Abstract
The NAR online Molecular Biology Database Collection is a public resource that contains links to the databases described in this issue of Nucleic Acids Research, previous NAR database issues, as well as a selection of other molecular biology databases that are freely available on the web and might be useful to the molecular biologist. The 2007 update includes 968 databases, 110 more than the previous one. Many databases that have been described in earlier issues of NAR come with updated summaries, which reflect recent progress and, in some instances, an expanded scope of these databases. The complete database list and summaries are available online on the Nucleic Acids Research web site http://nar.oxfordjournals.org/.
COMMENTARY
The current issue of the Nucleic Acids Research features 174 databases, of which 106 are new and 68 are updates of previously described databases. These new databases, as well as 15 ones described elsewhere, have been added to the NAR online Molecular Biology Database Collection (http://www.oxfordjournals.org/nar/database/a/), bringing the total list to 968. The geography of the database collection kept expanding and now includes the first database created by Bulgarian (and US) scientists (BANMOKI, http://www.ces.clemson.edu/compbio/databases/kinases, at No. 976). On the other hand, 11 databases featured in the previous release of the NAR database collection (1) have been dropped from the list. Some of these (Crow21, PIR-NREF) have been superseded by newer and more advanced databases. Three databases (DbCat, GenetPig and HugeMap) were discontinued owing to the demise of the French INFOBIOGEN centre [some other INFOBIOGEN databases migrated to the new web site http://urgi.versailles.inra.fr/ maintained by the Institut National de la Recherche Agronomique (INRA)]. The BIND project, which never lived up to its full promise, has gone commercial. In any case, these 11 databases comprise only a small fraction of total database list, which again held very nicely and showed surprising resilience.
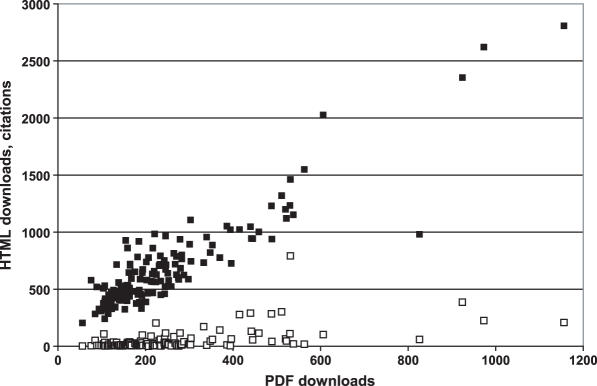
In the comment to the last year's release of the NAR database collection (1), I have discussed the citation rates for various papers in the 2004 NAR database issue and noted that the high-citation rate of certain databases reflects their worldwide acceptance as de facto standards of protein functional annotation [UniProt, http://www.uniprot.org, No. 318, Ref. 2], domain structure [http://www.sanger.ac.uk/Software/Pfam/, No. 210, Ref. 3] and biomedical terminology [Gene Ontology, http://www.geneontology.org/, No. 487, Ref. 4]. However, citation data can be biased; e.g. in many articles use of information from publicly available databases is acknowledged by providing their URLs, or not acknowledged at all. Besides, some databases could be cited on the web sites and in new or obscure journals, not covered by the ISI Citation Index. With this in mind, I have tried here to use additional metrics for assessing the popularity of the NAR database issue. First, I have checked the citations of the database papers listed on the Google Scholar web site, which reflects citations on the web sites. In addition, I have looked at the number of times that the full text of each paper (in PDF or HTML versions) was downloaded from the PubMed Central web site (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PMC). It should be mentioned that all papers in the NAR database issue are freely available for downloading from PubMed Central and NAR web sites; the numbers of downloads from both sites are believed to be somewhat similar. The NAR website http://nar.oxfordjournals.org/ already lists the most frequently downloaded and most cited papers of all time, which include three papers on the Pfam database published in NAR, respectively, in 2000, 2002 and 2004 (5–7), as well as two papers on SwiissProt (8,9) and one on the Protein Data Bank (10), the same databases whose descriptions topped the list of the most cited papers from the 2004 database issue (1). It would seem that these three metrics all reflect usage of the NAR database issue: the user typically starts by finding a database of interest in PubMed or some other bibliographic database, then proceeds to browse the full text in the HTML format. If the paper is interesting enough, s/he would download its text in the PDF format. Finally, if the database turns to be useful, it might be acknowledged with a formal citation. Indeed, the number of HTML downloads and PDF downloads for the same paper correlated very well; the number of PDF downloads was about one-third of the HTML downloads (Figure 1). Curiously, citation rate poorly correlated with the number of downloads. The two most obvious deviations were the 2004 Pfam paper (7) that is extremely well cited but moderately downloaded (791 citations, 1992 total downloads) and my own comment (11) that is much more often downloaded than it is cited (59 citations, 1806 downloads). I am glad to report that, with a single exception, all papers in the 2004 NAR database issue have now been cited at least two times (and downloaded at least 260 times). That single non-cited exception is the description of the ORFDB (http://orf.invitrogen.com/), the Invitrogen's collection of human and mouse ORF clones (12). This paper, which was never intended to be cited, has been nevertheless downloaded 983 times (including 207 times as a PDF) and apparently has served its purpose. Obviously, a list of downloads is an interesting and valuable tool for analyzing various trends in science. For example, of all papers in the 2006 NAR database issue, three of the top five downloads are all descriptions of microRNA databases, miRNAMap, miRBase and the Argonaute (13–15), which obviously reflects the explosive growth of this area. Highlighting such databases has always been and will remain the key goal of the NAR database issues and the NAR online Molecular Biology Database Collection.
Figure 1.
The total number of full-text HTML downloads (closed squares) and literature citations (open squares) as function of the number of the PDF downloads for 142 papers in the 2004 NAR database issue.
Acknowledgments
I thank Rich Roberts, Alex Bateman and my colleagues at NCBI for helpful comments and Jane Lomax and Michael Ashburner for assigning the GO terms to 236 databases in the current list. This study was supported by the Intramural Research Program of the US National Institutes of Health at the National Library of Medicine. Use of the PubMed Central download statistics has been made possible by permission from the Oxford University Press and the kind help of Sergey Krasnov (NCBI). The author's opinions do not necessarily reflect the views of the NCBI, NLM or the National Institutes of Health. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Galperin M.Y. The Molecular Biology Database Collection: 2006 update. Nucleic Acids Res. 2006;34:D3–D5. doi: 10.1093/nar/gkj162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman A., Birney E., Durbin R., Eddy S.R., Howe K.L., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bairoch A., Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res. 1999;27:49–54. doi: 10.1093/nar/27.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galperin M.Y. The Molecular Biology Database Collection: 2004 update. Nucleic Acids Res. 2004;32:D3–D22. doi: 10.1093/nar/gkh143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang F., Matrubutham U., Parvizi B., Yen J., Duan D., Mirchandani J., Hashima S., Nguyen U., Ubil E., Loewenheim J., et al. ORFDB: an information resource linking scientific content to a high-quality Open Reading Frame (ORF) collection. Nucleic Acids Res. 2004;32:D595–D599. doi: 10.1093/nar/gkh118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu P.W., Huang H.D., Hsu S.D., Lin L.Z., Tsou A.P., Tseng C.P., Stadler P.F., Washietl S., Hofacker I.L. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shahi P., Loukianiouk S., Bohne-Lang A., Kenzelmann M., Kuffer S., Maertens S., Eils R., Grone H.J., Gretz N., Brors B. Argonaute—a database for gene regulation by mammalian microRNAs. Nucleic Acids Res. 2006;34:D115–D118. doi: 10.1093/nar/gkj093. [DOI] [PMC free article] [PubMed] [Google Scholar]