Abstract
The PEDANT genome database provides exhaustive annotation of 468 genomes by a broad set of bioinformatics algorithms. We describe recent developments of the PEDANT Web server. The all-new Graphical User Interface (GUI) implemented in Java™ allows for more efficient navigation of the genome data, extended search capabilities, user customization and export facilities. The DNA and Protein viewers have been made highly dynamic and customizable. We also provide Web Services to access the entire body of PEDANT data programmatically. Finally, we report on the application of association rule mining for automatic detection of potential annotation errors. PEDANT is freely accessible to academic users at http://pedant.gsf.de.
INTRODUCTION
The PEDANT genome database was first announced in a short note entitled ‘PEDANTic genome analysis’ which appeared in Trends in Genetics in 1997 (1) and reported computational analysis of seven completely sequenced and two partial genomes available at that time. From the very beginning the main mission of PEDANT was defined as filling the gap between manually curated high quality protein sequence databases, such as UniProt/Swiss-Prot (2), and the enormous amounts of other protein sequences produced by genome sequencing projects at an ever increasing pace. Over the past decade the PEDANT genome database was produced by systematically applying an automatic annotation pipeline to genome data released in the public domain. These efforts resulted in one of the most comprehensive currently available genome databases which includes 468 organisms from all three domains of life. More than 1.76 million proteins sequences have been annotated.
The PEDANT software suite has been described in detail elsewhere (3). Here we report on three new features of the PEDANT Web server: (i) an all-new graphical user interface (GUI), (ii) availability of Web Services and (iii) rule-based detection of annotation errors.
New Graphical User Interface (GUI)
The current version of the PEDANT software familiar to most users was introduced several years ago and is known as version 2 (3). In 2006 we were deploying and testing the all-new version 3 which represents a complete re-implementation of the PEDANT software in the Java™ programing language. PEDANT3 is a platform-independent client–server application for enterprise-scale molecular sequence analysis with advanced features such as highly dynamic workflow-based process management, direct interface to computing grids, support for essentially all existing SQL database management systems, open architecture for easy integration of additional algorithms, and powerful scripting interfaces (D. Frishman et al., manuscript in preparation).
Migration of PEDANT genomes to the new version 3 is currently in progress and will be accomplished by mid-2007. So far we have completed a pilot study to annotate ∼40 genomes to demonstrate the advantages of PEDANT3. The new version of the PEDANT GUI can be viewed by selecting any genome with a ‘new’ icon next to it on the PEDANT home page.
The PEDANT GUI has been made user-configurable and supports four main types of windows: (i) sequence analysis: information about the complete analysis with links to ORF lists of the individual algorithms, (ii) gene report: information about a selected gene, (iii) contig report: information about a selected contig and (iv) genetic element report (where available): information about a selected nonprotein coding genetic element. On selection of a PEDANT3 genome, the analysis window is opened displaying a list of genes and their best-blast hits. The left-hand panel contains a context-dependent navigation tree that makes available different choices dependent on the particular view the user is working with.
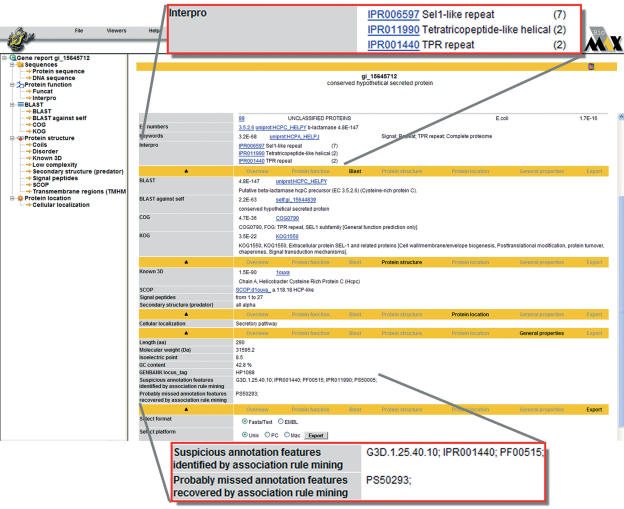
A report page can be displayed for each gene by selecting its code. The report page (Figure 1) is split into different sections (Overview, Protein function, Protein structure, Protein location, General properties and Export) and lists the analysis results for a particular gene product, which are also available for download in different formats. The DNA and Protein viewers have been completely re-designed. They can be extensively customized and are now based on dynamic panel architecture. The protein viewer displays several panels for various types of evidence that can be configured by selecting ‘preferences’ in the top panel. For example certain panels can be hidden, resized and the color scheme can be changed. The DNA viewer allows for easy navigation along the chromosomes and is capable of displaying an unlimited number of features at different zoom levels.
Figure 1.
Navigation tree and report page for the hypothetical secreted protein of Helicobacter pylori (gi_15645712). This figure also illustrates the application of association rule mining for finding potential annotation errors (see text). A number of domain assignments have been marked as suspicious. The InterPro domain IPR001440 and PFAM domain PF00515 were erroneously assigned to this gene product based on a weak similarity hit to a single TPR_1 repeat whereas the domain definition requires at least three repeat copies. Note that all other features marked as suspicious are in fact annotated correctly; the method does not detect annotation errors as such, but rather incompatible feature combinations which might include annotation errors.
Any PEDANT3 dataset can be searched using sequence IDs, sequences, PROSITE-like patterns or free text as the query option.
PEDANT Web Services
Web Services technology is becoming increasingly popular within the bioinformatics community as a means to exploit the large amounts of data, software programs and computing power available at various institutions (4,5). According to the World Wide Web Consortium (W3C) a Web Service is a software system designed to support interoperable machine-to-machine interactions over a network (http://www.w3.org/2002/ws/). This technology is based on the eXtensible Markup Language (XML) and open standards, and is platform and programing language independent. This enables clients for a particular service to be written in many languages, such as Java or Perl, irrespective of the language the service was written in.
A Web Service has an interface that is described in a machine processable format using the XML based Web Services Description Language (WSDL). WSDL provides a format for the description of a Web Service interface, including parameters and data types in sufficient detail for a programer to write a client application for that service. Tools are available for various programing languages to generate the required client classes, such as Apache Axis's WSDL2Java (http://ws.apache.org/axis/java/user-guide.html). The client programs interact with the Web Service using messages based on the Simple Object Access Protocol (SOAP). As with WSDL, SOAP messages are XML based, permitting the interoperability of Web Services. For the transport layer itself, Web Services typically use the Hypertext Transfer Protocol (HTTP), preventing problems sending the SOAP messages through firewalls.
Bioinformatics users can avoid keeping local copies of databases and software and use a client program instead to access remote databases and software via Web Services. The PEDANT Web Service allows the user to query the database in an automated way from client programs and workflows. We provide a number of data retrieval methods in our Data Retrieval Service (Table 1). For example, to fetch the functional and structural annotations of a particular protein, the client program can call the getReportbyDbCodeandContig method. All these methods are described in the WSDL file: http://mips.gsf.de/webservice/pedant2retrieval/services/DataRetrievalService?wsdl.
Table 1.
Methods available in the Data Retrieval Service
| Methods | Description |
|---|---|
| getDatabases | Returns a string array of the pedant2 databases available for data retrieval |
| getProteinSequencesbyDb | Takes the database name as a parameter and returns all protein codes and sequences in the db as a 2D string array |
| getReportbyDbCodeandContig | Takes the database name, protein code and contig as parameters and returns the report data as a ReportItem bean |
| areCodesUniqueWithinDb | Takes the database name as a parameter and returns a Boolean |
| getRawHitsbyDbCodeandMethod | Takes the database name, protein code and method as parameters and returns the code and raw output for that method as a 2D string array |
| getRawHitsbyDbCodeContigandMethod | Takes the database name, protein code, contig and method as parameters and returns the code and raw output for that method as a 2D string array |
| getProteinCodesbyDb | Takes the database name as a parameter and returns the protein codes as a string array |
| getProteinCodesandContigsbyDb | Takes the database name as a parameter and returns the protein codes and contigs as a 2D string array |
| getMethodsbyDb | Takes the database name as a parameter and returns the available methods as a string array |
Currently this Web Service only retrieves data from PEDANT2 analyses; the PEDANT3 Web Service is in preparation. The raw (unparsed) output from various bioinformatics methods can be obtained using the methods getRawHitsbyDbCodeandMethod and getRawHitsbyDbCodeContigandMethod. For completely sequenced genomes, a protein can be uniquely identified with just the database name and protein code. For unfinished genomes, where the proteins were predicted using Orpheus (6), it is necessary to provide the database name, protein code and contig name to uniquely identify a protein. A list of the bioinformatics methods available for each genome can be obtained by calling the getMethodsbyDb method.
We have generated a java client program using the Apache Axis software (WSDL2Java) which has an example class demonstrating how to make calls to the DataRetrievalService. The jar file is available at http://pedant.gsf.de/webservices.jsp. We have also included PEDANT Web Service client functionality in our PROMPT workbench as a use case to demonstrate the various advantages of the Web Services. PROMPT is a standalone application which enables a user to compare protein sequence sets, revealing statistically significant differences in their annotation features (7); (http://webclu.bio.wzw.tum.de/prompt).
Improving PEDANT annotation quality by data mining
Genome annotation produced by automatic pipelines such as PEDANT is notoriously prone to various kinds of errors (8). While every effort is being made to select the most reliable, carefully benchmarked bioinformatics tools and to avoid spurious similarity hits by setting conservative similarity thresholds, automatically produced function predictions are still not on a par with the results of manual curation of sequence data in high-quality databases such as UniProt (2). We estimate that only 5% of all known proteins have been manually annotated and, given the progress in superfast sequencing technologies (9), it is becoming increasingly clear that the overwhelming majority of sequence data will not be processed by human experts.
We have recently developed a technology to improve the quality of automatically generated annotation using data mining techniques (10). The entire body of annotation available in the PEDANT database can be considered as a collection of records of variable length, one for each gene product, containing functional and structural attributes, such as predicted functional categories, domain assignments and the like. Using a large collection of PEDANT records as input, we apply an unsupervised learning algorithm called association rule mining (11) to derive protein features that occur frequently together. Specifically, implications in the form ‘A&B ≥ C’ are derived which mean that the majority of proteins possessing features A and B also possess C. Some rules have the strength 1.0 which means that they are always fulfilled, while other rules may be true only in a certain percentage of cases. An exception from a reasonably strong rule will have features A and B, but not C, which may be caused either by over-annotation (features A or B ascribed erroneously), or by under-annotation (feature C missed).
We have shown that ∼70% of exceptions from strong association rules found in PEDANT data point to incompatible or missing annotation and thus may be instrumental in identifying annotation errors. However, since strong association rules detect not annotation errors as such but only incompatible feature combinations, it is not possible to automatically correct errors. Instead, we highlight suspicious features and add the features which were putatively missed in a separate section at the bottom of the report page (Figure 1). In a recent pilot project we applied this approach to analyze the PEDANT annotation of 10 model genomes—Arabidopsis thaliana, Aeropyrum pernix, Bacillus subtilis, Escherichia coli, Helicobacter pylori, Mycobacterium tuberculosis, Parachlamydia, Saccharomyces cerevisiae, Synechocystis and Thermoplasma acidophilum—containing a total of 55 123 protein entries. In the course of 2007 we intend on providing rule-based correction for all PEDANT genomes.
Acknowledgments
The authors are grateful to Richard Gregory for assistance with the Web Services. Irena Artamonova was supported by the EU Network of Excellence ‘Biosapiens’. Funding to pay the Open Access publication charges for this article was provided by the GSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Frishman D., Mewes H.W. PEDANTic genome analysis. Trends Genet. 1997;13:415–416. [Google Scholar]
- 2.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frishman D., Albermann K., Hani J., Heumann K., Metanomski A., Zollner A., Mewes H.W. Functional and structural genomics using PEDANT. Bioinformatics. 2001;17:44–57. doi: 10.1093/bioinformatics/17.1.44. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson M., Schoof H., Ernst R., Haase D. BioMOBY successfully integrates distributed heterogeneous bioinformatics Web Services. The PlaNet exemplar case. Plant Physiol. 2005;138:5–17. doi: 10.1104/pp.104.059170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillai S., Silventoinen V., Kallio K., Senger M., Sobhany S., Tate J., Velankar S., Golovin A., Henrick K., Rice P., et al. SOAP-based services provided by the European Bioinformatics Institute. Nucleic Acids Res. 2005;33:W25–W28. doi: 10.1093/nar/gki491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frishman D., Mironov A., Mewes H.W., Gelfand M. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 1998;26:2941–2947. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt T., Frishman D. PROMPT: a protein mapping and comparison tool. BMC Bioinformatics. 2006;7:331. doi: 10.1186/1471-2105-7-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galperin M.Y., Koonin E.V. Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1998;1:55–67. [PubMed] [Google Scholar]
- 9.Metzker M.L. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 10.Artamonova, Frishman G., Gelfand M.S., Frishman D. Mining sequence annotation databanks for association patterns. Bioinformatics. 2005;21:iii49–iii57. doi: 10.1093/bioinformatics/bti1206. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal R., Srikant R. Bocca Jorge B., Matthias Jarke, Carlo Zaniolo. Fast algorithms for mining association rules. Proceedings of the Twentieth International Conference on Very Large Data Bases; 1994. pp. 487–499. Morgan Kaufmann. [Google Scholar]