less intriguing, are the roles of phosphoinositide binding, phosphorylation and SUMOylation in the dynamics and function of septins.
Acknowledgments
We thank members of the Thorner Laboratory and the following colleagues for fruitful discussions and/or sharing unpublished data: T. Alber, B.J. Andrews, Y. Barral, V.J. Cid, D. Drubin, J. Dobbelaere, P. Grob, D. Kellogg, M. McMurray, J. Moffat, E. Nogales, S-S. Park, A. Rodal and K. Tatchell. This work was supported by a Long-Term Fellowship (LT00257/2001-M/4) from the Human Frontier Science Program Organization and a NATO Advanced Fellowship (to M.V.) and by NIH Research Grant GM21841 and facilities provided by the Berkeley campus Cancer Research Laboratory (to J.T.).
Glossary
- Incipient bud site
the cortical site at the periphery of an S. cerevisiae cell where the future bud will emerge.
- Septin patch or cap
a somewhat amorphous accumulation of septins at the incipient bud site that forms in G1 cells prior to bud emergence in which the septins are highly mobile (as determined by FRAP analysis).
- Septin disk or ring
a flattened life preserver-like structure, in which the septins are more concentrated at the edges than at the center, that forms at the time of bud emergence and in which septins still display a high rate of exchange.
- Septin collar
a tubular structure with flared ends (hourglass-shaped) whose walls are composed of arrays of highly ordered septin filament that persists at the bud neck for most of the cell cycle, which can be readily detected by electron microscopy in grazing thin-sections of the bud neck and in which septins are immobilized.
- Split septin rings
the two separate rings formed from the septin collar during cytokinesis, in which septins again display a high rate of exchange and which disassemble completely after cell division and prior to the next cell cycle; this splitting is thought to form a privileged compartment wherein factors essential for cytokinesis are concentrated.
- Medial cortex
the cytoskeletal structure that assembles in the middle of an S. pombe cell during M-phase that demarcates the site where cytokinesis will occur.
- Polarized versus isotropic bud growth
in S. cerevisiae, during initial emergence of the bud in late G1-phase, new membrane is deposited primarily at the bud tip, which results in polarized (apical) bud growth, and, if anisotropic growth persists, in a markedly elongated bud. In a normal cell cycle, shortly after bud emergence, bud growth switches to a mode in which membrane deposition is distributed more evenly (isotropic growth), resulting in a rounded or oval-shaped bud.
- Prospore envelope
the membranes that encase each of the four newly-forming haploid spores after the completion of meiosis II and during the early stages of sporulation.
References
- 1.Hartwell L. Genetic control of the cell division cycle in yeast. IV Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- 2.Neufeld TP, Rubin GM. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 3.Kinoshita M, et al. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 1997;11:1535–1547. doi: 10.1101/gad.11.12.1535. [DOI] [PubMed] [Google Scholar]
- 4.Hall PA, et al. Expression profiling the human septin gene family. J Pathol. 2005;206:269–278. doi: 10.1002/path.1789. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita M. The septins. Genome Biol. 2003;4:236. doi: 10.1186/gb-2003-4-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macara IG, et al. Mammalian septins nomenclature. Mol Biol Cell. 2002;13:4111–4113. doi: 10.1091/mbc.E02-07-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Field CM, et al. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frazier JA, et al. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinoshita M, et al. Self- and Actin-Templated Assembly of Mammalian Septins. Dev Cell. 2002;3:791–802. doi: 10.1016/s1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- 10.Versele M, et al. Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:4568–4583. doi: 10.1091/mbc.E04-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69:717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byers B, Goetsch L. Loss of the filamentous ring in cytokinesis-defective mutants of budding yeast. J Cell Biol. 1976;70:35. [Google Scholar]
- 13.Rodal AA, et al. Actin and septin ultrastructures at the budding yeast cell cortex. Mol Biol Cell. 2005;16:372–384. doi: 10.1091/mbc.E04-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gladfelter AS, et al. The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- 15.Faty M, et al. Septins: a ring to part mother and daughter. Curr Genet. 2002;41:123–131. doi: 10.1007/s00294-002-0304-0. [DOI] [PubMed] [Google Scholar]
- 16.Kusch J, et al. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 2002;16:1627–1639. doi: 10.1101/gad.222602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeVirgilio C, et al. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology. 1996;142:2897–2905. doi: 10.1099/13500872-142-10-2897. [DOI] [PubMed] [Google Scholar]
- 18.Fares H, et al. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J Cell Biol. 1996;132:399–411. doi: 10.1083/jcb.132.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berlin A, et al. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J Cell Biol. 2003;160:1083–1092. doi: 10.1083/jcb.200212016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tasto JJ, et al. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J Cell Biol. 2003;160:1093–1103. doi: 10.1083/jcb.200211126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kartmann B, Roth D. Novel roles for mammalian septins: from vesicle trafficking to oncogenesis. J Cell Sci. 2001;114:839–844. doi: 10.1242/jcs.114.5.839. [DOI] [PubMed] [Google Scholar]
- 22.Hsu SC, et al. Subunit composition, protein interactions, and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron. 1998;20:1111–1122. doi: 10.1016/s0896-6273(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 23.Beites CL, et al. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- 24.Spiliotis ET, et al. A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science. 2005;307:1781–1785. doi: 10.1126/science.1106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue J, et al. Phosphorylation of a new brain-specific septin, G-septin, by cGMP-dependent protein kinase. J Biol Chem. 2000;275:10047–10056. doi: 10.1074/jbc.275.14.10047. [DOI] [PubMed] [Google Scholar]
- 26.Field CM, Kellogg D. Septins: cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 1999;9:387–394. doi: 10.1016/s0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- 27.Cid VJ, et al. Orchestrating the cell cycle in yeast: sequential localization of key mitotic regulators at the spindle pole and the bud neck. Microbiology. 2002;148:2647–2659. doi: 10.1099/00221287-148-9-2647. [DOI] [PubMed] [Google Scholar]
- 28.Versele M, Thorner J. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J Cell Biol. 2004;164:701–715. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barral Y, et al. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- 30.Takizawa PA, et al. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 31.Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- 32.Wu JQ, et al. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 33.An H, et al. Requirements of fission yeast septins for complex formation, localization, and function. Mol Biol Cell. 2004;15:5551–5564. doi: 10.1091/mbc.E04-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt K, Nichols BJ. A barrier to lateral diffusion in the cleavage furrow of dividing mammalian cells. Curr Biol. 2004;14:1002–1006. doi: 10.1016/j.cub.2004.05.044. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, et al. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr Biol. 1999;9:1458–1467. doi: 10.1016/s0960-9822(00)80115-3. [DOI] [PubMed] [Google Scholar]
- 36.Casamayor A, Snyder M. Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function. Mol Cell Biol. 2003;23:2762–2777. doi: 10.1128/MCB.23.8.2762-2777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez-Escudero, I. et al. Reconstitution of the mammalian PI3K-PTEN-Akt pathway in yeast. Biochem. J. (in press)
- 38.Vrabioiu AM, et al. The majority of the Saccharomyces cerevisiae septin complexes do not exchange guanine nucleotides. J Biol Chem. 2004;279:3111–3118. doi: 10.1074/jbc.M310941200. [DOI] [PubMed] [Google Scholar]
- 39.Mortensen EM, et al. Cell Cycle-dependent Assembly of a Gin4-Septin Complex. Mol Biol Cell. 2002;13:2091–2105. doi: 10.1091/mbc.01-10-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen TQ, et al. The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J Cell Sci. 2000;113:3825–3837. doi: 10.1242/jcs.113.21.3825. [DOI] [PubMed] [Google Scholar]
- 41.Finger FP, et al. A role for septins in cellular and axonal migration in C. elegans. Dev Biol. 2003;261:220–234. doi: 10.1016/s0012-1606(03)00296-3. [DOI] [PubMed] [Google Scholar]
- 42.Larisch S. The ARTS connection: role of ARTS in apoptosis and cancer. Cell Cycle. 2004;3:1021–1023. [PubMed] [Google Scholar]
- 43.Mata J, et al. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 44.Kinoshita M. Assembly of mammalian septins. J Biochem (Tokyo) 2003;134:491–496. doi: 10.1093/jb/mvg182. [DOI] [PubMed] [Google Scholar]
- 45.Sheffield PJ, et al. Borg/Septin interactions and the assembly of mammalian septin heterodimers, trimers and filaments. J Biol Chem. 2003;278:3483–3488. doi: 10.1074/jbc.M209701200. [DOI] [PubMed] [Google Scholar]
- 46.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caviston JP, et al. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol Biol Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobbelaere J, et al. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4:345–357. doi: 10.1016/s1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt K, Nichols BJ. Functional interdependence between septin and actin cytoskeleton. BMC Cell Biol. 2004;5:43–55. doi: 10.1186/1471-2121-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dent J, et al. A prototypic platelet septin and its participation in secretion. Proc Natl Acad Sci U S A. 2002;99:3064–3069. doi: 10.1073/pnas.052715199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Surka MC, et al. The mammalian septin MSF localizes with microtubules and is required for completion of cytokinesis. Mol Biol Cell. 2002;13:3532–3545. doi: 10.1091/mbc.E02-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagata K, et al. Filament formation of MSF-A, a mammalian septin, in human mammary epithelial cells depends on interactions with microtubules. J Biol Chem. 2003;278:18538–18543. doi: 10.1074/jbc.M205246200. [DOI] [PubMed] [Google Scholar]
- 53.Kinoshita A, et al. Identification of septins in neurofibrillary tangles in Alzheimer’s disease. Am J Pathol. 1998;153:1551–1560. doi: 10.1016/S0002-9440(10)65743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ihara M, et al. Association of the cytoskeletal GTP-binding protein Sept4/H5 with cytoplasmic inclusions found in Parkinson’s disease and other synucleinopathies. J Biol Chem. 2003;278:24095–24102. doi: 10.1074/jbc.M301352200. [DOI] [PubMed] [Google Scholar]
- 55.Mendoza M, et al. GTP binding induces filament assembly of a recombinant septin. Curr Biol. 2002;12:1858–1863. doi: 10.1016/s0960-9822(02)01258-7. [DOI] [PubMed] [Google Scholar]
- 56.Larisch S, et al. A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nat Cell Biol. 2000;2:915–921. doi: 10.1038/35046566. [DOI] [PubMed] [Google Scholar]
- 57.Vega IE, Hsu SC. The septin protein Nedd5 associates with both the exocyst complex and microtubules and disruption of its GTPase activity promotes aberrant neurite sprouting in PC12 cells. Neuroreport. 2003;14:31–37. doi: 10.1097/00001756-200301200-00006. [DOI] [PubMed] [Google Scholar]
- 58.Hanai N, et al. Biochemical and cell biological characterization of a mammalian septin, Sept11. FEBS Lett. 2004;568:83–88. doi: 10.1016/j.febslet.2004.05.030. [DOI] [PubMed] [Google Scholar]
- 59.Robertson C, et al. Properties of SEPT9 isoforms and the requirement for GTP binding. J Pathol. 2004;203:519–527. doi: 10.1002/path.1551. [DOI] [PubMed] [Google Scholar]
- 60.Nogales E. Structural insight into microtubule function. Annu Rev Biophys Biomol Struct. 2001;30:397–420. doi: 10.1146/annurev.biophys.30.1.397. [DOI] [PubMed] [Google Scholar]
- 61.Longtine MS, Bi E. Regulation of septin organization and function in yeast. Trends Cell Biol. 2003;13:403–409. doi: 10.1016/s0962-8924(03)00151-x. [DOI] [PubMed] [Google Scholar]
- 62.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 64.Gladfelter AS, et al. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J Cell Biol. 2002;156:315–326. doi: 10.1083/jcb.200109062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gladfelter AS, et al. Genetic interactions among regulators of septin organization. Eukaryot Cell. 2004;3:847–854. doi: 10.1128/EC.3.4.847-854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiss EL, et al. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat Cell Biol. 2000;2:677–685. doi: 10.1038/35036300. [DOI] [PubMed] [Google Scholar]
- 67.Schmidt M, et al. Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol Biol Cell. 2003;14:2128–2141. doi: 10.1091/mbc.E02-08-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kadota J, et al. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Longtine MS, et al. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joberty G, et al. Borg proteins control septin organization and are negatively regulated by Cdc42. Nat Cell Biol. 2001;3:861–866. doi: 10.1038/ncb1001-861. [DOI] [PubMed] [Google Scholar]
- 71.Joberty G, et al. The Borgs, a new family of Cdc42 and TC10 GTPase-interacting proteins. Mol Cell Biol. 1999;19:6585–6597. doi: 10.1128/mcb.19.10.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang CS, Reed SI. Phosphorylation of the septin cdc3 in g1 by the cdc28 kinase is essential for efficient septin ring disassembly. Cell Cycle. 2002;1:42–49. [PubMed] [Google Scholar]
- 73.Huang D, et al. Mammalian Cdk5 is a functional homologue of the budding yeast Pho85 cyclin-dependent protein kinase. Proc Natl Acad Sci U S A. 1999;96:14445–14450. doi: 10.1073/pnas.96.25.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moffat J, Andrews B. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat Cell Biol. 2004;6:59–66. doi: 10.1038/ncb1078. [DOI] [PubMed] [Google Scholar]
- 75.Oegema K, et al. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J Cell Biol. 2000;150:539–552. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finger FP. One ring to bind them. Septins and actin assembly. Dev Cell. 2002;3:761–763. doi: 10.1016/s1534-5807(02)00371-4. [DOI] [PubMed] [Google Scholar]
- 77.Straight AF, et al. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol Biol Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Osman MA, et al. Iqg1p links spatial and secretion landmarks to polarity and cytokinesis. J Cell Biol. 2002;159:601–611. doi: 10.1083/jcb.200205084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schirmer EC, et al. Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol Biol Cell. 2004;15:2061–2072. doi: 10.1091/mbc.E02-08-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee S, et al. The ClpB/Hsp104 molecular chaperone-a protein disaggregating machine. J Struct Biol. 2004;146:99–105. doi: 10.1016/j.jsb.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 81.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 82.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 83.Nagata K, et al. Biochemical and cell biological analyses of a mammalian septin complex, Sept7/9b/11. J Biol Chem. 2004;279:55895–55904. doi: 10.1074/jbc.M406153200. [DOI] [PubMed] [Google Scholar]
- 84.Barral Y, et al. Nim1-related kinases coordinate cell-cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shulewitz MJ, et al. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Longtine MS, et al. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Booher RN, et al. Properties of the Saccharomyces cerevisiae wee1 and its differential regulation of p34cdc28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lew DJ. The morphogenesis checkpoint: how yeast cells watch their figures. Curr Opin Cell Biol. 2003;15:648–653. doi: 10.1016/j.ceb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 89.Hanrahan J, Snyder M. Cytoskeletal activation of a checkpoint kinase. Mol Cell. 2003;12:663–673. doi: 10.1016/j.molcel.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 90.Cid VJ, et al. Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. Mol Biol Cell. 2001;12:1645–1669. doi: 10.1091/mbc.12.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sakchaisri K, et al. Coupling morphogenesis to mitotic entry. Proc Natl Acad Sci U S A. 2004;101:4124–4129. doi: 10.1073/pnas.0400641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shulewitz, M.J. (2000) Septin assembly regulates cell cycle progression through activation of a protein kinase signaling pathway. Ph.D. Thesis, Univ. of California, Berkeley, 172pp
- 93.Theesfeld CL, et al. A monitor for bud emergence in the yeast morphogenesis checkpoint. Mol Biol Cell. 2003;14:3280–3291. doi: 10.1091/mbc.E03-03-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Castillon GA, et al. Septins have a dual role in controlling mitotic exit in budding yeast. Curr Biol. 2003;13:654–658. doi: 10.1016/s0960-9822(03)00247-1. [DOI] [PubMed] [Google Scholar]


