Abstract
The AthaMap database generates a map of cis-regulatory elements for the whole Arabidopsis thaliana genome. This database has been extended by new tools to identify common cis-regulatory elements in specific regions of user-provided gene sets. A resulting table displays all cis-regulatory elements annotated in AthaMap including positional information relative to the respective gene. Further tables show overviews with the number of individual transcription factor binding sites (TFBS) present and TFBS common to the whole set of genes. Over represented cis-elements are easily identified. These features were used to detect specific enrichment of drought-responsive elements in cold-induced genes. For identification of co-regulated genes, the output table of the colocalization function was extended to show the closest genes and their relative distances to the colocalizing TFBS. Gene sets determined by this function can be used for a co-regulation analysis in microarray gene expression databases such as Genevestigator or PathoPlant. Additional improvements of AthaMap include display of the gene structure in the sequence window and a significant data increase. AthaMap is freely available at http://www.athamap.de/.
INTRODUCTION
Bioinformatic tools in molecular biology can easily establish hypotheses for a directed design of experimental set-ups. Bioinformatic gene expression analysis is supported by increasing data on spatial and temporal gene expression and transcription factors (TFs). Gene transcription is mainly regulated by the binding of TFs to cis-regulatory sequences. The occurrence of a cis-sequence is the prerequisite for direct DNA binding that promotes or represses transcription of the gene. Eukaryotic regulation of gene expression is complex and involves synchronized binding of TFs to adjacent cis-regulatory sequences (1). A colocalization analysis of TF binding sites (TFBS) is useful to predict such combinatorial effects on gene expression. Furthermore, binding of TFs can coordinately regulate whole sets of genes.
Bioinformatic methods have been established to predict putative binding sites of TFs in DNA sequences. Web-based resources for detecting TF binding sites or cis-regulatory sequences in plant genes not restricted to Arabidopsis thaliana are Place, PlantCare, and TRANSFAC® (2–4). Genome-wide detection of binding sites can be performed online with the regulatory sequence analysis (RSA) tools (5). A similar genomic sequence search in Arabidopsis can be performed using Patmatch at TAIR (6,7). Pattern recognition programs such as MatInspector, Match or Patser utilize alignment matrices which are derived for example from random binding site selection experiments that determine a set of DNA sequences that can be bound by the same factor (8–10).
Using Patser, the AthaMap database was established for A.thaliana. This database generates a genome wide map of putative TF binding sites determined from alignment matrices (11). Web tools have been implemented for the detection of colocalizing cis-regulatory elements in the genome (12). Combinatorial elements based on known TF interactions have been identified. In addition to positional weight matrix-based detection of binding sites, experimentally verified binding sites were annotated as well (13). The last version of AthaMap contained the genomic positions of more than 8 × 106 putative TFBS for 88 TFs from 21 different families. Another resource for cis-regulatory sequences in A.thaliana is AGRIS (14,15). In contrast to AGRIS, AthaMap covers the whole A.thaliana genome and is mainly based on binding site detection by positional weight matrices.
Co-regulation of genes may be directed by similar combinations of cis-regulatory elements. For A.thaliana, several web-based services harbour gene expression data from microarray experiments and allow recovery of co-regulated genes. Such web-based services are for example TAIR, NASCArrays tools, Stanford Microarray Database, Botany Array Resource, GEO, and Genevestigator (6,16–21). For the detection of gene clusters with similar expression patterns, ACT, Botany Array Resource, CSB.DB, and Genevestigator can be used (19,21–24).
To enable discovery and analysis of common cis-regulatory elements annotated in AthaMap, a new Gene Analysis feature has been developed to allow comparative analysis of cis-elements in sets of co-transcribed genes. Similar expression patterns can also be determined by colocalizing TFBS. For this, the colocalization function has been improved for identification of gene sets harbouring similar combinations of TFBS. Furthermore, the data content in AthaMap has increased significantly and the gene structure is shown in the sequence display window.
INCREASED FUNCTIONALITY OF ATHAMAP
The Gene Analysis web tool
To identify and analyze co-regulated genes for common TFBS, the Gene Analysis web tool has been implemented. On the Gene Analysis page at AthaMap, a gene list can be entered by providing the locus identifier (AGI) of each gene separated by carriage returns. In addition to the gene list, the region of the genes to be analyzed needs to be specified as well. Therefore, the upstream and downstream borders of the analyzed regions relative to the annotated translation start point have to be entered. Because all matrix-based TF binding sites have a specific score between the threshold and maximum score defined by Patser, a restriction to higher conserved TFBS can be applied as well (12).
As an example for co-regulation, the Demo button displays three genes in the input area. By default, the genomic region for analysis ranges from −500 bp upstream to 50 bp downstream relative to the translation start point. No restriction to higher conserved TFBS is set. A search result table lists the TF binding sites in the analyzed genomic region in detail. It displays the gene, the name and the family of the transcription factor and the chromosomal position of its TFBS. In addition, the distance of the binding site relative to the translation start point and the orientation of the binding site relative to the gene are specified. A plus means that the TF binding site and the gene are in the same orientation. Furthermore, for matrix-based TFBS also the maximum score and threshold score of the screening matrix as well as the individual score of the TFBS as a measure for sequence conservation are given. All listed genes and positions of the TFBS are linked to the sequence window for single gene display in the genomic context of surrounding binding sites.
Because a gene may harbour more than one binding site for a specific TF, an overview table can be selected by using the ‘Show overview’ link. This results in a list of all gene-factor combinations having at least one binding site. The list shows the gene, the TF, the TF family, and the number of TFBS detected. The number of sites located upstream and downstream as well as their relative orientation to the start point of translation are given. This and all other tables can easily be copied and exported to a spreadsheet program for additional data processing.
Orchestrated regulation of genes involves binding of specific TFs to sets of genes. By selecting ‘Show factors that are common in genes’, the occurrence of binding sites among the whole set of genes from a Gene Analysis search is displayed. In this table, all TFs with identified TF binding sites in the gene set are shown. The table is sorted hierarchically by the total number of genes per TF. Further information given is the total number of sites detected among the set of genes. To estimate TFBS frequencies, the theoretical number of TF binding sites in the genomic regions analyzed is also shown. This number is based on a theoretical random distribution of the total annotated TF binding sites of the respective TF. The ratio between real occurrence of TFBS and theoretical occurrence shows whether particular TF binding sites are over- or underrepresented. Further valuable information can be extracted by selecting ‘Show all factors’. This extends the table by showing also all TF with binding sites that are absent in the analyzed gene regions.
A similar resource to inspect Arabidopsis promoter sets for cis-regulatory sequences is Athena (25). Important differences between AthaMap and Athena are the fixed promoter region of 3 kb in Athena and the flexible gene region selection in AthaMap. Furthermore, the data content is different. Athena binding sites are based on 105 TF consensus sequences from PLACE and AGRIS (2,14). In contrast, AthaMap is mainly based on alignment matrices of TFBS (11). This leads to a much higher TF binding site density in AthaMap. Athena has ∼30 TFBS in each promoter region of 3 kb (25). In comparison, AthaMap has a TF binding site density of ∼260 TFBS in a 3 kb region including the data update presented here.
Specific enrichment of drought-responsive elements in cold-induced genes
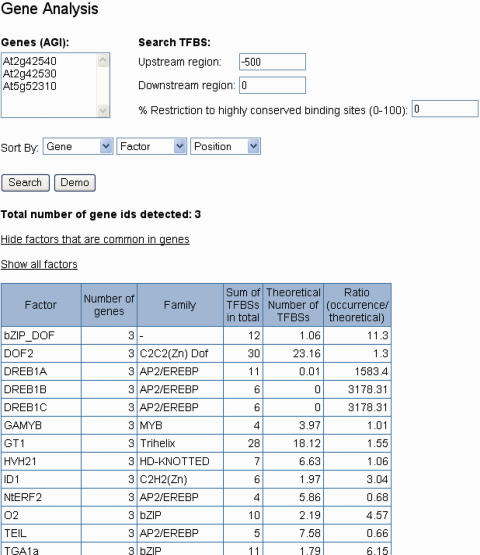
To demonstrate the functionality of the Gene Analysis web tool, three cold-inducible genes (cor15a: At2g42540, cor15b: At2g42530, and rd29A: At5g52310) were used as an example (26). The genomic region analyzed was first restricted to the upstream regions (−500 bp upstream, 0 bp downstream). The output of this Gene Analysis search is displayed in a detailed result list. The distribution of binding sites among the whole set of genes can be analyzed by selecting ‘Show factors that are common in genes’. Figure 1 shows that all three genes harbour DREB1A (CBF3), DREB1B (CBF1) and DREB1C (CBF2) binding sites in the upstream region. A P-value of 4.36 × 10−32 was determined for the occurence of 11 and more DREB1A binding sites within 500 bp of the upstream region of the three selected genes. This value was determined from the total number of 552 DREB1A TFBS identified in the genome (AthaMap documentation page), the total Arabidopsis genome sequence length of 119 186 497 bp, and the total 1500 bp analysed for DREB1A binding sites. For the 6 DREB1B and C TFBS the P-value is 6.21 × 10−20. It has been demonstrated, that these TFs, which are members of the AP2/EREBP family, can activate cold-induced genes by binding to the DRE/CRT cis-acting elements present in their promoter regions (27–29). The three sample genes are regulated by members of the AP2/EREBP transcription factor family, namely CBF/DREBs (26).
Figure 1.
Partial screenshot of a Gene Analysis result page. Only the first lines of the result table showing common binding sites in the gene set are displayed.
In a further analysis of these genes, the genomic region was restricted to 0 bp upstream and 500 bp downstream to determine whether AP2/EREBP binding site overrepresentation is specific to the upstream regions. In this analysis, no DREB1A (CBF3), DREB1B (CBF1) and DREB1C (CBF2) binding sites were identified (data not shown). This indicates specific accumulation of these binding sites in the upstream region of the three genes. This example demonstrates the application of the Gene Analysis function for a set of co-regulated genes.
Extended functionality of the Colocalization Analysis web tool
The AthaMap colocalization web tool permits the positional identification of putative combinatorial elements (12). In the earlier version of AthaMap, only positional information of a predicted combinatorial element on a chromosome was shown. Now, the locus IDs of the closest genes of all colocalizing TF binding sites and the relative distances to the translation start sites are identified. For colocalization analysis, either a TF from the complete list of TFs can be selected by factor name or a factor family can be choosen and one member of this family can be selected for colocalization analysis. A denominator in front of the factor name indicates how the TF binding sites were identified. A bar (–) precedes all TFs that were annotated by matrix-based searches (11). A double bar (=) is assigned to combinatorial elements (12). TFs with binding sites derived from experimentally verified single sites based on consensus sequences are preceded by an arrow (>) (13).
After a colocalization analysis, the resulting table specifies the chromosomal positions of the colocalizing binding sites and the locus ID of the nearest annotated gene. Furthermore, the distance relative to the start point of translation is given. Upstream positions are preceded by a minus. The locus IDs in this table are linked to the sequence display window showing the genomic context of the gene. Furthermore, an entire result list can easily be submitted to Gene Analysis for determination of further common cis-regulatory elements by using the respective link. Another link directly exports the gene list to the microarray gene expression analysis form in PathoPlant for the identification of co-expression during plant-pathogen interaction (30). Such a list of genes can also be used with other programs for co-expression analysis in microarray databases such as Genevestigator (21).
IMPROVED SEQUENCE DISPLAY AND INCREASED DATA CONTENT
Since the last update of AthaMap, a significant change in data display was implemented. In the earlier version, whole genes were displayed as underlined sequence stretches (11). Now, also gene structure elements, i.e. untranslated regions (UTRs), exons and introns, are identified. The annotation of gene structure is based on XML flatfiles downloaded from the TIGR web site (release 5.0) (31). These flatfiles were parsed using a Perl script and positional information for 5′- and 3′-UTRs, exons and introns were annotated to AthaMap. These regions are displayed in AthaMap with a colour code similar to the one used by TAIR (6). The colour code is explained on the sequence display window in AthaMap. The orientation of each gene is indicated below the sequence display window. Forward means that the gene is encoded on the annotated and displayed DNA strand, reverse means that the gene is encoded on the reverse complement strand. For further information on the respective gene, a short description is provided and direct links to TAIR, TIGR, and MIPS records are given (6,31,32).
The data content of AthaMap was increased with nine new alignment matrices derived from eight TFs, another eight TFs with single site-based binding sites and one combinatorial element. These putative binding sites were determined as reported earlier (11–13). Table 1 lists the number of new TF binding sites detected with each matrix and the reference for the alignment matrix. In the case of SPL3 and SPL8, the sequences for alignment matrix generation were obtained directly from the authors of the respective publication (Table 1). Sequences for new single site-based screenings are shown in Table 2. One new combinatorial element was annotated as well. Binding sites derived from both HOX2a matrices were used for determination of combinatorial HOX2a elements (33). AthaMap now contains 9 872 372 TF binding sites detected with alignment matrices, 94 963 TF binding sites detected with experimentally verified TFBS, and 359 867 combinatorial elements based on known TF interactions. The TFs annotated in AthaMap cover most plant TF families (34). Table 3 summarizes the TF families and the number of different TFs represented in AthaMap.
Table 1.
Matrix-based AthaMap data increase
| Factor | Family | Species | Number of sites | Reference for alignment matrix |
|---|---|---|---|---|
| AGL1 | MADS | A.thaliana | 108 045 | (35) |
| AGL2 | MADS | A.thaliana | 55 389 | (35) |
| SPL3 | SBP | A.thaliana | 74 680 | (36) |
| SPL8 | SBP | A.thaliana | 146 721 | (36) |
| HOX2a(1) | HD-HOX | Zea mays | 755 289 | (33) |
| HOX2a(2) | HD-HOX | Zea mays | 495 484 | (33) |
| bHLH66 | bHLH | Oryza sativa | 6811 | (37) |
| CBT | CAMTA | Oryza sativa | 887 | (38) |
| LEC2 | ABI3/VP1 | A.thaliana | 139 340 | (39) |
Number of putative binding sites detected with nine new alignment matrices for eight TFs in the A.thaliana genome.
Table 2.
Site-based AthaMap data increase
| Family | Factor | Synonyms | AGI | Screening sequences | Number of sites | Reference |
|---|---|---|---|---|---|---|
| AP2/EREBP | ERF7 | At3g20310 | GAGCCGCCA | 344 | (40) | |
| TINY2 | DREB3 | At5g11590 | GAGCCGCCAC | 659 | (41) | |
| CTACCGACAT | ||||||
| CTGCCGACAT | ||||||
| CTTCCGACAT | ||||||
| CTATCGACAT | ||||||
| CTACCGTCAT | ||||||
| bHLH | MYC2 | JIN1, ZBF1, bHLH6, RAP1 | At1g32640 | TATACGTGTC GACACGTGGC | 662 | (42) |
| CAMTA | SR1 | At2g22300 | AAACGCGGAA | 427 | (43) | |
| AAACGCGTAA | ||||||
| AAACGCGCAA | ||||||
| AACCGCGGAA | ||||||
| AACCGCGCAA | ||||||
| MYB | CCA1 | At2g46830 | GCTAACTCG | 309 | (44) | |
| LHY | At1g01060 | GCTAACTCG | 309 | (44) | ||
| WER | MYB66 | At5g14750 | TCTCCAACTG | 877 | (45) | |
| TACAACCGCA | ||||||
| TCTATGCCTG | ||||||
| TACGCATGCA | ||||||
| ACTAACGGTA | ||||||
| ACTAACAGTA | ||||||
| WRKY(Zn) | WRKY40 | At1g80840 | CTTTGACCAA | 641 | (46) |
A.thaliana TFs and screening sequences are listed with the corresponding core sequences being underlined. The number of sites annotated in AthaMap and the respective references are given.
Table 3.
Transcription factor families represented in AthaMap
| Family | Number of TFs |
|---|---|
| ABI3/VP1 | 3 |
| AP2/EREBP | 16 |
| bHLH | 3 |
| bZIP | 11 |
| C2C2 (Zn) Dof | 1 |
| C2C2 (Zn) GATA | 4 |
| C2H2(Zn) | 2 |
| CAMTA | 2 |
| CBF | 1 |
| E2F/DP | 6 |
| GARP | 1 |
| GARP/ARR-B | 2 |
| GATA | 1 |
| HD-HOX | 1 |
| HD-KNOTTED | 1 |
| HD-PHD | 1 |
| HD-ZIP | 6 |
| MADS | 5 |
| MYB | 14 |
| NAC | 5 |
| SBP | 6 |
| TBP | 1 |
| TCP | 2 |
| Trihelix | 4 |
| WRKY(Zn) | 4 |
| Total | 103 |
In case of the CAT and TATA box binding proteins (CBF, TBP), the alignment matrices were extracted from the PlantProm database (47).
Acknowledgments
The authors would like to thank Peter Huijser for providing binding sequences of SPL3 and SPL8. This work was carried out in the Intergenomics Center Braunschweig (http://www.intergenomics.de) and was supported by the German Federal Ministry for Education and Research (BMBF grant No. 031U110C/031U210C). Funding to pay the Open Access publication charges for this article was provided by BMBF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Remenyi A., Scholer H.R., Wilmanns M. Combinatorial control of gene expression. Nat. Struct. Mol. Biol. 2004;11:812–815. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]
- 2.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouze P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan T., Yoo D., Berardini T.Z., Mueller L.A., Weems D.C., Weng S., Cherry J.M., Rhee S.Y. PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res. 2005;33:W262–W266. doi: 10.1093/nar/gki368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertz G.Z., Stormo G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 10.Kel A.E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steffens N.O., Galuschka C., Schindler M., Bülow L., Hehl R. AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res. 2004;32:D368–D372. doi: 10.1093/nar/gkh017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steffens N.O., Galuschka C., Schindler M., Bülow L., Hehl R. AthaMap web tools for database-assisted identification of combinatorial cis-regulatory elements and the display of highly conserved transcription factor binding sites in Arabidopsis thaliana. Nucleic Acids Res. 2005;33:W397–W402. doi: 10.1093/nar/gki395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bülow L., Steffens N.O., Galuschka C., Schindler M., Hehl R. AthaMap: from in silico data to real transcription factor binding sites. In Silico Biol. 2006;6:0023. [PubMed] [Google Scholar]
- 14.Davuluri R.V., Sun H., Palaniswamy S.K., Matthews N., Molina C., Kurtz M., Grotewold E. AGRIS: arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palaniswamy S.K., James S., Sun H., Lamb R.S., Davuluri R.V., Grotewold E. AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 2006;140:818–829. doi: 10.1104/pp.105.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craigon D.J., James N., Okyere J., Higgins J., Jotham J., May S. NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gollub J., Ball C.A., Sherlock G. The stanford microarray database: a user's guide. Methods Mol. Biol. 2006;338:191–208. doi: 10.1385/1-59745-097-9:191. [DOI] [PubMed] [Google Scholar]
- 18.Ball C.A., Awad I.A., Demeter J., Gollub J., Hebert J.M., Hernandez-Boussard T., Jin H., Matese J.C., Nitzberg M., Wymore F., et al. The Stanford Microarray Database accommodates additional microarray platforms and data formats. Nucleic Acids Res. 2005;33:D580–D582. doi: 10.1093/nar/gki006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. The Botany array resource: e-northerns, expression angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 20.Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manfield I.W., Jen C.H., Pinney J.W., Michalopoulos I., Bradford J.R., Gilmartin P.M., Westhead D.R. Arabidopsis Co-expression Tool (ACT): web server tools for microarray-based gene expression analysis. Nucleic Acids Res. 2006;34:W504–W509. doi: 10.1093/nar/gkl204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jen C.H., Manfield I.W., Michalopoulos I., Pinney J.W., Willats W.G., Gilmartin P.M., Westhead D.R. The Arabidopsis co-expression tool (ACT): a WWW-based tool and database for microarray-based gene expression analysis. Plant J. 2006;46:336–348. doi: 10.1111/j.1365-313X.2006.02681.x. [DOI] [PubMed] [Google Scholar]
- 24.Steinhauser D., Usadel B., Luedemann A., Thimm O., Kopka J. CSB.DB: a comprehensive systems-biology database. Bioinformatics. 2004;20:3647–3651. doi: 10.1093/bioinformatics/bth398. [DOI] [PubMed] [Google Scholar]
- 25.O'Connor T.R., Dyreson C., Wyrick J.J. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21:4411–4413. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
- 26.Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004;38:982–993. doi: 10.1111/j.1365-313X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- 27.Narusaka Y., Nakashima K., Shinwari Z.K., Sakuma Y., Furihata T., Abe H., Narusaka M., Shinozaki K., Yamaguchi-Shinozaki K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–148. doi: 10.1046/j.1365-313x.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 28.Gilmour S.J., Zarka D.G., Stockinger E.J., Salazar M.P., Houghton J.M., Thomashow M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 29.Stockinger E.J., Gilmour S.J., Thomashow M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl Acad. Sci. USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bülow L., Schindler M., Choi C., Hehl R. PathoPlant®: A database on plant-pathogen interactions. In Silico Biol. 2004;4:529–536. [PubMed] [Google Scholar]
- 31.Haas B.J., Wortman J.R., Ronning C.M., Hannick L.I., Smith R.K., Jr, Maiti R., Chan A.P., Yu C., Farzad M., Wu D., et al. Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol. 2005;3:7. doi: 10.1186/1741-7007-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoof H., Ernst R., Nazarov V., Pfeifer L., Mewes H.W., Mayer K.F. MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource for plant genomics. Nucleic Acids Res. 2004;32:D373–D376. doi: 10.1093/nar/gkh068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirch T., Bitter S., Kisters-Woike B., Werr W. The two homeodomains of the ZmHox2a gene from maize originated as an internal gene duplication and have evolved different target site specificities. Nucleic Acids Res. 1998;26:4714–4720. doi: 10.1093/nar/26.20.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riechmann J.L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 35.Huang H., Tudor M., Su T., Zhang Y., Hu Y., Ma H. DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. Plant Cell. 1996;8:81–94. doi: 10.1105/tpc.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birkenbihl R.P., Jach G., Saedler H., Huijser P. Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005;352:585–596. doi: 10.1016/j.jmb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Xue G.P. A CELD-fusion method for rapid determination of the DNA-binding sequence specificity of novel plant DNA-binding proteins. Plant J. 2005;41:638–649. doi: 10.1111/j.1365-313X.2004.02323.x. [DOI] [PubMed] [Google Scholar]
- 38.Choi M.S., Kim M.C., Yoo J.H., Moon B.C., Koo S.C., Park B.O., Lee J.H., Koo Y.D., Han H.J., Lee S.Y., et al. Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.) J. Biol. Chem. 2005;280:40820–40831. doi: 10.1074/jbc.M504616200. [DOI] [PubMed] [Google Scholar]
- 39.Braybrook S.A., Stone S.L., Park S., Bui A.Q., Le B.H., Fischer R.L., Goldberg R.B., Harada J.J. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl Acad. Sci. USA. 2006;103:3468–3473. doi: 10.1073/pnas.0511331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song C.P., Agarwal M., Ohta M., Guo Y., Halfter U., Wang P., Zhu J.K. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell. 2005;17:2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei G., Pan Y., Lei J., Zhu Y.X. Molecular cloning, phylogenetic analysis, expressional profiling and in vitro studies of TINY2 from Arabidopsis thaliana. J. Biochem. Mol. Biol. 2005;38:440–446. doi: 10.5483/bmbrep.2005.38.4.440. [DOI] [PubMed] [Google Scholar]
- 42.Yadav V., Mallappa C., Gangappa S.N., Bhatia S., Chattopadhyay S. A basic helix–loop–helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell. 2005;17:1953–1966. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang T., Poovaiah B.W. A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 2002;277:45049–45058. doi: 10.1074/jbc.M207941200. [DOI] [PubMed] [Google Scholar]
- 44.Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl Acad. Sci. USA. 2005;102:10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koshino-Kimura Y., Wada T., Tachibana T., Tsugeki R., Ishiguro S., Okada K. Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant Cell Physiol. 2005;46:817–826. doi: 10.1093/pcp/pci096. [DOI] [PubMed] [Google Scholar]
- 46.Xu X., Chen C., Fan B., Chen Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. 2006;18:1310–1326. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shahmuradov I.A., Gammerman A.J., Hancock J.M., Bramley P.M., Solovyev V.V. PlantProm: a database of plant promoter sequences. Nucleic Acids Res. 2003;31:114–117. doi: 10.1093/nar/gkg041. [DOI] [PMC free article] [PubMed] [Google Scholar]