Abstract
In contrast to the majority of tetrameric SSB proteins, the recently discovered SSB proteins from the Thermus/Deinoccus group form dimers. We solved the crystal structures of the SSB protein from Thermus aquaticus (TaqSSB) and a deletion mutant of the protein and show the structure of their ssDNA binding domains to be similar to the structure of tetrameric SSBs. Two conformations accompanied by proline cis–trans isomerization are observed in the flexible C-terminal region. For the first time, we were able to trace 6 out of 10 amino acids at the C-terminus of an SSB protein. This highly conserved region is essential for interaction with other proteins and we show it to adopt an extended conformation devoid of secondary structure. A model for binding this region to the χ subunit of DNA polymerase III is proposed. It explains at a molecular level the reason for the ssb113 phenotype observed in Escherichia coli.
INTRODUCTION
Bacterial single-stranded DNA–binding (SSB) proteins play an important role in DNA metabolism. They protect the vulnerable single-stranded DNA (ssDNA) from nucleolytic digestion and prevent hairpin formation, thus keeping ssDNA in a suitable conformation for the action of enzymes involved in processes like DNA replication, repair and recombination. Beyond this preparation of the ssDNA for proper enzymatic function, SSB proteins are involved in physical interactions with a couple of proteins that play an important role in those pathways (1–6). The presence of SSB proteins is essential for the survival of bacteria (7) and they are believed to belong to the minimal set of genes required for cellular life (8).
The primary structure of bacterial SSB proteins contains three distinct regions. The N-terminal DNA-binding domain, which forms an oligonucleotide/oligosaccharide-binding (OB)-fold (9), is succeeded by a glycine and proline-rich region and the acidic, highly conserved region formed by the last 10 amino acids. Almost all bacterial SSB proteins are active as tetramers where four OB-folds act together in the binding of ssDNA. The interaction of the four OB-folds seems to be essential for in vivo function, since a mutant SSB protein of Escherichia coli (EcoSSB) that only forms homodimers is not able to take over the function of EcoSSB in the E.coli cell (10).
Only recently dimeric SSB proteins have been discovered in the Thermus/Deinococcus group of bacteria (11–13). The protomers of these proteins are nearly twice the size of the tetrameric bacterial SSB proteins and contain two OB-folds per monomer (14). This observation suggests a gene duplication event in the evolution of the Thermus/Deinococcus group of bacteria (13). Therefore, the cooperation of four OB-folds in the function of bacterial SSB proteins seems to be a common principle.
While the structures of the DNA-binding domains of SSB proteins from E.coli (15–17), Mycobacterium tuberculosis (18), Mycobacterium smegmatis (19) and Deinococcus radiodurans (14) have been solved, there is little structural information about the C-terminal region of SSB proteins. Since the glycine- and proline-rich region of EcoSSB seems to be disordered (15,20), the highly conserved region formed by the last 10 amino acids could not be resolved in the crystals. However, it was shown that the negatively charged C-terminal region is essential for the survival of E.coli cells (21) and interacts with proteins involved in DNA replication, repair and recombination. Among these proteins are the χ subunit of the DNA polymerase III (4,5), exonuclease I (2), uracil DNA glycosylase (3) and priA helicase (1). In contrast, the glycine- and proline-rich region is not essential for the survival of E.coli cells and it was speculated to act as a spacer (21).
Here, we describe the crystal structures of wild-type SSB protein from the thermophilic, eubacterial organism, Thermus aquaticus (TaqSSB) and a mutant form of the protein that lacks the glycine- and proline-rich region (TaqSSBΔ228-252 R190A). In both structures the main part of the protein consists of an elongated β-barrel domain with two adjacent α-helices, which together form two OB-folds (9). For the wild-type protein we show that the glycine- and proline-rich C-terminal part of the protein can adopt two different conformations. In the structure of the deletion mutant, which lacks this region, we were able to trace 6 out of 10 C-terminal amino acids that are essential for protein–protein interactions. These residues are highly disordered and devoid of secondary structure. We present a model that shows the C-terminal region of SSB protein docked to the χ subunit of E.coli DNA polymerase III (22).
MATERIALS AND METHODS
Buffers and reagents
All materials were of the highest purity available and were obtained from Sigma and Fluka. T.aquaticus strain ATCC25104 was purchased from German Collection of Microorganisms and Cell Cultures (DSMZ).
Cloning of the Thermus aquaticus ssb gene
PCR amplification of the ssb gene was performed with heat disrupted T.aquaticus cells and primers (MWG Biotech) 5′-GGAGACCATGGCTCGAGGCCTGAACCA-3′ for forward priming and 5′-GAAGAGGATCCTCAAAACGGCAAATCCTCCTC-3′ for reverse priming of the complete ssb gene. PCR was carried out using Pfu DNA polymerase (Stratagene), 200 μM dNTP and 0.5 μM of each primer with an annealing temperature of 54°C. After removal of nucleotides and proteins (PCR Purification Kit, Qiagen) the PCR product was digested with NcoI and BamHI and cloned into the NcoI and BamHI sites of plasmid pET15b (Novagen) to generate pETTaqSSB. All DNA constructs were verified by sequencing of both DNA strands of the gene (MWG Biotech).
Construction of mutant TaqSSB proteins
Site-directed mutagenesis was performed using the QuikChange mutagenesis kit (Stratagene). For construction of the deletion mutant TaqSSBΔ228–252 a NotI restriction site was introduced at bp 675–681 and an Ndel site downstream of the Taqssb stop-codon generating pETTaqSSBentry. After digestion of this construct with NotI and NdeI a double-stranded DNA cassette (MWG Biotech) encoding TaqSSB amino acids 253–264 was introduced to generate pETTaqSSBΔ228–252. Thus TaqSSBΔ228–252 is a fusion protein in which amino acids 1–227 of TaqSSB are followed by the last 12 amino acids of TaqSSB. TaqSSBΔ228–252 R190A was constructed by site-directed mutagenesis of pETTaqSSBΔ228–252, exchanging the codon of arginine 190 by an alanine codon.
Expression and purification of proteins
As the TaqSSB gene contains codons rarely used in E.coli, protein expression of TaqSSB and TaqSSB mutants was performed in the E.coli strain Rosetta (DE3) pLysS (Novagen). A 900 mL overnight culture grown in standard Luria–Bertani (LB) medium in the presence of 100 μg/mL ampicillin, 30 μg/mL chloramphenicol and 1% glucose at 37°C was used to inoculate 10 L of LB medium in an aerated fermenter and the cells were grown at 37°C. An aliquot of 100 mL of the overnight culture was used for plasmid preparation and subsequent DNA sequencing to confirm the sequence of the expressed gene shortly before induction. After growing the cells to A600 nm=1.0 protein expression was induced by addition of 1 mM IPTG and was continued for 5 h. After harvesting the cells were washed with buffer W [0.2 M NaCl, 50 mM Tris–HCl pH 8.3, 1 mM EDTA and 10% (w/v) sucrose], resuspended in buffer S [buffer W containing 15 mM spermidin] and stored at −80°C.
Wild-type TaqSSB, TaqSSBΔ228–252 and TaqSSBΔ228–252 R190A proteins were purified as follows. After thawing the cells, one tablet of ‘complete EDTA free’ protease inhibitor (Roche) per 50 mL, 0.04% sodiumdesoxycholat and 0.1 mM PMSF were added. Sonication and removal of cell debris by preparative ultracentrifugation was followed by precipitation of non-thermophilic proteins by heat treatment at 65°C for 4 min. Proteins in the supernatant of a second preparative ultracentrifugation step were precipitated at 4°C by a drop by drop addition of polyethylenimine pH 6.9 up to a final concentration of 0.4%, incubated for 10 min and pelleted. The pellet was washed with TGE400 [400 mM NaCl, 50 mM Tris–HCl pH 8.3, 1 mM EDTA and 20% (v/v) glycerol] and extracted with TGE700 [700 mM NaCl, 50 mM Tris–HCl pH 8.3, 1 mM EDTA and 20% (v/v) glycerol] and insoluble material was removed by centrifugation. The clear supernatant was precipitated with 25% (w/v) (NH4)2SO4 at 4°C, the pellet was redissolved in buffer L [2 M NaCl, 50 mM Tris–HCl pH 7.5, 1 mM EDTA and 10% (v/v) glycerol] and loaded onto a BlueSepharose column (23) equilibrated in the same buffer. After washing the column with B1 (buffer L containing 50 mM urea), the column was eluted with buffer B2 (buffer L containing 5 M urea). Fractions were analyzed by SDS–PAGE (24) and TaqSSB fractions were pooled, diluted to A280 nm = 4 and dialyzed against buffer Q1 [100 mM NaCl, 20 mM Tris–HCl pH 7.6, 1 mM EDTA, 10% (v/v) glycerol]. The supernatant was loaded onto a ResourceQ column (GE Healthcare Life Sciences) equilibrated in the same buffer. After washing with buffer Q1, a linear salt gradient (100 mM–500 mM NaCl in buffer Q1) was used to elute proteins. Fractions were analyzed by SDS–PAGE and TaqSSB fractions were pooled, concentrated using Vivaspin concentrators (Vivascience), dialyzed against storage buffer [1 M NaCl, 20 mM KPi pH 7.4, 1 mM EDTA, 60% (v/v) glycerol] and stored at −20°C. The purified proteins were at least 98% pure as judged from Coomassie Brilliant blue stained SDS-gels.
Crystallization
Both wild-type and mutant protein crystals were grown at 20°C by vapor diffusion using the sitting drop geometry. For wild-type TaqSSB 1.5 μL of protein (3.8 mg/mL in 10 mM Tris–HCl pH 8.0) and reservoir solutions were mixed, the latter contained 100 mM Imidazol pH 7.4, 35% (v/v) 2-ethoxyethanol and 200 mM calcium acetate. TaqSSBΔ228–252 R190A mutant (3.6 mg/mL in 10 mM Tris–HCl pH 8.0) has crystallized in similar conditions with reservoir containing 100 mM Imidazol pH 6.2, 35% (v/v) 2-ethoxyethanol and 220 mM calcium acetate. Triangle-shaped crystal plates of wild-type TaqSSB (0.3 × 0.3 × 0.05 mm3) and rectangular crystal plates of TaqSSBΔ228–252 R190A mutant (0.45 × 0.3 × 0.05 mm3) appeared within 2–3 days. Prior to flash cooling, crystals were rinsed in the corresponding reservoir solution.
Diffraction data collection, structure determination and refinement
Diffraction data were collected at the Microfocus Beamline ID13, ESRF and MPG/GBF beamline BW6, DESY using MAR and ADSC-Q4 CCD detectors (see Table 1 for details). High-redundancy data used for tracing flexible protein regions were collected at Bruker AXS B.V. (Delft, Netherlands) using an X8PROTEUM system with MICROSTAR x-ray source, flexible kappa goniostat and Pt135 CCD detector. The synchrotron data were processed with XDS programme package (25) and the high-redundancy data collected at the rotating anode generator were processed with PROTEUM2 software (Bruker AXS).
Table 1.
Crystallographic data and refinement statistics for wild-type TaqSSB and the TaqSSBΔ228-252 R190A mutant
| Protein | TaqSSB wild-type | TaqSSBΔ228–252 R190A |
|---|---|---|
| PDB code | 2ihe | 2ihf |
| Crystal parameters | ||
| Space group | C2 | C2221 |
| Cell parameters: a, b, c, α, β, γ | 69.8, 80.7, 56.5, 90, 124.4, 90 | 67.6, 80.4, 97.8, 90, 90, 90 |
| Data collection | ||
| Beamline/source | ID13, ESRF | BW6, DESY |
| Wave length (Å) | 0.976 | 1.07 |
| Resolution of data (Å) | 20.0–2.1 | 20.0–1.9 |
| No. of observations/unique reflections | 74 521/14 759 | 391 536/21 312 |
| Completeness (total/higha)% | 96.8/95.6 | 99.6/98.9 |
| Redundancy | 5.1 | 18.4 |
| <I/σ(I)>(total/higha) | 12.2/5.1 | 20.8/8.4 |
| Rsym (total/higha)% | 8.0/33.4 | 10.3/36.7 |
| Refinement statistics | ||
| Resolution range (Å) | 20.0–2.1 | 20.0–1.9 |
| Included amino acids | 2–111, 127–161, 170–204, 214–228 | 1–111, 126–161, 169–204, 214–227, 253–260 |
| No. of protein atoms | 1588 | 1650 |
| No. of waters | 124 | 185 |
| Rwork/Rfreeb% | 20.9/25.3 | 22.2/26.1 |
| r.m.s. deviation for bonds (Å)/angles (deg) | 0.007/1.3 | 0.007/1.4 |
aThe high resolution shell is 2.2–2.1 Å for TaqSSB wild-type and 2.0–1.9 Å for TaqSSBΔ228–252 R190A.
bFor the Rfree calculations 5% of the total number of reflections were used.
The structure of wild-type TaqSSB was determined by molecular replacement (26) using coordinates of SSB from D.radiodurans (14), pdb code: 1se8) as an initial model. Refinement was continued with CNS (27) and SHELX (28) and included simulated annealing and individual B-factor refinement. During cyclic rounds of refinement and manual rebuilding using programmes O (29) and COOT (30), solvent molecules were included in the molecular model. The structure determination of TaqSSBΔ228–252 R190A was initiated by a round of rigid body refinement using the protein part of the wild-type TaqSSB coordinates and continued by the same building/refinement procedure. The final electron density maps are shown in Figures 1 and 3.
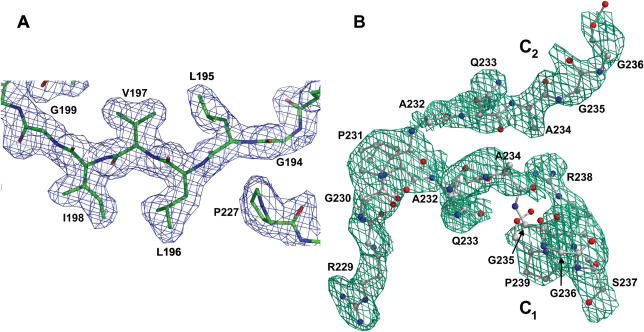
Figure 1.
(A) A portion of the 2Fo–Fc electron density in the region including residues G194–G199 and P227 of wild-type TaqSSB. The map is contoured at the 1.2σ level. (B) 2.2σ Fo–Fc electron density omit map superimposed with the final model of the glycine-rich C-terminal part of wild-type TaqSSB including residues P231–P239 in conformation C1 and residues P231–G236 in conformation C2.
The final models display good stereochemistry (see Table 1). Structures were overlaid with the programme COOT and adjusted manually. The coordinates and structure factor amplitudes for wild-type TaqSSB and TaqSSBΔ228–252 R190A structures have been deposited in the Protein Data Bank (PDB ID codes 2ihe and 2ihf, respectively).
Tracing flexible protein regions
To facilitate tracing of the glycine-rich C-terminal part of wild-type TaqSSB we have collected 100% complete high-redundancy diffraction data using the home source X8PROTEUM (Bruker) with the flexible kappa goniostat. These data allowed tracing two conformations of the C-terminal part up to the residue 239 (C1) and 236 (C2) (Figure 1B). The 20-fold redundancy was necessary to obtain a traceable electron density map for the above residues at low sigma cutoffs (Figure 1B). Further improvement of the data quality did not reveal additional details in electron density maps. In case of TaqSSBΔ228–252 R190A the model of the flexible C-terminal part starting from residue L253 was built by the following iterative procedure. The model including residues 1–227 was refined by ARP/wARP (31) from CCP4i programme package (32) using 18-fold redundant synchrotron data set (Table 1). The ARP/wARP model phases and figures of merit were used in the following density modification procedure by DM programme (32). During the procedure of density modification the region occupied by the existing protein model and additional region covering all possible directions for the next two C-terminal residues were protected by the mask prepared by MAMA and MAPMAN programmes (33,34). The electron density map obtained after the density modification was used to trace one or two additional residues. The new residues were included into the protein model and the same steps were repeated for the next two residues. Such a procedure allowed building the C-terminal residues up to E260 after which the maps have become untraceable.
RESULTS AND DISCUSSION
Structure of wild-type TaqSSB
The 3D structure of the single-stranded DNA-binding protein from T.aquaticus (TaqSSB) was determined at 2.3 Å resolution. Figure 2 shows an overall view of the structure. From 264 amino acid residues of the TaqSSB sequence this structure contains residues 2–239 excluding residues 112–126, 162–169 and 205–213 where no electron density was observed. The main part of the protein comprises an elongated β-barrel domain with two adjacent α-helices, which is formed by two OB-folds (9) covalently linked by a flexible loop. The flexibility of the loop is reflected in the lack of electron density for OB-fold connector residues 112–126. The hydrophobic core of the molecule contains 39 residues. Most of these residues (29) are highly conserved in OB-folds of bacterial SSB proteins, which correlates with the high degree of structural conservation in the molecular architecture of SSB proteins. Interestingly residue H54 is part of the conserved hydrophobic core amino acids. It has been shown that the corresponding residue (H55) of the SSB protein from E.coli (EcoSSB) is crucial for the formation of the biological active tetramer (35). Mutating H55 of EcoSSB to Y or K destabilizes the OB–OB interaction to such an extent that only monomeric species are observed in solution (36). A number of hydrophobic residues are partially exposed and most of them form a vast hydrophobic patch on one side of the β-barrel in the region where two monomers associate to form the biological dimer (see below). Another source of molecular structure stabilization is provided by a vast network of main-chain hydrogen bonds connecting β-strands into the β-barrel and a number of surface hydrogen bonds and salt bridges between the side chains of charged and polar residues. There are 16 side-chain hydrogen bonds in TaqSSB that stabilize the mutual arrangement of the protein's secondary structure elements. Most of the residues participating in these interactions are not conserved, which may imply that the corresponding hydrogen bonds were acquired to increase the protein stability in the thermophilic organism. This assumption is supported by the fact that some of the non-conserved surface hydrogen bonds tend to cluster into more extended networks (like interactions E102–R81–E83–R100 or R218–Q177–E220). Formation of such clusters is known to contribute to the stabilization of protein structures from thermophilic and extreme thermophilic organisms (37,38).
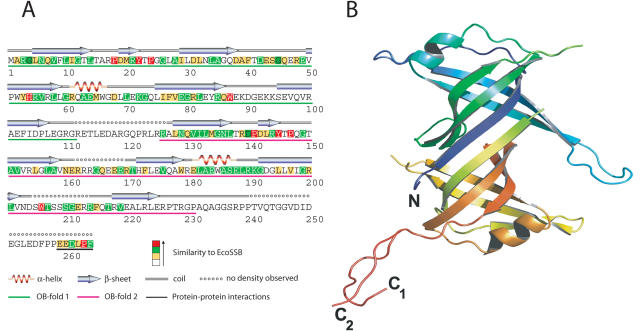
Figure 2.
(A) Amino acid sequence and secondary structure of the SSB protein from Thermus aquaticus. The N-terminal (underlined in green) and the C-terminal (underlined in red) OB-folds are connected by a flexible loop and form together an elongated β-barrel domain with two adjacent α-helices. The amino acids are coloured according to their similarity to the corresponding residues in EcoSSB. (B) Ribbon representation of the overall structure of wild-type TaqSSB. The colour changes from blue to red from N- to C-terminus, respectively. The two conformations of the glycine-rich C-terminal region are labelled with C1 and C2.
A close structural analog of TaqSSB is SSB from D.radiodurans (DraSSB) (14) (the r.m.s.d. value for the Cα atoms of the two structures is 1.3 Å). The main differences are observed in the region of the N-terminal α-helix, and in the loop regions. The part of loop 3, which is missing in the DraSSB structure, was traceable in TaqSSB although the electron density is weak. On the other hand the OB-fold connecting region structured as β-hairpin in DraSSB (residues 109–128 in DraSSB) appears to be very flexible and completely disordered in TaqSSB, as indicated by the complete lack of electron density.
Structure of the TaqSSB C-terminal part and of TaqSSBΔ228–252 R190A
Although the structure of a couple of bacterial SSB proteins has been solved (14–19,39), no structural information was obtained about the glycine-rich region and the ultimate C-terminal part. In the case of wild-type TaqSSB, the electron density maps calculated using high-redundancy x-ray diffraction data allowed tracing of two alternative conformations for the glycine-rich region. Chain tracing could be extended to residue 239 (C1) and 236 (C2) at low sigma cutoffs (Figures 1B and 3). The transition between C1 and C2 is coupled with the cis–trans isomerization of proline P231. Due to the high degree of flexibility, no information could be obtained about the structure of the last 25 amino acids. For EcoSSB the last 10 amino acids have been shown to be crucial for the interaction with other proteins involved in DNA replication, repair and recombination (1–5,14) and to be essential for the survival of the bacterial cell (21).
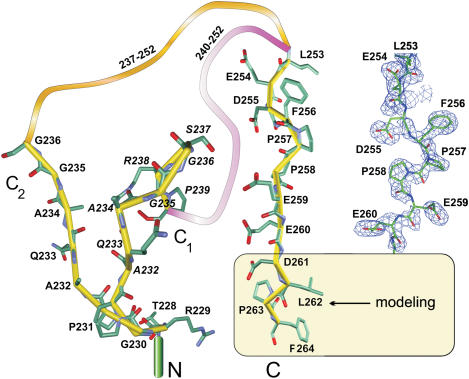
Figure 3.
Summary of the structural information on the flexible C-terminal part of TaqSSB obtained from wild-type TaqSSB and TaqSSBΔ228-252 R190A mutant crystal structures. The ball-and-stick model on white background represents residues resulting from the tracing of experimental electron density maps. Residues drawn on light yellow background had no experimental electron density and were obtained by a crystallographic energy minimization procedure. These residues were not included in the final structural model. The Cα-trace of the above mentioned regions is shown in yellow. Orange and magenta ribbons represent the unresolved flexible parts following residues G236 and P239 in the two conformations of the glycine-rich region. A portion of the 0.8σ 2Fo–Fc composite omit map for residues L253–E260 from TaqSSBΔ228-252 R190A obtained as described in ‘Materials and Methods’ section is shown on the right.
Since this region is separated from the well-structured DNA-binding domain by the flexible glycine-rich region, it was not possible to trace the electron density to the very end of the protein. Therefore, we constructed the deletion mutant TaqSSBΔ228–252, lacking the unstructured glycine-rich region, in which the last 12 amino acids are directly fused to the DNA-binding domain. Unfortunately, this protein formed twinned crystals that were not suitable for x-ray analysis. Mutation of arginine 190 to alanine (TaqSSBΔ228–252 R190A) resolved this problem. Since R190 is involved in intermolecular interactions of wild-type TaqSSB proteins in the crystal, the crystal packing of the TaqSSBΔ228–252 R190A mutant was completely different and no tendency to form twinned crystals could be observed. The TaqSSBΔ228–252 R190A mutant contains 239 amino acids in the sequence. To ease comparison of the structures of wild-type TaqSSB and TaqSSBΔ228–252 R190A the numbering of the residues in the C-terminal part of both proteins is kept identical. Thus, in the sequence of the mutant L253 directly follows P227. The sequence of the mutant thus contains amino acids numbered 1–227 and 253–264. Residues 112–125, 162–168, 205–213 and 261–264 are excluded from the model due to lack of electron density. Except for loop 1, loop 3 and loop 7, the structure of the main part is practically identical for mutant and wild-type TaqSSB (r.m.s.d. for Cα atoms is 0.8 Å). The observed differences in the loop regions are most likely a consequence of the different crystal packings. Due to the lack of the glycine-rich region parts of the C-terminus, which has been elusive to structure determination before, can be observed and are found to be highly disordered and devoid of secondary structure. In the mutant protein structure, electron density extends almost to the C-terminus. Model building was possible only up to the residue E260 at low sigma values (see Figure 3), as electron density becomes very weak and disconnected closer to the C-terminus. Tracing was not possible for the four C-terminal amino acids. The summary of C-terminal structural information obtained from both wild-type TaqSSB and mutant structures is presented in Figure 3. Weak electron density and a considerable amount of noise in those regions reflect a high degree of conformational variability, which implies that the observed structures represent only a small fraction of all possible conformations that can exist in solution. Nevertheless, these results contain important information, since the C-terminal region is responsible for the interaction of SSB with a couple of different proteins such as the χ subunit of DNA polymerase III (4,5), exonuclease I (2) and uracil DNA glycosylase (3). The structural flexibility of this region might be a prerequisite for its functional versatility. The fact that the glycine-rich region is disordered and protrudes from the DNA-binding domain suggests that the last 10 amino acids should be easily accessible for different SSB interaction partners, which differ considerably in size. We tested the protein–protein interaction behaviour of TaqSSB using the χ subunit of E.coli DNA polymerase III as a representative interaction partner. The affinity of this interaction (unpublished data) is similar to the affinity between the χ protein and the cognate EcoSSB (5). By mutational analysis, we showed that for both EcoSSB (5) and TaqSSB (unpublished data) the C-terminus is responsible for this interaction.
Docking of TaqSSB C-terminus to the χ subunit of DNA polymerase III
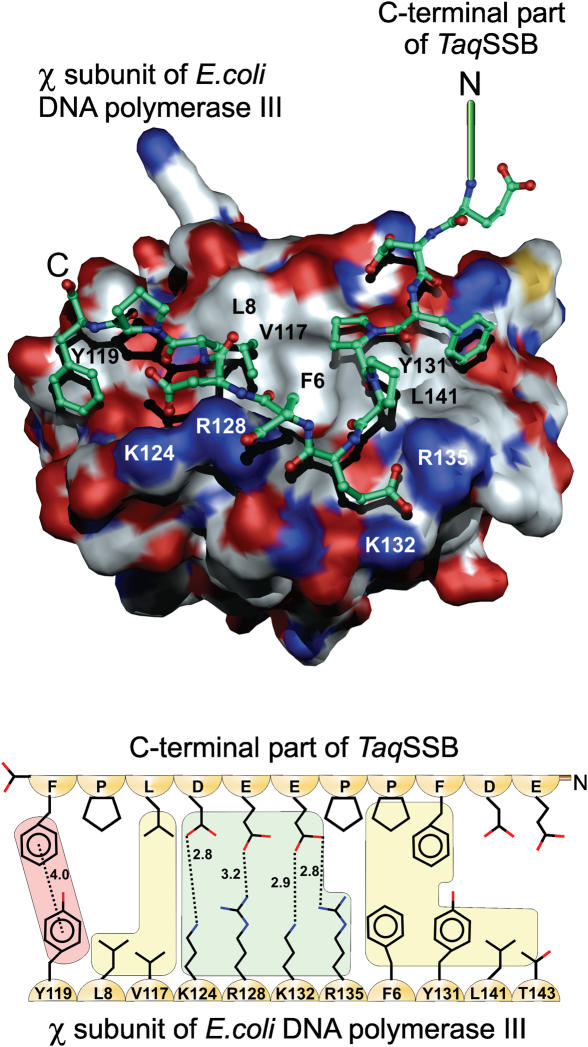
To examine how such an unstructured region could interact with the χ protein, we docked the peptide E254–F264 in the potential χ−SSB binding site proposed by Gulbis et al. (22). The structure of the χ protein was taken from the crystal structure of the χψ subassembly of the E.coli DNA polymerase clamp-loader complex (pdb code: 1em8). The starting peptide conformation was that of the peptide in the TaqSSBΔ228–252 R190A structure. The docking was performed using a combination of flexible peptide docking procedures (40) and molecular dynamics calculations followed by the molecular mechanics energy minimization (27). The result of the docking is presented in Figure 4. The final geometry of the peptide has good stereochemistry and its conformation is unstrained. The highly conserved positively charged residues K124, R128, K132 and R135 of the χ protein interact with the negatively charged and highly conserved residues E259, E260 and D261 from TaqSSB. These interactions lead to the formation of four hydrogen bonds. The side chain of TaqSSB E259 is accommodated in the groove on the surface between K132 and R135 of χ. Residues F256, P257 and L262 of TaqSSB make hydrophobic interactions with residues L141, T143, V117 and L8 of χ. The terminal F264 of TaqSSB makes a stacking interaction with Y119 of χ. This stacking interaction is facilitated by the kink in the amino acid chain, which is caused by the presence of the preceding proline residue. These ultimate two amino acid residues are extremely well conserved in bacterial SSB proteins. In the case of EcoSSB, the mutant strain E.coli ssb113 has been described, in which the penultimate proline residue is replaced by a serine. The importance of this residue is emphasized by the fact that the mutation is conditionally lethal. Cells producing the mutant protein are rendered hyper-sensitive to UV radiation and high temperatures (41,42). In our model the penultimate proline residue is not directly involved in the protein–protein interaction, but is essential for bringing the C-terminal phenylalanine in the correct conformation to form a stacking interaction with Y119 of the χ subunit of E.coli DNA polymerase III.
Figure 4.
Results of docking the TaqSSB C-terminus to the χ subunit of E.coli DNA polymerase III (pdb code: 1em8). In the upper part of the figure the colour scheme on the surface of χ corresponds to the atom charge: positive (blue), negative (red) and neutral (white). The lower part shows a schematic representation of the interactions between the TaqSSB C-terminus and χ. The coloured background highlights different interactions: stacking (magenta), hydrogen bonds (light green) and hydrophobic (light yellow). The distances between the interacting residues are given in Å.
Biological assembly
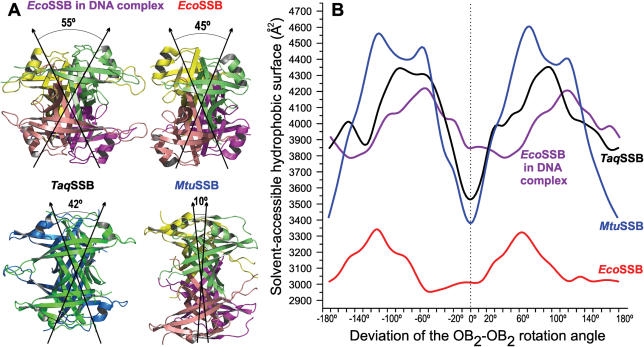
The structure of bacterial SSB proteins is an arrangement of four OB-folds. In the TaqSSB monomer two of these folds are covalently linked and two monomers form an OB2–OB2 dimer with 2-fold symmetry. In homotetrameric SSB proteins, like EcoSSB and MtuSSB (SSB from M.tuberculosis), two OB-folds form a dimer and two of these dimers are arranged in a 2-fold symmetry to form the tetramer with a structure similar to that of the TaqSSB dimer. In TaqSSB the extensive contact area covering 1397 Å2 surface of each monomer has a flat topology and comprises predominantly hydrophobic and some polar groups. Comparison of the mutual arrangement of TaqSSB monomers with arrangements of the corresponding homodimers of EcoSSB and MtuSSB reveals that in TaqSSB the subunits are rotated with respect to each other more than in MtuSSB but less than in EcoSSB (Figure 5A). Such a rotational variability in the subunit arrangement of SSBs from different species implies that the precise orientation of the two subunits may not be crucial for the protein function. This is supported by modelling of the TaqSSB DNA complex (see below), which shows that the difference in orientation of TaqSSB OB2 subunits compared to EcoSSB does not produce any hindrance for DNA binding. Moreover, even larger changes of the OB2–OB2 rotation angle do not create sterical problems for DNA binding. Nevertheless, revealing the factors defining the OB2–OB2 rotation angle will help understanding the diversity in quaternary structures of SSB proteins from different organisms.
Figure 5.
(A) OB2–OB2 orientations in EcoSSB in complex with DNA (pdb code: 1eyg), EcoSSB (pdb code: 1kaw), TaqSSB and MtuSSB (pdb code: 1ue6). (B) Solvent-accessible hydrophobic surface as a function of OB2–OB2 rotation angle calculated with CCP4i program package (32). In each case 0° corresponds to the native orientation.
Analysis of the OB2–OB2 interface of TaqSSB, EcoSSB and MtuSSB shows that the total buried hydrophobic areas are 1664 Å2, 735 Å2 and 2259 Å2, respectively. Using a rough estimate of the free energy of the hydrophobic effect, which is about +0.105 kJ/mol for each Å2 of accessible hydrophobic surface transferred from non-polar solvent to water (43), the gain in free energy resulting from the formation of SSB OB4 assemblies would be 174 kJ/mol for TaqSSB, 77 kJ/mol for EcoSSB and 237 kJ/mol for MtuSSB. In the interface, the hydrophobic residues are oriented in such a way that the hydrophobic patches of interacting molecules form an extensive intermolecular hydrophobic core. This core together with intramolecular cores of the interacting molecules creates the joint, extended hydrophobic core of the biologically active OB4 unit, which is packed so tightly that it appears like a single molecule. This interaction can be additionally stabilized by electrostatic interactions or hydrogen bonds (like hydrogen bonds A2 (main chain)–D105, Q7–Q7, Q130–Q130 in TaqSSB or salt bridge K7–E80 in EcoSSB). Formation of joint, extended hydrophobic cores was previously detected in structures of ribosomal proteins and their complexes (38,44–46) and it was found that such a type of interaction is common for some proteins within the ribosome. To check whether the efficiency of the joint, extended hydrophobic core formation correlates with the OB2–OB2 rotation angle, we calculated the degree of hydrophobic interaction as a function of that angle. The ‘native’ OB2–OB2 angles in structures from TaqSSB, EcoSSB and MtuSSB were considered to be zero. Then for each protein one of the OB2 subunits was rotated with respect to the other around the pseudo-2-fold axis. The rotation range was varied from 0° to 360° in 2° steps and the solvent-accessible hydrophobic surface was calculated for each step. The resulting angular dependencies are presented in Figure 5B. For both TaqSSB and MtuSSB, the native OB2–OB2 orientations correspond to global minima of the accessible hydrophobic surface. The minima have sharp parabolic profiles and a deviation of 10° leads to a noticeable increase in accessible hydrophobic surface. In case of EcoSSB the curve profile in the vicinity of the native orientation is not parabolic but rather box shaped with a broad minimum. If we assume that the efficiency of the joint, extended hydrophobic core formation would determine the native OB2–OB2 orientation then the curve profile would mean that EcoSSB possesses a degree of rotational flexibility around the pseudo-2-fold axis to allow at least 10° rotations. Such rotational flexibility is observed in the crystal structures of an EcoSSB–ssDNA complex and of the free protein, where the OB2–OB2 angles differ by 10° (16,47) (Figure 5A). Rotational flexibility in the case of EcoSSB is also supported by the above free energy considerations: estimates of the free energy of the OB4 unit formation show 2–3 times less gain for EcoSSB compared to TaqSSB or MtuSSB. The same solvent-accessible hydrophobic surface calculations for an EcoSSB–ssDNA complex (with DNA omitted) show that the native orientation is located in a different region of the broadened minimum. The changes in the broadened minimum and in the absolute values of the solvent-accessible hydrophobic surface are due to reorientation of the hydrophobic surface residues interacting with DNA bases. Fine tuning of the orientation of the EcoSSB OB2 units within the broadened minimum of orientation angles is then accomplished by additional amino acid interactions and by interactions with ssDNA. The corresponding profiles for TaqSSB and MtuSSB suggest that both these proteins do not change their quaternary structure upon DNA binding.
Modelling the TaqSSB–DNA complex
In fluorescence titrations we found that at high salt conditions the binding of ssDNA to TaqSSB and EcoSSB is very similar (data not shown). Thus, a TaqSSB–DNA complex was modelled by docking the ssDNA structure from an EcoSSB–ssDNA complex (47) (pdb code: 1eyg) to TaqSSB followed by energy minimization of the resulting structure. There are 48 amino acid residues interacting with ssDNA in this model, 13 of them are conserved between TaqSSB and EcoSSB. In a detailed analysis of the amino acids involved in binding ssDNA in this model we did not observe differences in the interaction of the N- and C-terminal OB-fold that could lead to a weaker binding of the N-terminal fold, as found for the related DraSSB (14). The model indicates that ssDNA binds to TaqSSB in a similar way as to EcoSSB. The interactions include hydrogen bonds, Coulomb interactions, hydrophobic contacts and stacking interactions. The modelling shows that with minor adjustments of TaqSSB side chains most stacking interactions described for EcoSSB (47) are preserved. The difference in the mutual orientation of the protein subunits in the biological dimer of TaqSSB and the EcoSSB tetramer does not lead to any hindrance for ssDNA binding. Modelling of the ssDNA onto the TaqSSB structure was not possible with the glycine-rich region being in conformation C1, whereas conformation C2 is compatible with ssDNA binding.
SUMMARY
We have solved the structures of the single-stranded DNA binding protein from the thermophilic organism T.aquaticus (TaqSSB) and its deletion mutant TaqSSBΔ228–252 R190A. Both structures showed the main part of the protein to consist of two OB-folds (9) covalently linked by a flexible loop. In the flexible glycine-rich region we observe a bifurcation of the Cα trace at P231 existing as both cis- and trans-isomers. Due to the flexibility of this region, the C-terminus of the wild-type protein could not be resolved. The highly conserved last 10 amino acids play an important role in interactions of SSB proteins with other proteins involved in DNA replication, repair and recombination (1–5,13,21). In the structure of the deletion mutant, in which the glycine-rich region is missing, we could show this highly conserved C-terminal region to be highly disordered and devoid of secondary structure. This might be a prerequisite for the interaction with different protein interfaces presented by different interaction partners.
Docking the structural model obtained for the last 11 amino acids of TaqSSB to the putative SSB binding site of the χ subunit of DNA polymerase III from E.coli (22) showed that the last nine amino acids are able to make a tight interaction with χ. Besides the formation of four hydrogen bonds between highly conserved negatively charged residues of TaqSSB and positively charged amino acids of χ, a stacking interaction between the ultimate phenylalanine residue of TaqSSB and a tyrosine residue of χ can be formed. This interaction is facilitated by the penultimate amino acid of TaqSSB, a proline, which creates a kink in the peptide chain to position the phenylalanine for stacking with the tyrosine of χ. In EcoSSB a mutation of this proline to serine renders the E.coli cells UV- and temperature-sensitive (42) and reduces the affinity of EcoSSB to other proteins (2,4).
All bacterial SSB proteins function as a combination of four OB-folds, either as a molecular tetramer with a single OB-fold per molecule or, in the case of the Thermus/Deinococcus group, as a molecular dimer. Whereas the overall structure of the SSB proteins is similar, independent of their oligomeric composition, there exists a significant difference in the angles formed by the two OB2 units interacting to form the biologically active proteins. Hydrophobic interaction calculations show that the formation of an extended hydrohobic core determines the relative orientation of the OB2 units. For EcoSSB our calculations predicted a rotational flexibility which in turn is reflected by the differences between free and ssDNA bound structures (16,47).
Acknowledgments
We thank Lidia Litz for excellent technical assistance, Dr Joachim Greipel for valuable discussions and Dr Anita Coetzee (Bruker, Delft) for help with the X8PROTEUM high-redundancy data collection and valuable discussions. This work was supported by a HiLF grant of Hannover Medical School to U. Curth and by grant MA1081/5-3 from Deutsche Forschungsgemeinschaft. Funding to pay the Open Access publication charges for this article was provided by Hannover Medical School.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cadman C.J., McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genschel J., Curth U., Urbanke C. Interaction of E.coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biol. Chem. 2000;381:183–192. doi: 10.1515/BC.2000.025. [DOI] [PubMed] [Google Scholar]
- 3.Handa P., Acharya N., Varshney U. Chimeras between single-stranded DNA-binding proteins from Escherichia coli and Mycobacterium tuberculosis reveal that their C-terminal domains interact with Uracil DNA Glycosylases. J. Biol. Chem. 2001;276:16992–16997. doi: 10.1074/jbc.M100393200. [DOI] [PubMed] [Google Scholar]
- 4.Kelman Z., Yuzhakov A., Andjelkovic J., O'Donnell M. Devoted to the lagging strand—the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witte G., Urbanke C., Curth U. DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–4440. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuzhakov A., Kelman Z., O'Donnell M. Trading places on DNA—a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 7.Porter R., Black S., Pannuri S., Carlson A. Use of the Escherichia coli ssb gene to prevent bioreactor takeover by plasmidless cells. BioTechnology. 1990;8:47–51. doi: 10.1038/nbt0190-47. [DOI] [PubMed] [Google Scholar]
- 8.Mushegian A.R., Koonin E.V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murzin A.G. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landwehr M., Curth U., Urbanke C. A dimeric mutant of the homotetrameric single-stranded DNA binding protein from Escherichia coli. Biol. Chem. 2002;383:1325–1333. doi: 10.1515/BC.2002.151. [DOI] [PubMed] [Google Scholar]
- 11.Dabrowski S., Olszewski M., Piatek R., Brillowska-Dabrowska A., Konopa G., Kur J. Identification and characterization of single-stranded-DNA-binding proteins from Thermus thermophilus and Thermus aquaticus—new arrangement of binding domains. Microbiology. 2002;148:3307–3315. doi: 10.1099/00221287-148-10-3307. [DOI] [PubMed] [Google Scholar]
- 12.Eggington J.M., Haruta N., Wood E.A., Cox M.M. The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC Microbiol. 2004;4:2. doi: 10.1186/1471-2180-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witte G., Urbanke C., Curth U. Single-stranded DNA-binding protein of Deinococcus radiodurans: a biophysical characterization. Nucleic Acids Res. 2005;33:1662–1670. doi: 10.1093/nar/gki310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein D.A., Eggington J.M., Killoran M.P., Misic A.M., Cox M.M., Keck J.L. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc. Natl Acad. Sci. USA. 2004;101:8575–8580. doi: 10.1073/pnas.0401331101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto T., Morimoto Y., Shibata N., Kinebuchi T., Shimamoto N., Tsukihara T., Yasuoka N. Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis. J. Biochem. (Tokyo) 2000;127:329–335. doi: 10.1093/oxfordjournals.jbchem.a022611. [DOI] [PubMed] [Google Scholar]
- 16.Raghunathan S., Ricard C.S., Lohman T.M., Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9Å resolution. Proc. Natl Acad. Sci. USA. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webster G., Genschel J., Curth U., Urbanke C., Kang C., Hilgenfeld R. A common core for binding single-stranded DNA: structural comparison of the single-stranded DNA-binding proteins (SSB) from E.coli and human mitochondria [published errata appear in FEBS Lett October 6, 1997; 415(3):351 and October 27, 1997;416(3):387] FEBS Lett. 1997;411:313–316. doi: 10.1016/s0014-5793(97)00747-3. [DOI] [PubMed] [Google Scholar]
- 18.Saikrishnan K., Jeyakanthan J., Venkatesh J., Acharya N., Sekar K., Varshney U., Vijayan M. Structure of Mycobacterium tuberculosis single-stranded DNA-binding protein. Variability in quaternary structure and its implications. J. Mol. Biol. 2003;331:385–393. doi: 10.1016/s0022-2836(03)00729-0. [DOI] [PubMed] [Google Scholar]
- 19.Saikrishnan K., Manjunath G.P., Singh P., Jeyakanthan J., Dauter Z., Sekar K., Muniyappa K., Vijayan M. Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologous SSBs: biological implications of structural plasticity and variability in quaternary association. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2005;61:1140–1148. doi: 10.1107/S0907444905016896. [DOI] [PubMed] [Google Scholar]
- 20.Savvides S.N., Raghunathan S., Futterer K., Kozlov A.G., Lohman T.M., Waksman G. The C-terminal domain of full-length E.coli SSB is disordered even when bound to DNA. Protein Sci. 2004;13:1942–1947. doi: 10.1110/ps.04661904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curth U., Genschel J., Urbanke C., Greipel J. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 1996;24:2706–2711. doi: 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulbis J.M., Kazmirski S.L., Finkelstein J., Kelman Z., O'Donnell M., Kuriyan J. Crystal structure of the chi:psi subassembly of the Escherichia coli DNA polymerase clamp–loader complex. Eur. J. Biochem. 2004;271:439–449. doi: 10.1046/j.1432-1033.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 23.Böhme H.J., Kopperschlägel G., Schulz J., Hofmann E. Affinity chromatography of phosphofructokinase using cibachrome blue F36-A. J. Chromat. 1972;69:209–214. doi: 10.1016/s0021-9673(00)83103-9. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 26.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 27.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Sheldrick G.M., Schneider T.R. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 29.Jones T.A., Zou J.-Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr., Sect. A: Found. Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Lamzin V.S., Perrakis A., Wilson K.S. Int. Tables for Crystallography. Kluwer Academic Publishers: Dordrecht; 2001. pp. 720–722. [Google Scholar]
- 32.Potterton E., Briggs P., Turkenburg M., Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 33.Kleywegt G.J., Jones T.A. xdlMAPMAN and xdlDATAMAN—programmes for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr.,Sect. D: Biol. Crystallogr. 1996;52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 34.Kleywegt G.J., Jones T.A. Software for handling macromolecular envelopes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1999;55:941–944. doi: 10.1107/s0907444999001031. [DOI] [PubMed] [Google Scholar]
- 35.Williams K.R., Murphy J.B., Chase J.W. Characterization of the structural and functional defect in the E.coli single-stranded DNA binding protein encoded by the ssb1 gene. J. Biol. Chem. 1984;259:11804–11811. [PubMed] [Google Scholar]
- 36.Curth U., Bayer I., Greipel J., Mayer F., Urbanke C., Maass G. Amino acid 55 plays a central role in tetramerization and function of Escherichia coli single-stranded DNA binding protein. Eur. J. Biochem. 1991;196:87–93. doi: 10.1111/j.1432-1033.1991.tb15789.x. [DOI] [PubMed] [Google Scholar]
- 37.Georlette D., Damien B., Blaise V., Depiereux E., Uversky V.N., Gerday C., Feller G. Structural and functional adaptations to extreme temperatures in psychrophilic, Mesophilic, and Thermophilic DNA Ligases. J. Biol. Chem. 2003;278:37015–37023. doi: 10.1074/jbc.M305142200. [DOI] [PubMed] [Google Scholar]
- 38.Nevskaya N., Tishchenko S., Fedorov R., Al-Karadaghi S., Liljas A., Kraft A., Piendl W., Garber M., Nikonov S. Archaeal ribosomal protein L1: the structure provides new insights into RNA binding of the L1 protein family. Struct. Fold. Des. 2000;8:363–371. doi: 10.1016/s0969-2126(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 39.Didonato M., Sri Krishna S., Schwarzenbacher R., McMullan D., Jaroszewski L., Miller M.D., Abdubek P., Agarwalla S., Ambing E., Axelrod H., et al. Crystal structure of a single-stranded DNA-binding protein (TM0604) from Thermotoga maritima at 2.60 Å resolution. Proteins. 2006;63:256–260. doi: 10.1002/prot.20841. [DOI] [PubMed] [Google Scholar]
- 40.Jones G., Willett P., Glen R.C., Leach A.R., Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 41.Chase J.W., L'Italien J.J., Murphy J.B., Spicer E.K., Williams K.R. Characterization of the E.coli ssb113 mutant single-stranded DNA binding protein. Cloning of the gene, DNA and protein sequence analysis, HPLC, peptide mapping and DNA binding studies. J. Biol. Chem. 1984;259:805–814. [PubMed] [Google Scholar]
- 42.Vales L.D., Chase J.W., Murphy J.B. Effect of ssbA1 and lexC113 mutations on lambda prophage induction, bacteriophage growth, and cell survival. J Bacteriol. 1980;143:887–896. doi: 10.1128/jb.143.2.887-896.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulz G.E., S.R.H. Principles of protein structure. New York, Heidelberg, Berlin: Springer-Verlag; 1979. [Google Scholar]
- 44.Fedorov R., Nevskaya N., Khairullina A., Tishchenko S., Mikhailov A., Garber M., Nikonov S. Structure of ribosomal protein L30 from Thermus thermophilus at 1.9 Å resolution: conformational flexibility of the molecule. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1999;55:1827–1833. doi: 10.1107/s0907444999010227. [DOI] [PubMed] [Google Scholar]
- 45.Leijonmarck M., Liljas A. Structure of the C-terminal domain of the ribosomal protein L7/L12 from Escherichia coli at 1.7 Å. J. Mol. Biol. 1987;195:555–579. doi: 10.1016/0022-2836(87)90183-5. [DOI] [PubMed] [Google Scholar]
- 46.Liljas A. Structural studies of ribosomes. Progr. Biophys. Mol. Biol. 1982;40:161–228. doi: 10.1016/0079-6107(82)90013-x. [DOI] [PubMed] [Google Scholar]
- 47.Raghunathan S., Kozlov A.G., Lohman T.M., Waksman G. Structure of the DNA binding domain of E.coli SSB bound to ssDNA. Nature Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]