Abstract
Prediction and elucidation of pharmacogenetic effects is important for facilitating the development of personalized medicines. Knowledge of polymorphism-induced and other types of drug-response variations is needed for facilitating such studies. Although databases of pharmacogenetic knowledge, polymorphism and toxicogenomic information have appeared, some of the relevant data are provided in separate web-pages and in terms of relatively long descriptions quoted from literatures. To facilitate easy and quick assessment of the relevant information, it is helpful to develop databases that provide all of the information related to a pharmacogenetic effect in the same web-page and in brief descriptions. We developed a database, Pharmacogenetic Effect Database (PharmGED), for providing sequence, function, polymorphism, affected drugs and pharmacogenetic effects. PharmGED can be accessed at http://bidd.cz3.nus.edu.sg/phg/ free of charge for academic use. It currently contains 1825 entries covering 108 disease conditions, 266 distinct proteins, 693 polymorphisms, 414 drugs/ligands cited from 856 references.
INTRODUCTION
Individual response to drugs often differs significantly and these drug-response variations are frequently associated with polymorphisms of pharmacologically related proteins (1–5). Pharmacogenetic study of these proteins and their regulatory sites is important for the understanding of molecular mechanism of drug responses and for the development of personalized medicines (1,6–9). Resources that provide information about molecular mechanism of drug-response variations are useful for facilitating pharmacogenomics study and the development of personalized medicine (10). There have been calls and efforts for developing such resources (11) and the related informatics tools (12,13). A number of freely accessible web-based resources have been developed for providing information about genetic and clinical pharmacogenetic information (14), polymorphisms in drug-related proteins (15–20) and toxicogenomics data (21).
Although pharmacogenetic knowledge, polymorphism and toxicogenomic information are provided in these databases, some of the reported pharmacogetic effects are given in web-pages separate from that of other important information such as protein and drug information, and are often given by relatively long descriptions quoted from literatures. To facilitate easy and quick assessment of the relevant information, it is helpful to develop databases that provide all the information related to a pharmacogenetic effect in the same web-page. We developed Pharmacogenetic Effect Database (PharmGED) with the aim to provide the information about the effects of a particular protein polymorphism, non-coding region mutation, splicing alteration or expression variation on the response of a particular drug. It currently contains 1825 entries covering 108 disease conditions, 266 distinct proteins, 693 polymorphisms, 414 drugs/ligands cited from 856 references.
DATABASE STRUCTURE AND ACCESS
PharmGED has a web interface at http://bidd.cz3.nus.edu.sg/phg/. The entries of this database were derived from a comprehensive search of published literatures (via Medline) by using a similar search and evaluation procedure as we have used for developing other databases of drug-related proteins (15–18). Entries of this database are searchable by several methods. These methods include the search of protein name, drug/ligand name, disease name (extracted from the related terms described in the relevant publications) and drug class (derived based on the related terms described in the relevant publications). Full list of protein names, drug/ligand names, disease names and drug classes are separately provided in the PharmGED main web-page for facilitating the search of particular entries.
Moreover, keyword-based text search is also supported. The search is case insensitive and wildcards are supported. In a query, a user can specify full name or any part of the name in a text field. Wild character of ‘*’ and ‘?’ is allowed in text field. Here, ‘?’ represents any single character and ‘*’ represents a string of characters of any length. For example, input of ‘dehydrogenase’ in the field of protein/gene name enables the finding of all entries containing ‘dehydrogenase’ in the protein name, such as 3-oxo-5-alpha-steroid 4-dehydrogenase 2, Alcohol dehydrogenase 1B, Alcohol dehydrogenase 1C, Fatty aldehyde dehydrogenase, Glucose-6-phosphate 1-dehydrogenase, NAD(P)H dehydrogenase (quinone) 1, etc. On the other hand, input of Fatty*dehydrogenase enables the finding of all dehydrogenases whose names start with ‘Fatty’.
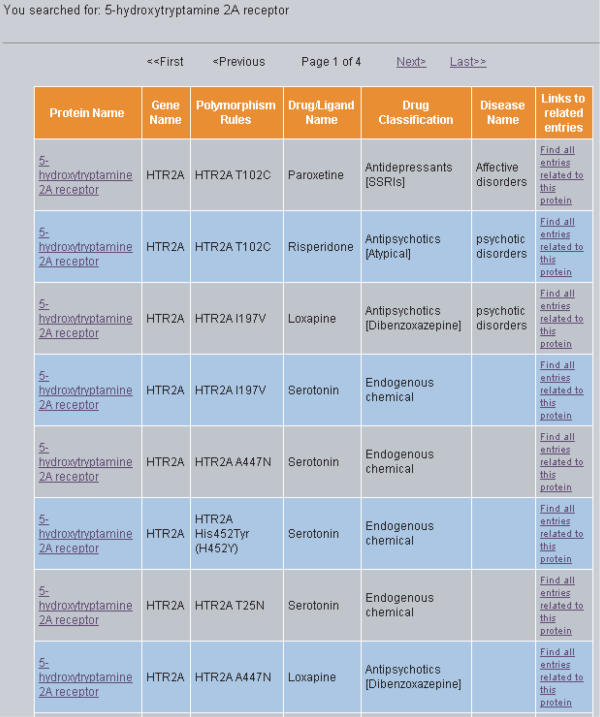
The result of a typical search is illustrated in Figure 1. In this interface, all entries that satisfy the specified search criteria are listed along with protein name, polymorphism rules, drug/ligand name, drug classification, disease name and links to other related entries in this database. More detailed information of an entry can be obtained by clicking the corresponding protein name. The result is displayed in an interface shown in Figure 2. From this interface, one finds the accession number, name, sequence and function of protein, pharmacogenetic polymorphism, affected drugs and drug class, corresponding disease condition and pharmacogenetic effect. Moreover, the information about the related references and links to the literature database PUBMED (22) is also provided.
Figure 1.
The interface displaying a search result on PharmGED. All entries that satisfy the specified search criteria are listed along with protein name, polymorphism rules, drug/ligand name, drug classification, disease name and links to other related entries in this database.
Figure 2.
Interface displaying the detailed information of an entry in PharmGED.
POTENTIAL APPLICATIONS
Established links between polymorphisms of drug-related proteins and individual drug responses have been used in combination with genetic studies as indicators for predicting individual variations of drug response (23–27). Based on the statistical analysis of the data of polymorphisms and variation of drug response of the participating patients, simple rules may be derived in some cases for predicting individual variations of drug response from polymorphism data (23,24,26,28,29). These simple rules may be collected and used for developing a computer prediction system in a fashion similar to that of the HIV drug-resistant genotype interpretation systems (30).
Table 1 gives examples of the drug-related proteins in PharmGED with available information about pharmacogenetic polymorphism and drug-response variation from which a reasonably accurate rule have been derived in the literature for predicting responses to a specific drug or drug group. The reported percentage of patients who have a polymorphism and showed the expected effect is also given. Based on the test of the patients described in these reports, most of these rules are capable of predicting drug responses at success rates of 50–100%, which are not too much lower than and in many cases comparable to the accuracies of 81–97% for predicting HIV drug resistance mutations from the HIV-resistant genotype interpretation systems (30). This suggests that these simple rules have certain level of capacity for facilitating the prediction of pharmacogenetic effects and they may be used as the basis for developing more sophisticated interpretation systems similar to those of the HIV-resistant genotype interpretation systems (30).
Table 1.
Prediction of specific drug responses from the polymorphisms of ADME-associated proteins by using simple rules
| Protein | Drugs and treatment/action | Drug responses | Polymorphism rules and year of report | Number of patients with polymorphism | Reported percentage of patients who have a polymorphism and showed the expected effect |
|---|---|---|---|---|---|
| Cytochrome P450 1A2 | Antipsychotic agents for schizophrenia patients | Tardive dyskinesia | Bsp120I (C→A) polymorphism in CYP1A2 gene, 2000 (38) | 85 | 69 |
| Cytochrome P450 2D6 | Neuroleptic agents for chronic schizophrenic patients | Tardive dyskinesia | CYP2D6*4 genotype, 1998 (39) | 13 | 81 |
| UDP-glucuronysltransferase | Capecitabine/irinotecan for the treatment of metastatic colorectal cancer | Greater antitumor response with low toxicity | UGT1A7*2/*2 genotype, 2005 (27) | 6 | 100 |
| UGT1A7*3/*3 genotype, 2005 (27) | 7 | 100 | |||
| UDP-glucuronysltransferase I | Tranilast for the prevention of restenosis following coronary revascularization | Hyperbilirubinemia | Homozygosity for a (TA)7-repeat element within the promotor region of UGT1A1 gene, 2004 (23) | 146 | 40 |
| N-acetyltransferase 2 | Isoniazid for the prophylaxis and treatment of tuberculosis | ADRs such as peripheral neuritis, fever and hepatic toxicity | SA type (NAT2*6/*6, NAT2*6/*7 and NAT2*7/*7), 2002 (25) | 6 | 83 |
| Tryptophan hydroxylase | Fluvoxamine for the treatment of depression | Antidepressant response | A218C A/C phenotypes, 2001 (40) | 107 | 76 |
| A218C C/C phenotypes, 2001 (40) | 70 | 81 | |||
| A218C A/A phenotypes, 2001 (40) | 40 | 65 | |||
| Norepinephrine transporter | Milnacipran for the treatment of depression | Antidepressant response | T allele of the NET T182C polymorphism, 2004 (26) | 50 | 72 |
| Serotonin transporter | Serotonin reuptake inhibitors for the treatment of depression | Antidepressant response | s/s genotype of serotonin transporter gene promoter region, 2000–2004 (1,2,41,42) | 11–72 | 54% at sixth week |
| s/l genotype of serotonin transporter gene promoter region, 2000–2004 (1,2,41,42) | 2–47 | 55% at sixth week | |||
| l/l genotype of serotonin transporter gene promoter region, 2000–2004 (1,2,41,42) | 4–16 | 48% at sixth week | |||
| Multidrug resistance-associated protein 1 | Epileptic drugs for the treatment of epilepsy | Drug-resistant epilepsy | ABCB1 C3435T C/C genotype, 2003 (43) | 73 | 75 |
| ABCB1 C3435T C/T genotype, 2003 (43) | 169 | 63 | |||
| ABCB1 C3435T T/T genotype, 2003 (43) | 73 | 53 | |||
| Multidrug resistance-associated protein 1 | Combination therapy of nelfinavir, efavirenz and nucleoside reverse transcriptase inhibitors for HIV-1 infected children | Virologic response by week 8 | MDR1 C3435T C/C genotype, 2005 (44) | 31 | 59 |
| MDR1 C3435T C/T genotype, 2005 (44) | 33 | 91 |
PharmGED and other databases (14–21) can be potentially used for facilitating the generation of these rules. For instance, one entry of PharmGED describes that patients using classical neuropleptic such as fluphenazine, haloperidol have a higher incidence (81%) of Tardive dyskinesia if they are of the genotype CYP2D6*4. The corresponding polymorphism can be obtained from a link provided in the database. These data combined with other information in PharmaGED can be used to generate the rule for detecting this pharmacogenetic effect described in Table 1. In a second example, another entry of PharmGED describes that the polymorphism C3435T (Ile1145Ile) of protein MDR1-3435 variant is associated with different virologic response of nelfinavir in HIV-1 infected children. Fifty-nine percent of the 31 C/C genotype and 91% of the 33 C/T genotype show virologic response at eighth week, respectively. These data can then be used to generate the rule for this pharmacogenetic effect as described in Table 1.
CONCLUDING REMARKS
Knowledge about protein polymorphisms and drug responses appears to have reached a meaningful level for facilitating pharmacogenetic study and for predicting various types of individual variations of drug responses. Specialized pharmacogenetics databases serve as convenient resources for obtaining the relevant information. With the rapid development of genomics (31), pharmacokinetics (32–35) and pharmacogenomics (6,8,9), more information about drug-related proteins, polymorphisms and variations of drug responses are expected to become available. Moreover, progress in the study of proteomics (36) and pathways (37) related to drug-related proteins will further facilitate our understanding of the mechanism of individual variations in drug response.
Acknowledgments
This work was supported in part by grants from Singapore ARF R-151-000-031-112. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Evans W.E., Relling M.V. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 2.Lindpaintner K. Pharmacogenetics and the future of medical practice. J. Mol. Med. 2003;81:141–153. doi: 10.1007/s00109-002-0416-5. [DOI] [PubMed] [Google Scholar]
- 3.Marzolini C., Paus E., Buclin T., Kim R.B. Polymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevance. Clin. Pharmacol. Ther. 2004;75:13–33. doi: 10.1016/j.clpt.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Evans W.E., Hon Y.Y., Bomgaars L., Coutre S., Holdsworth M., Janco R., Kalwinsky D., Keller F., Khatib Z., Margolin J., et al. Preponderance of thiopurine S-methyltransferase deficiency and heterozygosity among patients intolerant to mercaptopurine or azathioprine. J. Clin. Oncol. 2001;19:2293–2301. doi: 10.1200/JCO.2001.19.8.2293. [DOI] [PubMed] [Google Scholar]
- 5.Marshall A. Laying the foundations for personalized medicines. Nat. Biotechnol. 1997;15:954–957. doi: 10.1038/nbt1097-954. [DOI] [PubMed] [Google Scholar]
- 6.Altman R.B., Klein T.E. Challenges for biomedical informatics and pharmacogenomics. Annu. Rev. Pharmacol. Toxicol. 2002;42:113–133. doi: 10.1146/annurev.pharmtox.42.082401.140850. [DOI] [PubMed] [Google Scholar]
- 7.Kalow W. Pharmacogenetics, pharmacogenomics, and pharmacobiology. Clin. Pharmacol. Ther. 2001;70:1–4. doi: 10.1067/mcp.2001.116714. [DOI] [PubMed] [Google Scholar]
- 8.Marshall A. Getting the right drug into the right patient. Nat. Biotechnol. 1997;15:1249–1252. doi: 10.1038/nbt1197-1249. [DOI] [PubMed] [Google Scholar]
- 9.Nicholls H. Improving drug response with pharmacogenomics. Drug Discov. Today. 2003;8:281–282. doi: 10.1016/s1359-6446(03)02650-3. [DOI] [PubMed] [Google Scholar]
- 10.Zheng C.J., Sun L.Z., Han L.Y., Ji Z.L., Chen X., Chen Y.Z. Drug ADME-associated protein database as a resource for facilitating pharmacogenomics research. Drug Dev. Res. 2004;62:134–142. [Google Scholar]
- 11.Gurwitz D., Lunshof J.E., Altman R.B. A call for the creation of personalized medicine databases. Nature Rev. Drug Discov. 2006;5:23–26. doi: 10.1038/nrd1931. [DOI] [PubMed] [Google Scholar]
- 12.Vyas H., Summers R. An information-driven approach to pharmacogenomics. Pharmacogenomics. 2005;6:473–480. doi: 10.2217/14622416.6.5.473. [DOI] [PubMed] [Google Scholar]
- 13.Freimuth R.R., Stormo G.D., McLeod H.L. PolyMAPr: programs for polymorphism database mining, annotation, and functional analysis. Hum. Mutat. 2005;25:110–117. doi: 10.1002/humu.20123. [DOI] [PubMed] [Google Scholar]
- 14.Klein T.E., Chang J.T., Cho M.K., Easton K.L., Fergerson R., Hewett M., Lin Z., Liu Y., Liu S., Oliver D.E., et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 15.Ji Z.L., Han L.Y., Yap C.W., Sun L.Z., Chen X., Chen Y.Z. Drug Adverse Reaction Target Database (DART): proteins related to adverse drug reactions. Drug Saf. 2003;26:685–690. doi: 10.2165/00002018-200326100-00002. [DOI] [PubMed] [Google Scholar]
- 16.Ji Z.L., Sun L.Z., Chen X., Zheng C.J., Yao L.X., Han L.Y., Cao Z.W., Wang J.F., Yeo W.K., Cai C.Z., et al. Internet resources for proteins associated with drug therapeutic effects, adverse reactions and ADME. Drug Discov. Today. 2003;8:526–529. doi: 10.1016/s1359-6446(03)02742-9. [DOI] [PubMed] [Google Scholar]
- 17.Sun L.Z., Ji Z.L., Chen X., Wang J.F., Chen Y.Z. ADME-AP: a database of ADME associated proteins. Bioinformatics. 2002;18:1699–1700. doi: 10.1093/bioinformatics/18.12.1699. [DOI] [PubMed] [Google Scholar]
- 18.Chen X., Ji Z.L., Chen Y.Z. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002;30:412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozawa N., Shimizu T., Morita R., Yokono Y., Ochiai T., Munesada K., Ohashi A., Aida Y., Hama Y., Taki K., et al. Transporter database, TP-Search: a web-accessible comprehensive database for research in pharmacokinetics of drugs. Pharm. Res. 2004;21:2133–2134. doi: 10.1023/b:pham.0000048207.11160.d0. [DOI] [PubMed] [Google Scholar]
- 20.Iida A., Saito S., Sekine A., Takahashi A., Kamatani N., Nakamura Y. Japanese single nucleotide polymorphism database for 267 possible drug-related genes. Cancer Sci. 2006;97:16–24. doi: 10.1111/j.1349-7006.2006.00142.x. [DOI] [PubMed] [Google Scholar]
- 21.Salter A.H. Large-scale databases in toxicogenomics. Pharmacogenomics. 2005;6:749–754. doi: 10.2217/14622416.6.7.749. [DOI] [PubMed] [Google Scholar]
- 22.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosford D.A., Lai E.H., Riley J.H., Xu C.F., Danoff T.M., Roses A.D. Pharmacogenetics to predict drug-related adverse events. Toxicol. Pathol. 2004;32:9–12. doi: 10.1080/01926230490424743. [DOI] [PubMed] [Google Scholar]
- 24.Ranganathan P., Eisen S., Yokoyama W.M., McLeod H.L. Will pharmacogenetics allow better prediction of methotrexate toxicity and efficacy in patients with rheumatoid arthritis? Ann. Rheum. Dis. 2003;62:4–9. doi: 10.1136/ard.62.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiratsuka M., Kishikawa Y., Takekuma Y., Matsuura M., Narahara K., Inoue T., Hamdy S.I., Endo N., Goto J., Mizugaki M. Genotyping of the N-acetyltransferase2 polymorphism in the prediction of adverse drug reactions to isoniazid in Japanese patients. Drug Metab. Pharmacokinet. 2002;17:357–362. doi: 10.2133/dmpk.17.357. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida K., Takahashi H., Higuchi H., Kamata M., Ito K., Sato K., Naito S., Shimizu T., Itoh K., Inoue K., et al. Prediction of antidepressant response to milnacipran by norepinephrine transporter gene polymorphisms. Am. J. Psychiatr. 2004;161:1575–1580. doi: 10.1176/appi.ajp.161.9.1575. [DOI] [PubMed] [Google Scholar]
- 27.Carlini L.E., Meropol N.J., Bever J., Andria M.L., Hill T., Gold P., Rogatko A., Wang H., Blanchard R.L. UGT1A7 and UGT1A9 polymorphisms predict response and toxicity in colorectal cancer patients treated with capecitabine/irinotecan. Clin. Cancer Res. 2005;11:1226–1236. [PubMed] [Google Scholar]
- 28.Anttila S., Illi A., Kampman O., Mattila K.M., Lehtimaki T., Leinonen E. Interaction between NOTCH4 and catechol-O-methyltransferase genotypes in schizophrenia patients with poor response to typical neuroleptics. Pharmacogenetics. 2004;14:303–307. doi: 10.1097/00008571-200405000-00005. [DOI] [PubMed] [Google Scholar]
- 29.Kajinami K., Brousseau M.E., Ordovas J.M., Schaefer E.J. Interactions between common genetic polymorphisms in ABCG5/G8 and CYP7A1 on LDL cholesterol-lowering response to atorvastatin. Atherosclerosis. 2004;175:287–293. doi: 10.1016/j.atherosclerosis.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Zazzi M., Romano L., Venturi G., Shafer R.W., Reid C., Dal Bello F., Parolin C., Palu G., Valensin P.E. Comparative evaluation of three computerized algorithms for prediction of antiretroviral susceptibility from HIV type 1 genotype. J. Antimicrob. Chemother. 2004;53:356–360. doi: 10.1093/jac/dkh021. [DOI] [PubMed] [Google Scholar]
- 31.Debouck C., Metcalf B. The impact of genomics on drug discovery. Annu. Rev. Pharmacol. Toxicol. 2000;40:193–207. doi: 10.1146/annurev.pharmtox.40.1.193. [DOI] [PubMed] [Google Scholar]
- 32.Eddershaw P.J., Beresford A.P., Bayliss M.K. ADME/PK as part of a rational approach to drug discovery. Drug Discov. Today. 2000;5:409–414. doi: 10.1016/s1359-6446(00)01540-3. [DOI] [PubMed] [Google Scholar]
- 33.Li A.P. Screening for human ADME/Tox drug properties in drug discovery. Drug Discov. Today. 2001;6:357–366. doi: 10.1016/s1359-6446(01)01712-3. [DOI] [PubMed] [Google Scholar]
- 34.Lin J.H., Lu A.Y. Role of pharmacokinetics and metabolism in drug discovery and development. Pharmacol. Rev. 1997;49:403–449. [PubMed] [Google Scholar]
- 35.Mobley C., Hochhaus G. Methods used to assess pulmonary deposition and absorption of drugs. Drug Discov. Today. 2001;6:367–375. doi: 10.1016/s1359-6446(01)01691-9. [DOI] [PubMed] [Google Scholar]
- 36.Dove A. Proteomics: translating genomics into products? Nat. Biotechnol. 1999;17:233–236. doi: 10.1038/6972. [DOI] [PubMed] [Google Scholar]
- 37.Scharpe S., De Meester I. Peptide truncation by dipeptidyl peptidase IV: a new pathway for drug discovery? Verh. K. Acad. Geneeskd Belg. 2001;63:5–32. discussion 32–33. [PubMed] [Google Scholar]
- 38.Basile V.S., Ozdemir V., Masellis M., Walker M.L., Meltzer H.Y., Lieberman J.A., Potkin S.G., Alva G., Kalow W., Macciardi F.M., et al. A functional polymorphism of the cytochrome P450 1A2 (CYP1A2) gene: association with tardive dyskinesia in schizophrenia. Mol. Psychiatr. 2000;5:410–417. doi: 10.1038/sj.mp.4000736. [DOI] [PubMed] [Google Scholar]
- 39.Kapitany T., Meszaros K., Lenzinger E., Schindler S.D., Barnas C., Fuchs K., Sieghart W., Aschauer H.N., Kasper S. Genetic polymorphisms for drug metabolism (CYP2D6) and tardive dyskinesia in schizophrenia. Schizophr. Res. 1998;32:101–106. doi: 10.1016/s0920-9964(98)00038-3. [DOI] [PubMed] [Google Scholar]
- 40.Serretti A., Zanardi R., Rossini D., Cusin C., Lilli R., Smeraldi E. Influence of tryptophan hydroxylase and serotonin transporter genes on fluvoxamine antidepressant activity. Mol. Psychiatr. 2001;6:586–592. doi: 10.1038/sj.mp.4000876. [DOI] [PubMed] [Google Scholar]
- 41.Smits K.M., Smits L.J., Schouten J.S., Stelma F.F., Nelemans P., Prins M.H. Influence of SERTPR and STin2 in the serotonin transporter gene on the effect of selective serotonin reuptake inhibitors in depression: a systematic review. Mol. Psychiatr. 2004;9:433–441. doi: 10.1038/sj.mp.4001488. [DOI] [PubMed] [Google Scholar]
- 42.Rau T., Wohlleben G., Wuttke H., Thuerauf N., Lunkenheimer J., Lanczik M., Eschenhagen T. CYP2D6 genotype: impact on adverse effects and nonresponse during treatment with antidepressants—a pilot study. Clin. Pharmacol. Ther. 2004;75:386–393. doi: 10.1016/j.clpt.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 43.Siddiqui A., Kerb R., Weale M.E., Brinkmann U., Smith A., Goldstein D.B., Wood N.W., Sisodiya S.M. Association of multidrug resistance in epilepsy with a polymorphism in the drug-transporter gene ABCB1. N. Engl. J. Med. 2003;348:1442–1448. doi: 10.1056/NEJMoa021986. [DOI] [PubMed] [Google Scholar]
- 44.Saitoh A., Singh K.K., Powell C.A., Fenton T., Fletcher C.V., Brundage R., Starr S., Spector S.A. An MDR1-3435 variant is associated with higher plasma nelfinavir levels and more rapid virologic response in HIV-1 infected children. AIDS. 2005;19:371–380. doi: 10.1097/01.aids.0000161766.13782.2f. [DOI] [PubMed] [Google Scholar]