Abstract
Pcf11 and Clp1 are subunits of cleavage factor IA (CFIA), an essential polyadenylation factor in Saccahromyces cerevisiae. We have determined the structure of a ternary complex of Clp1 together with ATP and the Clp1-binding region of Pcf11. Clp1 contains three domains, a small N-terminal β sandwich domain, a C-terminal domain containing a novel α/β-fold and a central domain that binds ATP. The arrangement of the nucleotide binding site is similar to that observed in SIMIBI-class ATPase subunits found in other multisubunit macromolecular complexes. However, despite this similarity, nucleotide hydrolysis does not occur. The Pcf11 binding site is also located in the central domain where three highly conserved residues in Pcf11 mediate many of the protein–protein interactions. We propose that this conserved Clp1–Pcf11 interaction is responsible for maintaining a tight coupling between the Clp1 nucleotide binding subunit and the other components of the polyadenylation machinery. Moreover, we suggest that this complex represents a stabilized ATP bound form of Clp1 that requires the participation of other non-CFIA processing factors in order to initiate timely ATP hydrolysis during 3′ end processing.
INTRODUCTION
The 3′ end processing of mRNAs is an essential step in the maturation of eukaryotic transcripts. In this orchestrated process, an initial site-specific cleavage in the 3′-untranslated region (3′-UTR) of the nascent mRNA is followed by the addition and subsequent trimming of a homopolymeric polyadenylate tail. The assembly of processing factors at the poly(A) site stimulates transcriptional termination through interaction with the transcription machinery, whilst addition and trimming of the poly(A) tail is important for regulating mRNA stability, efficient nuclear export and subsequent translation, reviewed (1–3).
The 3′ end processing events in Saccharomyces cerevisiae are co-ordinated by a series of conserved RNA sequence elements located in the 3′-UTR around the site of transcript cleavage (1,4). The first of these is the cleavage/poly(A) site itself, that comprises a pyrimidine base followed by a run of adenines (PyAn) (5). A series of UA repeats located at a variable number of nucleotides 5′ to the cleavage site constitutes the efficiency element (EE) (6) and an A-rich sequence located ∼20 nt 5′ to the cleavage site is termed the positioning element (PE) (7,8). The polyadenylation site is also frequently surrounded by upstream and downstream U-rich regions that affect the efficiency of cleavage and polyadenylation (9). In mammals, the 3′-UTRs of mRNAs contain similar, but not identical conserved recognition elements. The major difference being the presence of a GU- and U-rich elements located 10–30 nt downstream of the polyadenylation site (10,11).
In S.cerevisiae, at least 24 proteins have been identified that are required in combination with the 3′-UTR RNA sequence elements to carry out efficient and high-fidelity transcript cleavage, polyadenylation and trimming. These 24 proteins constitute three multiprotein complexes; cleavage factor IA (CFIA), cleavage and polyadenylation factor (CPF), poly(A) nuclease (PAN) together with poly(A) polymerase (Pap1) and three other associated factors Hrp1, Pab1 and Pbp1. CFIA is required for both transcript cleavage and polyadenylation (12). The complex is comprised of four core subunits Rna14, Rna15, Pcf11 and Clp1 but can be co-purified with the associated factor, Hrp1 (13,14). CPF is also required in both the cleavage and polyadenylation reactions. It contains a core of eight subunits Cft1, Cft2, Ysh1, Pta1 Mpe1, Pfs2, Fip1 and Yth1 (15). However a larger complex has been identified holo-CPF that contains an additional six subunits Ref2, Pti1, Swd2, Glc7, Ssu72 and Syc1 (16). Pap1 is required for addition of the poly(A) tail and PAN, consisting of Pan2 nuclease and Pan3 regulatory proteins, is the 3′–5′ PAN responsible for trimming newly formed poly(A) tails (17,18). PAN activity is also regulated by the Pbp1 and Pab1 associated factors (19).
Although the molecular mechanism of 3′ end processing is poorly understood, several of the protein–RNA interactions involved in the process has been identified. The CPF subunits Cft1, Cft2 and Yth1 bind the U-rich RNA sequences around the poly(A) site (9,20,21), Hrp1 binds to the EE (22,23) and through association with CFIA helps tether Rna15 to the PE (24).
Alongside these protein–RNA interactions, numerous protein–protein interactions between CFIA subunits have been identified together with inter-complex CF1A–CPF interactions indicating a large degree of crosstalk between these complexes. Specifically, Rna15 is tightly associated with Rna14 (13,14,23,25). Rna14 can further associate with Pcf11 and Hrp1 (14) and also binds the Pfs2 and Cft1 subunits of CPF (26,27). Pcf11 binds the CPF components Cft1, Cft2, Ysh1 and Pta1 (27) as well as being associated with all of the other CFIA subunits (14). The binding sites of Rna14/Rna15 and Clp1 have been mapped by two-hybrid analysis to a central region within Pcf11 that also contains two putative zinc-finger motifs (28–30). Located at the N-terminus, Pcf11 contains a CTD interaction domain (CID) that binds to the phosphorylated heptapeptide repeats within the C-terminal domain of the largest subunit of RNA polymerase II and couples 3′ end processing to transcription (30–34).
The Clp1 component of CFIA displays a degree of evolutionary conservation and homologues have been identified in eukaryotes ranging from yeasts to humans (35). This conserved sequence includes a Walker A motif (36), typically found in adenine or guanine nucleotide binding proteins suggesting that Clp1 function may involve nucleotide binding or catalysis. In S.cerevisiae it has been demonstrated that Clp1 binds to Pcf11 but not to the other CFIA subunits (14) and that Clp1 also associates with the Ysh1 subunit of CPF (27) proposed to be the endonuclease responsible for transcript cleavage (37–39). Similarly, in the humans, hClp1 is associated with hPcf11 as part of the CFIIm (mammalian cleavage factor II) complex were it is proposed that Clp1 function may involve bridging CFIIm to the other mammalian 3′ end processing complexes CPSF (CPF), and CFIm (cleavage factor I) (35).
Here, we report the structure of a ternary complex of Clp1-ATP and the Clp1-binding region of Pcf11. Because key residues from Pcf11 that are involved in the protein–protein interface are highly conserved across eukaryotes it follows that this Clp1–Pcf11 interaction is a feature common to all 3′ end processing complexes. The ATP is bound to Clp1 at a site containing a canonical P-loop together with the switch regions commonly associated with SIMIBI-class ATP/GTPases (40). Nevertheless, we have been unable to detect Clp1 mediated nucleotide hydrolysis even though the geometry of the ATP-binding site and orientation of the switch loops share a strong similarity with the active site of ATPase subunits found as part of other multimeric complexes. In light of these data, we present a model where CFIA-CPF association is required to stimulate the ATPase activity of Clp1 in order to transmit conformational effects through 3′ end processing factors bound at the polyadenylation signal sequences.
MATERIALS AND METHODS
Protein expression and purification
The DNA sequence coding for full-length Clp1, residues 1–445, together with residues 454–563 of Pcf11 were isolated by PCR amplification from the S.cerevisiae genome. Clp1 was inserted into the upstream polylinker of pACYCDuet-1 (Novagen) resulting in its fusion to an N-terminal hexa-histidine sequence. Pcf11 (454–563) was then inserted 3′ to Clp1 in the downstream polylinker, without addition of extra tagging sequences. The nucleotide sequence of the expression clone was verified by automated DNA sequencing. The Clp1–Pcf11 (454–563) complex was expressed in the Escherichia coli strain BL21(DE3) and purified from clarified crude cell extracts by immobilized metal-ion-affinity, ion-exchange and size exclusion chromatography. Selenium was incorporated into the proteins by expressing the complex in E.coli B834(DE3) on defined media were seleno-methionine was substituted for methionine. The protein concentration was determined from the absorbance at 280 nm using molar extinction coefficients derived by summing the contributions from tyrosine and tryptophan residues, Clp1–Pcf11 (454–563); 74 700.
Nucleotide assays
In order to assess the nucleotide content of Clp1, samples were precipitated by addition of perchloric acid to 0.6% and the supernatant containing the released nucleotide removed and neutralized by addition of 175 mM sodium acetate (pH 8.0). Extracted nucleotides were applied to a SB-C18 column (ZORBAX) equilibrated in 100 mM K2HPO4/KH2PO4, 8% acetonitrile, 10 mM tetrabutylammonium bromide (TBAB) (pH 6.5) and the column retention time of the nucleotide under these isocratic elution conditions was measured, monitoring the column eluent at 260 nm. The column retention time of individual adenosine and guanosine nucleoside di and triphosphates was also determined under these conditions enabling the protein bound nucleotide to be identified by comparison of its retention time with that of the standards. Under these conditions, retention times of standards and samples were highly reproducible. In order to further confirm the species of bound nucleotide, UV-absorbance spectra were recorded during the elution of the peak and these spectra compared with equivalent spectra of the standards.
Crystallization
Complexes of Clp1 and Pcf11 fragments were dialysed into 150 mM NaCl, 20 mM Tris (pH 7.8), 0.5 mM Tris(2-Carboxyethyl) phosphine hydrochloride (TCEP) and concentrated to ∼150 μM. The protein samples were crystallized by the hanging drop vapour diffusion method, where 1 μl of protein solution was mixed with 1 μl of precipitant solution, then suspended above a reservoir of the same solution. Several Clp1–Pcf11 complexes produced crystals but the best conditions were as follows: Clp1–Pcf11 (454–563) and 12% PEG 8000, 0.1 M HEPES (pH 7.5) at 18°C. Crystals typically appeared in 1–3 days and grew to a maximum size of ∼150 × 150 × 150 μm in 5–7 days. Prior to data collection crystals prepared from native or seleno-methionine substituted protein were transferred to a solution of the mother liquor containing 20% glycerol and flash frozen at 100 K.
Structure determination
The X-ray structure was determined using crystals of the Clp1–Pcf11 (454–563) complex belonging to the spacegroup P212121 and containing two copies of the complex in the asymmetric unit. Data were collected at 100 K from crystals of seleno-methionine substituted protein on station 14.2 at the SRS, Daresbury, UK and processed using HKL software suite (41). Twelve selenium atoms were located using SOLVE (42) from a SAD dataset recorded at the peak of the selenium absorbance edge. Initially, phases derived from the refined positions of the selenium atoms were calculated at 3.8 Å, overall FOM of 0.37. Identification of the NCS operator and application of 2-fold averaging together with density modification in RESOLVE (43) significantly improved the phases resulting in an overall FOM of 0.78. The map was further improved by extending the phases to 2.95 Å in DM (44) using a dataset collected on the same crystal at a high energy remote wavelength.
A model containing residues 18 to 445 of Clp1 and residues 478–498 of Pcf11 was manually built into the improved electron density map using the program ‘O’ (45). Copy B of the Clp1–Pcf11 complex was then generated using the NCS operators. Further improvements were made to the model and the ATP molecules added during subsequent rounds of refinement/automatic water searching procedure ARP/REFMAC (46–48). Initial refinement employed tight NCS restraints that were gradually removed. The final round of refinement was performed without NCS restraints, but employed TLS refinement, implemented in REFMAC. During the course of the refinement the stereo-chemical quality of the protein model was constantly monitored with PROCHECK (49). Details of the data collection phasing and refinement statistics are presented in Table 1.
Table 1.
Statistics of data collection, phasing and refinement
| Se peak (SAD) | Se remote (refinement) | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions a, b, c (Å) | 89.0, 95.0, 181.4 | 89.0, 95.0, 181.4 |
| Solvent content (%) | 56 | 56 |
| Wavelength (Å) | 0.979 | 0.970 |
| Unique reflections | 37 240 | 29 786 |
| Resolution range (Å) | 20–3.5 | 20–2.95 |
| Rsym (%) | 10.1 (26.0) | 11.2 (44.4) |
| I/σ(I) | 18.9 (7.7) | 9.7 (2.1) |
| Completeness (%) | 99.9 (99.5) | 90.2 (60.3) |
| Redundancy | 7.6 (7.0) | 4.6 (2.9) |
| Phasing | ||
| No. sites | 12 | |
| Mean FOM solve (12–3.8 Å) | 0.37 | |
| No copies in AU | 2 | |
| NCS correlation coefficient | 0.94 | |
| Mean FOM (resolve) (12–3.8 Å) | 0.78 | |
| Refinement | ||
| Resolution (Å) | 2.95 | |
| Rwork/Rfree (%) | 24.6/30.4 | |
| No residues/atoms | ||
| Clp1 | 856/6824 | |
| Pcf11 | 47/398 | |
| ATP | 2/60 | |
| Mg2+σ; | 2 | |
| Water | 147 | |
| B-factors (Å2) | ||
| Clp1 (chainA/B) | 57.1/46.4 | |
| Pcf11 (chainC/D) | 45.8/42.8 | |
| ATP | 41.3 | |
| Mg | 20.8 | |
| Water | 33.5 | |
| R.M.S.D | ||
| Bond lengths (Å) | 0.006 | |
| Bond angles (deg) | 1.113 | |
RESULTS
Clp1 binds ATP
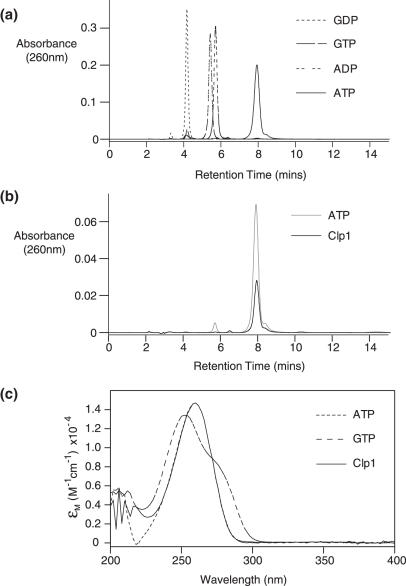
Primary sequence analysis of Clp1 revealed the presence of a Walker A motif, suggesting a capacity to bind to either an adenine- or guanine-based nucleotide. As the specificity of nucleotide binding was unknown, we determined which nucleotides are bound when Clp1 was expressed and purified from E.coli, either in isolation or in complex with Pcf11. A reverse phase high-performance liquid chromatography (HPLC) analysis of the nucleotide content of the Clp1–Pcf11 complex is shown in Figure 1. The data from this experiment and the equivalent experiment with free Clp1 (Supplementary Data) demonstrate that the only nucleotide extracted from Clp1 either free or bound to Pcf11 is ATP. Guanine-based nucleotides bound to Clp1 were not observed, indicating a preference for an adenine base. Moreover, bound ADP was never detected suggesting that the ATP bound to Clp1 is stable to hydrolysis or that hydrolysis to ADP causes the nucleotide to dissociate from Clp1.
Figure 1.
Nucleotide binding assays. (a) C-18 HPLC elution profiles recorded at 260 nm for GDP, GTP, ADP and ATP nucleotide standards. The retention times were 4.2, 5.4, 5.8 and 8.0 min, respectively. (b) The HPLC trace of nucleotide extracted from the Clp1–Pcf11 complex together with an elution profile of an ATP nucleotide control. Both peaks elute at 8.0 min. (c) Ultraviolet (UV) absorbance spectra recorded as each nucleotide GTP, ATP and Clp1–Pcf11 extracted peak eluted. The spectra are expressed in molar extinction in order to normalize for concentration.
Clp1 structure
We attempted to crystallize a variety of co-expressed Clp1–Pcf11 complexes using Pcf11 fragments that surround the proposed Clp1-binding site, residues 456–490 (30) and in some cases containing the putative zinc fingers located just N-terminal and C-terminal to this region. However, the best quality crystals were produced using Clp1 together with residues 454–563 from Pcf11 and without the addition of ATP to the crystallization drop. We have determined the crystal structure of this complex using a single-wavelength anomalous dispersion experiment measured at the selenium edge. The structure has been refined to a resolution of 2.95 Å with good refinement statistics, Table 1.
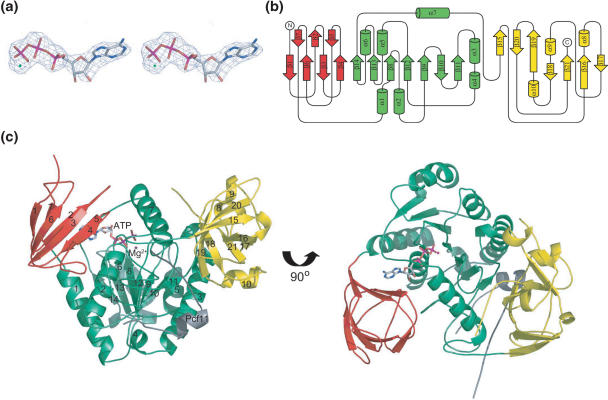
Inspection of the structure reveals Clp1 contains three distinct domains, a large central domain flanked by smaller N- and C-terminal domains (CtD). Electron density can clearly be observed for an ATP molecule bound in a cavity within the central domain (Figure 2a). A diagram describing the topological arrangement of secondary structures within Clp1 is presented in Figure 2b together with a cartoon representation of the complex in Figure 2c. The N-terminal domain (NtD) encompasses residues T18 to T102. There is no visible electron density N-terminal to T18 in the structure and so evidently these first 17 residues are disordered. The domain consists of a seven-stranded β sandwich (β1–β7) interspersed with long loops and the degree of curvature in the strands means the sandwich is quite globular, ∼20 Å in diameter. The interior of the sandwich is packed with bulky aliphatic and aromatic side-chains including W34 and W85 that make up the hydrophobic core of the domain. A search of the PDB database for structural homology using DALI (50) revealed no significant similarity to any other β sandwich domains. Inspection of a sequence alignment of Clp1 from a variety of eukaryotes (Figure 3) shows that within the NtD, the main region of sequence homology between Clp1 orthologues is in the linker between strands β3 and β4, residues 51 to 60. This region of the NtD is packed against the central domain mainly through interactions with α2. As a result of these inter-domain packing interactions, the NtD entirely covers the cavity in the central domain where the ATP is bound rendering the nucleotide binding site inaccessible while the protein is in this conformation (Figure 4a).
Figure 2.
The structure of Clp1. (a) Stereo view of a F0 − Fc map around the bound nucleotide. The calculated amplitudes were derived from the model before inclusion of ATP and prior to refinement. The electron density is contoured at 3σ. (b) Diagram showing the topological arrangement of secondary structure elements in Clp1. The three domains, NtD, Central and CtD are coloured in red green and yellow, respectively and secondary structure elements are labelled sequentially. (c). A cartoon representation of the Clp1–ATP–Pcf11 complex. The colouring of the Clp1 domains is the same as in (B), the ATP is shown in stick representation and Pcf11 is shown in grey.
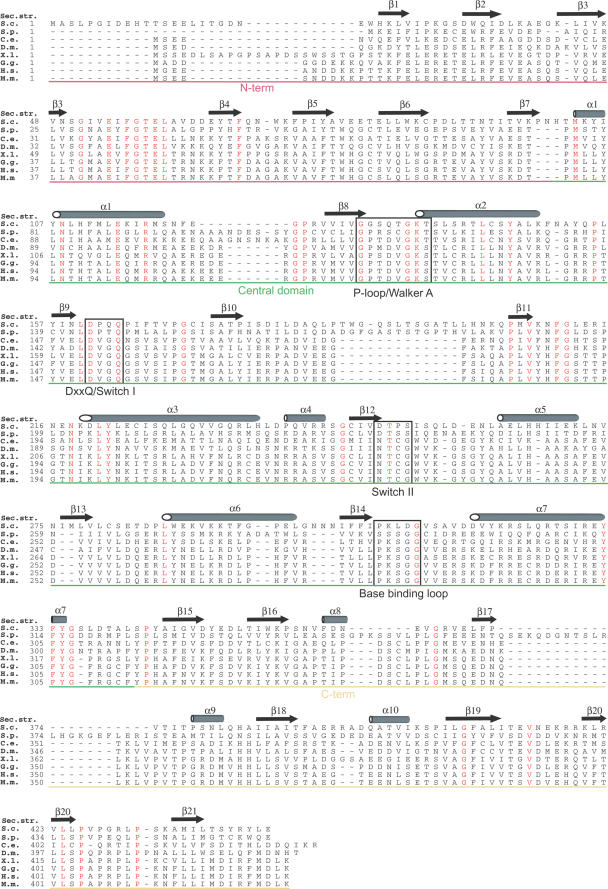
Figure 3.
Clp1 sequence alignment. A sequence alignment of Clp1 proteins from across the eukaryotic kingdom. S. c. S.cerevisiae (swissprot accession no., Q08685), S. p. Schizosaccharomyces pombe (swissprot accession no., Q10299), C. e. Caenorhabditis elegans (swissprot accession no., P52874), D. m. Drosophila melanogaster (swissprot accession no., Q9V6Q1), X. l. Xenopus laevis (swissprot accession no., Q6NS21), G. g. Gallus gallus (swissprot accession no., Q5ZJL4), H. s. Homo sapiens (swissprot accession no., Q92989), M. m. Mus musculus (swissprot accession no., Q99LI9). The position of secondary structure elements observed in the S.cerevisiae Clp1 structure are shown above the alignment, residues that are identical in all species are highlighted in red and sequence motifs involved in ATP-binding and catalysis are boxed. The regions corresponding to each domain, coloured as in Figure 2, are indicated.
Figure 4.
The ATP-binding site. (a) Packing of the Clp1 NtD against the central domain. The central domain (green) is shown in a surface representation with the bound ATP shown in sticks. The NtD is shown as a ribbon representation (red). (b) Nucleotide binding loops. ATP is shown in stick representation with the associated magnesium ion shown as a blue sphere. The central domain of Clp1 is shown as a green ribbon whilst the P-loop, Switch I, Switch II and base-binding loops are shown in stick representation in light red, yellow, blue and grey, respectively. (c) A schematic diagram displaying the protein-ligand contacts observed at the Clp1-ATP-binding site. The ATP is shown in stick representation and Clp1 residues are represented by rectangles. The residues in Clp1 that make contacts with the ATP from the central domain are highlighted in green and those from the NtD are highlighted in red. Hydrogen bonding interactions were the donor and acceptor inter-atomic distance is less than 3.3 Å are indicated with black dashed lines.
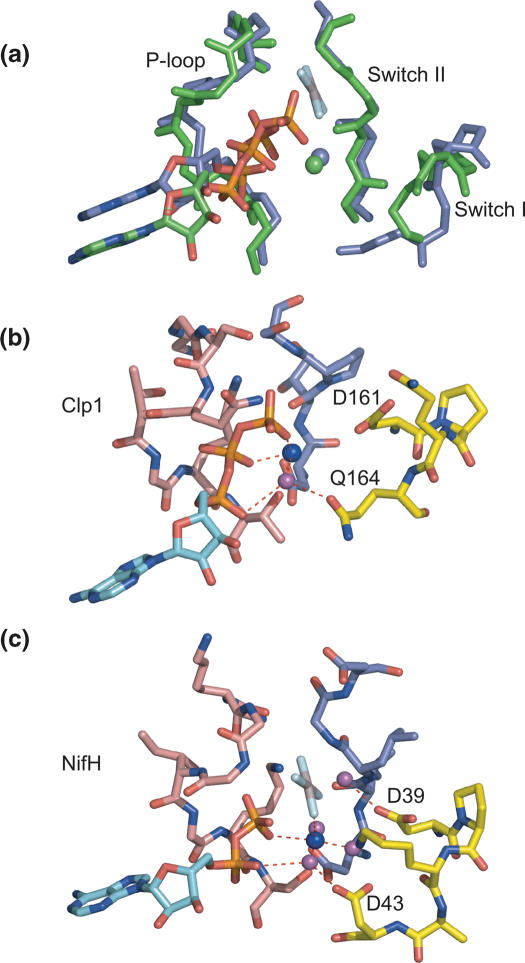
The central domain comprises a central seven-stranded β sheet (β8–β14) surrounded by seven α helices (α1–α7) and is a member of the SIMBI class of NTPases (40). The structure contains four recognizable structural motifs associated with nucleotide binding and hydrolysis. The most prominent is the P-loop or Walker A motif (GxxxxGKT/S) located in the loop between β8 and α2, spanning G130 to T137. A comparison with other structures using DALI reveals that the overall topology of the structure differs significantly from all other SIMBI family members solved to date. The most similar fold that was identified is the signal recognition particle (SRP) GTPase (Z-score 11.0, r.m.s.d. 3.2 for 141 Cα positions and 19% sequence identity, pdb code 1ng1). The SRP GTPase contains a βαβα extension to the central β-sheet termed the IBD (insertion box domain) that is important in the process of nucleotide binding and release (51,52). The Clp1 topology resembles the SRP GTPase topology in that β9, β10, β11, α3 form a similar extension to the central β-sheet. The greatest structural similarities between Clp1 and ATPases are with the NifH Fe subunit of the Azobacter vinelandii nitrogenase (Z-score 8.6, r.m.s.d. 3.0 for 143 Cα positions, 9% sequence identity, pdb code 1 × d8), (53) and the E.coli detoxification pump protein ArsA (Z-score 8.5, r.m.s.d. 3.3 for 148 Cα positions, 11% sequence identity, pdb code 1f48), (54,55).
NifH and ArsA are both ATPase components of macromolecular complexes that utilize ATP hydrolysis to transmit protein conformational changes (56,57). Together with a P-loop, and analogous to G-proteins, these ATPases contain loops referred to as switch I and switch II regions. The switch loops are associated with the nucleotide and residues within them are involved in ATP hydrolysis together with transduction of hydrolysis-dependent conformational changes. Besides the P-loop, Clp1 contains three other regions associated with nucleotide binding (Figure 4b). The first is a DxxQ sequence, residues 161 to 164, conserved among Clp1 family members. This motif is located in the loop between β9 and β10 in the β sheet extension and corresponds to the switch I region. The second is a DTPS sequence, residues 251 to 254, located between β12 and α5 that corresponds to the switch II region. Finally, residues P308 to G312 resemble the mobile base-binding loop seen in ATPases and GTPases. However, in Clp1 although the loop closes around the base enabling van der Waals contacts to be made between the main-chain and the base no base specific hydrogen bonding is mediated by this loop.
The CtD (residues P343 to E445) forms another mixed α/β globular domain, slightly larger than the NtD. The structure is made up from a highly twisted seven-stranded antiparallel beta sheet (β15–β21) interspersed with three short alpha helices. In this case, a search of the PDB for structural homology showed no significant similarity to any other family of structures revealing it to be a novel fold. Inspection of the Clp1 sequence alignment shows that the strongly conserved residues in the CtD are contained in the loop between β20 and β21. Examination of the structure reveals that this conserved loop forms part of a hydrophobic cleft that interacts with Pcf11 consistent with idea that these residues are important for the Pcf1–Clp1 interaction.
The ATP-binding site
The details of the hydrogen bonding interactions made between the bound ATP and residues in the P-loop together with K321 located at the N-terminal end of α7 and D33 and K72 from the NtD are illustrated in Figure 4c. In detail, the γ-phosphate makes hydrogen bonds with the side-chains of Q133 and K136 in the P-loop, as well as the ɛ amino group of K321. The β-phosphate is exclusively hydrogen bonded to residues in the P-loop including both the main-chain and the side-chain of T137. The β-phosphate also makes further contacts with the backbone amides of T134, G135 and K136. The α-phosphate makes fewer contacts but is hydrogen bonded to the main-chain of T137 and to both the side-chain hydroxyl and main-chain amide of S138. The only interaction with the ribose is a hydrogen bonding interaction between the 2′ OH of the sugar and the backbone carbonyl of K72 located in the NtD. The ɛ amino group of K72 although not directly involved in hydrogen bonding interactions with the ATP contacts the main-chain carbonyl of A315 at the C-terminus of the base-binding loop. This interaction helps to stabilize the conformation of the base-binding loop and so facilitates the Van der Waals interactions between the main-chain atoms in the loop, the adenine base and the side-chain of P74 in the NtD. A further contribution to base-binding is made by D33 in the NtD where the side-chain aspartate makes a hydrogen bonding interaction with the exocyclic N6 amino group of the adenine base. This interaction may mediate the specificity of purine base recognition, as the guanine O6 carbonyl oxygen in the equivalent position would be unable to adopt the same hydrogen bonding arrangement.
The P-loop, Switch I and Switch II loops of Clp1 display a degree of structural similarity with equivalent regions of NifH, illustrated by the structural overlap shown in Figure 5a. In NifH, residues in the switch loops do not contact the bound nucleotide directly but rather are involved in the co-ordination of the Mg2+ ion. Similarly, electron density that corresponds to an equivalent Mg2+ is present in the Clp1 structure, and residues in the switch loops are also involved in metal ion co-ordination. A result of this network of interactions is that the switch loops are brought together with the P-loop around the Mg2+ ion (Figure 5b). This network contains two axial ligands that coordinate the Mg2+ ion. These are a γ-phosphate oxygen together with the side-chain γ-oxygen of the P-loop T137. The side-chain of T137 also donates its hydrogen to the carboxylate of D251 in Switch II, effectively tethering together the Switch II and P-loops. This P-loop-Switch II interaction is a common feature conserved amongst many ATPases and GTPases but moreover the nature of the Mg2+ ion co-ordination in Clp1 is very similar to the Mg2+ co-ordination arrangement observed in the NifH–ADP–AlF4 − complex (Figure 5c). In Clp1, only two equatorial ligands, coordinating the Mg2+ ion are apparent, presumably due to the limited resolution. These equatorial ligands are β-phosphate oxygen and a single water molecule. In NifH–ADP–AlF4 − and in other SIMIBI-class ATPases where structures have been determined these same two interactions are conserved. However, in the NifH–ADP–AlF4 − complex two further equatorially positioned water molecules that make up the usual octahedral geometry of the magnesium co-ordination are also present. The three water molecules that are co-ordinated to the Mg2+ ion in NifH are hydrogen bonded by α-phosphate oxygen and by the main-chain and side-chains of residues in the Switch I and Switch II loops. In Clp1, the equatorially co-ordinated water molecule makes equivalent interactions as those observed for one of the NifH bound waters. In this case donating one hydrogen to the side-chain Oɛ1 of Q164 in the Switch I loop and the other to α-phosphate oxygen. Of the other potential Mg2+ ligands and/or hydrogen bond acceptors in the Switch regions the nearest are the carboxylate side-chain of the conserved D161 in Switch I and the main-chain carbonyl of T252 from the Switch II. In both cases the functional groups are located around 4 Å away from the Mg2+ ion so do not make interactions while the molecule is in this conformation. However, the equivalent residues in NifH, the D39 carboxylate and V126 carbonyl, mediate hydrogen bonding interactions with the two remaining equatorially liganded water molecules. Considering the structural similarity, it is likely that these residues have the same function in Clp1. Moreover, in the NifH–ADP–AlF4 − complex the carboxylate of D39 is hydrogen bonded to the in-line attacking water molecule indicating its importance in ATP hydrolysis and suggesting that D161 could perform the same catalytic function in Clp1.
Figure 5.
The switch loops and Mg2+ co-ordination. (a) An overlap of the Clp1-ATP (green) and NifH-AlF4 (blue) structures around the P-loop, Switch I and switch II nucleotide binding regions. The backbone representation for each molecule is shown together with a stick representation for the ATP or ADP-AlF4. (b) Interactions of the magnesium ion with the switch loops of Clp1. Stick representations of the P-loop, Switch I and Switch II are shown in light red, yellow and blue, respectively. The adenine base of the ATP is shown in cyan and the magnesium ion is shown as a blue sphere. The water molecule co-ordinated by the magnesium ion and hydrogen bonded by the side-chain of Q164 together with α phosphate oxygen is shown as a magenta sphere. D161 in Switch I is also indicated. (c) Interactions of the magnesium ion with the switch loops of NifH-ADP-AlF4. The colouring scheme is the same as in (B). D43 and D39 are indicated.
Pcf11 and the Clp1–Pcf11 interface
Although diffraction data were collected from crystals of a complex containing Clp1 and residues 454–563 of Pcf11, inspection of the experimental electron density maps at the protein–protein interface showed density only for residues I477–T498 of Pcf11 in one copy of the asymmetric unit and residues K475–S499 in the other. In both copies, these residues are entirely bound by Clp1 on the opposite surface from the ATP-binding site. Presumably, the remainder of Pcf11 not bound to Clp1 is disordered in this crystal form. The bound peptide backbone adopts a mainly extended conformation with only a short turn of alpha helix between residues D486 to A491 (Figure 6a). The residues N-terminal to the alpha helix, Q478–S485 are bound in a cleft between the central domain and CtD of Clp1. The alpha helical turn causes the remainder; residues S487–T498, to loop back into a shallow groove where they interact entirely with residues from the central domain.
Figure 6.
The Pcf11 binding site. (a) A surface representation of Clp1, The domains are coloured as in Figure 2, Pcf11 is shown as a cartoon representation and the highly conserved residues R480, W482 and W489 that make interactions with Clp1 are shown in the stick representation. (b) A schematic diagram displaying the protein–protein contacts observed at the Clp1–Pcf11 interface. Pcf11 residues are represented with ellipses, Clp1 residues by rectangles. The residues in Pcf11 that make contacts with Clp1 are highlighted in blue, hydrogen bonding interactions are shown as black dashed lines and hydrophobic interactions in green. (c) Details of the conserved arginine 480 interactions. R480 and the residues from the Clp1 central domain that are involved in hydrogen bonding interactions are shown in stick representation. The domains are coloured as in Figure 2. (d) A sequence alignment of the Clp1-binding region from Pcf11. The residues identical to the S.cerevisiae sequence are boxed and the residues that are conserved in all organisms are coloured red.
The extended conformation of Pcf11 both bound in the central domain-CtD cleft and in the shallow groove allows for extensive hydrogen bonding interactions. These are mediated by both the side-chains of Pcf11 residues as well as main-chain amide and carbonyls. A schematic diagram of the protein–protein interactions at the Pcf11–Clp1 interface is presented in Figure 6b. Examination of the central domain-CtD cleft reveals that it is lined on one side by the loop that connects β10 to β11 in the central domain β sheet. This loop constitutes a peptide-binding loop containing residues Q191 to A197, all of which are involved in hydrogen bonding interactions with residues Q478-N481 of Pcf11. In particular, the side-chains of Q478 and R480 make numerous contacts with both main-chain and the side-chains of hydrophilic residues located on the β10–β11 loop. The hydrogen bonding arrangement of R480 is illustrated in Figure 6c. Here the arginine side-chain makes contacts with the main-chain carbonyl of S192 together with the side-chain of Q191 from the β10-β11 loop. The side-chain of R480 is also hydrogen bonded to the main-chain carbonyls of Y332 and E331 that are located on the opposite side of the cleft on the final turn of α7.
Inspection of a sequence alignment of the Clp1-binding region in Pcf11 with other yeast, fish and mammalian homologues (Figure 6d), reveals that R480 is completely conserved among all these organisms. The extent of the hydrogen bonding network surrounding R480 combined with this degree of sequence conservation suggests that R480 is required to make critical contacts in the protein–protein interface, necessary for the stability of the complex. The other completely conserved residues observed in the alignment are W482 and W489. In the case of W482, there are no hydrogen bonding interactions between this residue and Clp1. However, there is a small hydrophobic cavity located in the CtD at the edge of the central domain-CtD cleft and the aromatic side-chain of W482 is swung out of the cleft into this cavity were it is surrounded by the aliphatic side-chains of L406, P426 and V427.
The other conserved tryptophan in Pcf11 is W489, which is located in the short alpha helix of Pcf11. The aromatic side-chain extends from the outside of the helix into a pocket on the surface of the Clp1 central domain. The tryptophan ring makes hydrophobic contacts with both the side-chain of M206 and the aliphatic portion of R236. This interaction is further stabilized by hydrogen bonding interactions between the side-chain Nɛ2 of W489 and the main-chain carbonyl of Q204 and by the main-chain carbonyl of W489 with the NH1 on the side-chain of R236.
DISCUSSION
The Clp1–Pcf11 interaction
Pcf11 interacts directly with all the other CFIA components (14) and links the cleavage-polyadenylation machinery to the transcriptional elongation complex by coupling CFIA to the phosphorylated CtD of RNA polymerase (30–32). Furthermore, Pcf11 also binds to several components of other 3′ end processing complexes (27) so it seems likely that part of Pcf11 function is to coordinate cross talk between CFIA, RNA polymerase and the other polyadenylation factors. The nature of the Pcf11 interaction with the CtD of RNA polymerase is well understood (33,34) and now our data have provided the molecular details of the interaction of Pcf11 with another one of its binding partners, Clp1.
The Clp1-binding domain of Pcf11 consists of a region of around 25 amino acids (residues 475–499). This short stretch of sequence is located between the two putative zinc-finger motifs found in the primary sequence of Pcf11 and overlaps the Clp1-binding domain that has previously been proposed from yeast two-hybrid analyses (30). As the fragment of Pcf11 used to form the complex was much larger, spanning residues 454 to 563 and we observe no interaction with residues outside this region, it seems likely that this peptide forms a large proportion if not the entire Clp1-binding interface of Pcf11.
The protein–protein interface is made up from both polar and apolar interactions and for such a short stretch of sequence the total amount of buried surface at the interface is reasonably extensive (1580 Å2). Moreover, this region of Pcf11 contains three conserved residues R480, W482 and W489 that are all involved in hydrogen bonding and/or hydrophobic interactions at the protein–protein interface. Taken together, these observations show that this Clp1–Pcf11 interaction is not transient and that the Clp1–Pcf11 heterodimer is a stable subcomplex of CFIA. This finding is supported by the fact that the complex readily co-purifies despite the fact that neither component when expressed individually in E.coli is particularly stable (data not shown).
The strong conservation of R480, W482 and W489 among species, combined with the observation that all three of these residues make extensive contributions to the Pcf11–Clp1 interface, suggests that the interactions made by these residues occur in the CFIA of other yeasts as well as in the 3′ end processing complexes of higher eukaryotes. In the mammalian system, there is no equivalent of CFIA. hPcf11 and hClp are the only characterized components of the CFIIm complex and CstF77 and CstF64, the mammalian homologues of Rna14 and Rna15 are combined with a third protein CstF50 forming CstF (cleavage stimulation factor). However, our data now suggest that although the subunit composition of yeast and mammalian 3′ end processing complexes may have diverged, it is likely that many of the interactions at the Clp1–Pcf11 interface have been preserved. Furthermore, since Pcf11 is extensively involved at the interface between the complexes involved in 3′ end processing it seems likely that this highly conserved Clp1–Pcf11 interaction is required to ensure the efficient and timely recruitment of the ATPase activity of Clp1 to the polyadenylation machinery.
Function of bound ATP
The exact nature of ATP-binding and/or hydrolysis in the 3′ end processing reaction remains unclear. It has been demonstrated that in vitro reconstitution of the 3′ end cleavage and polyadenylation reaction is stimulated by ATP (12) and that the cleavage reaction alone can proceed in the presence of other nucleotides, CTP and 3′ dATP (58). Furthermore, it has been shown that ATP is required for CFII to bind to RNA through the Cft2 protein (59). In light of these observations, it seems likely that ATP is required for several steps of the cleavage and polyadenylation reaction, but to date an ATPase enzyme has not been identified within the 3′ end processing machinery. Our data now reveal that Clp1 is an ATP-binding protein that could provide ATPase activity in this system. The structure contains ATP bound at a P-loop and the conformation of the switch regions in the nucleotide binding site together with co-ordination of the magnesium ion is similar to that observed in other SIMIBI-class catalytically competent ATPases, most notably NifH. However, although the Clp1 ATP-binding site appears capable of supporting nucleotide hydrolysis, in an in vitro assay we have been unable to detect any ATPase activity either using Clp1 alone or Clp1–Pcf11 complexes in combination with other components of CFI, with or without the addition of RNA (C. G. Noble and I. A. Taylor, unpublished data). This lack of enzymatic turnover combined with the inaccessibility of the bound ATP molecule poses the question of whether the nucleotide can be hydrolysed in vivo or whether it functions purely as a structural element of the Clp1 protein. What is clear is that turnover of bound ATP would require significant conformational rearrangements in the protein in order for catalysis to occur together with ADP dissociation and ATP re-binding.
In support of the idea that Clp1 does have ATPase activity it is interesting to examine the structurally related NifH protein. In this system, NifH lacks ATPase activity in isolation and in fact nucleotide hydrolysis activity only becomes strongly stimulated through the interaction of NifH with the Mo-Fe subunit of the nitrogenase (56). Similarly, two other related SIMIBI ATPases, MinD and Soj also require the interaction of other components of a macromolecular complex to stimulate ATP hydrolysis; MinE in the MinCDE cell-division system (60) and SpoJ in the Soj bacterial chromosome segregation system (61). Another common factor in all of these complexes is that the energy of ATP hydrolysis is coupled to the transduction of conformational changes through the macromolecular complex. For instance in NifH the conformational changes that result from ATP hydrolysis direct the transfer of electrons from NifH to the Mo-Fe subunit of the nitrogenase.
In light of these observations, it seems plausible that activation of Clp1 ATPase activity might require the interaction with a further 3′ end processing-factor. As we do not observe a stimulation of Clp1 ATPase activity upon the addition of the other CFI subunits, the question arises of which other 3′ end processing-factor might be required. Other than Pcf11, the only other factor demonstrated to bind to Clp1 is the Ysh1 subunit of CPF (27), proposed to be the endonuclease responsible for transcript cleavage (37–39). Since both CFIA and CPF are required for the cleavage reaction we propose that the association of the CFIA and CPF complexes bound at polyadenylation signal sequences is required to trigger nucleotide hydrolysis, perhaps through an Ysh1–Clp1 interaction. This notion that Clp1 might link 3′ end processing complexes together has a precedence in the mammalian system where it has been proposed that Clp1 is involved in bridging CFIIm to the other mammalian 3′ end processing complexes CPSF and CFIm (35).
The function of Clp1 mediated nucleotide hydrolysis in 3′-end processing remains obscure. However, in the other structurally related ATPases that forms part of multisubunit complexes, hydrolysis results in conformational changes that are transmitted between component subunits and actuate a variety of diverse effects. Therefore, the idea that Clp1 mediated ATP hydrolysis might be used to transduce conformational effects through complexes bound at the polyadenylation site is an attractive proposition.
COORDINATES
The coordinates of Clp1–ATP–Pcf11 (454–563) have been deposited in the Protein Data Bank under accession number 2NPI.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
The authors thank Guy Dodson and Steve Smerdon for critical reading of the manuscript and the MRC, UK for support. Funding to pay the Open Access publication charges for this article was provided by MRC, UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell. Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Edmonds M. A history of poly(A) sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 4.Guo Z., Sherman F. 3′ end-forming signals of yeast mRNA. Trends Biochem. Sci. 1996;21:477–481. doi: 10.1016/s0968-0004(96)10057-8. [DOI] [PubMed] [Google Scholar]
- 5.Heidmann S., Obermaier B., Vogel K., Domdey H. Identification of pre-mRNA polyadenylation sites in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:4215–4229. doi: 10.1128/mcb.12.9.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graber J.H., Cantor C.R., Mohr S.C., Smith T.F. Genomic detection of new yeast pre-mRNA 3′ end-processing signals. Nucleic Acids Res. 1999;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russo P., Li W.Z., Hampsey D.M., Zaret K.S., Sherman F. Distinct cis-acting signals enhance 3′ endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 1991;10:563–571. doi: 10.1002/j.1460-2075.1991.tb07983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Z., Sherman F. 3′ end-forming signals of yeast mRNA. Mol. Cell. Biol. 1995;15:5983–5990. doi: 10.1128/mcb.15.11.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dichtl B., Keller W. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 2001;20:3197–3209. doi: 10.1093/emboj/20.12.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil A., Proudfoot N.J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 11.Salisbury J., Hutchison K.W., Graber J.H. A multispecies comparison of the metazoan 3′-processing downstream elements and the CstF-64 RNA recognition motif. BMC Genomics. 2006;7:55. doi: 10.1186/1471-2164-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., Moore C. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell. Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler M.M., Zhao J., Moore C.L. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. Separation into two components that are required for both cleavage and polyadenylation of mRNA 3′ ends. J. Biol. Chem. 1996;271:27167–27175. doi: 10.1074/jbc.271.43.27167. [DOI] [PubMed] [Google Scholar]
- 14.Gross S., Moore C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl Acad. Sci. USA. 2001;98:6080–6085. doi: 10.1073/pnas.101046598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preker P.J., Ohnacker M., Minvielle-Sebastia L., Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nedea E., He X., Kim M., Pootoolal J., Zhong G., Canadien V., Hughes T., Buratowski S., Moore C.L., Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 17.Brown C.E., Tarun S.Z., Jr, Boeck R., Sachs A.B. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeck R., Tarun S., Jr, Rieger M., Deardorff J.A., Muller-Auer S., Sachs A.B. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 19.Mangus D.A., Evans M.C., Agrin N.S., Smith M., Gongidi P., Jacobson A. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol. 2004;24:5521–5533. doi: 10.1128/MCB.24.12.5521-5533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barabino S.M., Ohnacker M., Keller W. Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 2000;19:3778–3787. doi: 10.1093/emboj/19.14.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dichtl B., Blank D., Sadowski M., Hubner W., Weiser S., Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 2002;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S., Hyman L.E. A specific RNA-protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res. 1998;26:4965–4974. doi: 10.1093/nar/26.21.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessler M.M., Henry M.F., Shen E., Zhao J., Gross S., Silver P.A., Moore C.L. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross S., Moore C.L. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 2001;21:8045–8055. doi: 10.1128/MCB.21.23.8045-8055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noble C.G., Walker P.A., Calder L.J., Taylor I.A. Rna14-Rna15 assembly mediates the RNA-binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res. 2004;32:3364–3375. doi: 10.1093/nar/gkh664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohnacker M., Barabino S.M., Preker P.J., Keller W. The WD-repeat protein pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J. 2000;19:37–47. doi: 10.1093/emboj/19.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyburz A., Sadowski M., Dichtl B., Keller W. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Res. 2003;31:3936–3945. doi: 10.1093/nar/gkg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amrani N., Minet M., Wyers F., Dufour M.E., Aggerbeck L.P., Lacroute F. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amrani N., Minet M., Le Gouar M., Lacroute F., Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadowski M., Dichtl B., Hubner W., Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Licatalosi D.D., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 32.Barilla D., Lee B.A., Proudfoot N.J. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meinhart A., Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 34.Noble C.G., Hollingworth D., Martin S.R., Ennis-Adeniran V., Smerdon S.J., Kelly G., Taylor I.A., Ramos A. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nature Struct. Mol. Biol. 2005;12:144–151. doi: 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- 35.de Vries H., Ruegsegger U., Hubner W., Friedlein A., Langen H., Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callebaut I., Moshous D., Mornon J.P., de Villartay J.P. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan K., Calvo O., Manley J.L. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominski Z., Yang X.C., Marzluff W.F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Leipe D.D., Wolf Y.I., Koonin E.V., Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Terwilliger T.C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terwilliger T.C. Automated structure solution, density modification and model building. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- 44.Cowtan K.D., Main P. Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr. D Biol. Crystallogr. 1996;52:43–48. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]
- 45.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr., A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 46.Lamzin V.S., Wilson K.S. Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 47.Morris R.J., Perrakis A., Lamzin V.S. ARP/wARP and automatic interpretation of protein electron density maps. Meth. Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Laskowski R.A., Moss D.S., Thornton J.M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 50.Holm L., Sander C. Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freymann D.M., Keenan R.J., Stroud R.M., Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 52.Freymann D.M., Keenan R.J., Stroud R.M., Walter P. Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP. Nature Struct. Biol. 1999;6:793–801. doi: 10.1038/11572. [DOI] [PubMed] [Google Scholar]
- 53.Schindelin H., Kisker C., Schlessman J.L., Howard J.B., Rees D.C. Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997;387:370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- 54.Zhou T., Radaev S., Rosen B.P., Gatti D.L. Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation. J. Biol. Chem. 2001;276:30414–30422. doi: 10.1074/jbc.M103671200. [DOI] [PubMed] [Google Scholar]
- 55.Zhou T., Radaev S., Rosen B.P., Gatti D.L. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J. 2000;19:4838–4845. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgess B.K., Lowe D.J. Mechanism of molybdenum nitrogenase. Chem. Rev. 1996;96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 57.Gatti D., Mitra B., Rosen B.P. Escherichia coli soft metal ion-translocating ATPases. J. Biol. Chem. 2000;275:34009–34012. doi: 10.1074/jbc.R000012200. [DOI] [PubMed] [Google Scholar]
- 58.Minvielle-Sebastia L., Beyer K., Krecic A.M., Hector R.E., Swanson M.S., Keller W. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J., Kessler M.M., Moore C.L. Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J. Biol. Chem. 1997;272:10831–10838. doi: 10.1074/jbc.272.16.10831. [DOI] [PubMed] [Google Scholar]
- 60.Ma L., King G.F., Rothfield L. Positioning of the MinE binding site on the MinD surface suggests a plausible mechanism for activation of the Escherichia coli MinD ATPase during division site selection. Mol. Microbiol. 2004;54:99–108. doi: 10.1111/j.1365-2958.2004.04265.x. [DOI] [PubMed] [Google Scholar]
- 61.Leonard T.A., Butler P.J., Lowe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer–a conserved biological switch. EMBO J. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]