Abstract
The GeneTrees phylogenomics system pursues comparative genomic analyses from the perspective of gene phylogenies for individual genes. The GeneTrees project has the goal of providing detailed evolutionary models for all protein-coding gene components of the fully sequenced genomes. Currently, a database of alignments and trees for all protein sequences for 325 fully sequenced and annotated prokaryote genomes is available. The prokaryote database contains 890 000 protein sequences organized into over 100 000 alignments, each described by a phylogenetic tree. An original homology group discovery tool assembles sets of related proteins from all versus all pairwise alignments. Multiple alignments for each homology group are stored and subjected to phylogenetic tree inference. A graphical web interface provides visual exploration of the GeneTrees database. Homology groups can be queried by sequence identifiers or annotation terms. Genomes can be browsed visually on a gene map of each chromosome or plasmid. Phylogenetic trees with support values are displayed in conjunction with the associated sequence alignment. A variety of classes of information can be selected to label the tree tips to aid in visual evaluation of annotation and gene function. This web interface is available at http://genetrees.vbi.vt.edu.
INTRODUCTION
The amount of genomic sequence data and information regarding protein structure and function continues to grow. Genomic biology flourishes through a continual reinterpretation of sequence data with the aid of new information streams and analysis methods. One approach that provides deeper understanding of available genomes sequences is comparative genomics. Comprehensive sets of aligned protein or DNA homologs have proved useful for years (1,2). More recent developments have emphasized phylogenetic tree models to further illuminate homology relationships (3–6). Here we describe a new phylogenomics database which complements these existing resources, first by its taxonomic focus on the fully sequenced bacterial genomes and second by unique features of the way it presents this valuable form of data integration.
We have developed a phylogenomics analysis platform, which we call ‘GeneTrees’, for automating large-scale comparative studies. The GeneTrees website provides evolutionary models for all genomic components of fully sequenced bacterial genomes describing the pattern of common ancestry (i.e. homology) observed among sequences. We distinguish between orthologous and paralogous relationships sensu Fitch (7). Orthology, the sharing of homologs in two species since the common ancestral species, is expected to track conserved function fairly well, while paralogy, the relationship between gene copies generated by gene duplication, is expected frequently to be accompanied by functional divergence of genes (8,9). The reality of lateral gene transfer in the prokaryotes adds another dimension to gene relationships (10). A phylogenomics system should enable users to make the best possible use of these evolutionary patterns among genes when inferring the functions of genes known by sequence alone by their relationship to genes of known function (9).
The GeneTrees database is structured as a list of taxa (species or within-species variants such as strains) using the GenBank taxonomy, the complete set of protein sequences annotated for these genomes and a set of homology models. Each homology model consists of a multiple sequence alignment for protein subregions plus the evolutionary trees inferred from the alignment using one or more algorithms. The web interface presents integrated visualization of phylogenetic models with their underlying sequence data. We expect the system to aid comparative genomics, genome annotation and a wide range of evolutionary investigations.
COMPUTING GeneTrees
Populating the GeneTrees database begins by downloading full genomes from NCBI (ftp://ftp.ncbi.nih.gov/genomes/Bacteria). Currently only protein sequences are analyzed, though the positions of proteins on chromosomes are used for a genome map visualization. Discovering and analyzing homology groups involves several steps. First, low-level pairwise homology statements are assembled using all versus all comparison, typically using BLASTP with an expect value threshold of 1E-6 (11). A program we have written uses pairwise alignments to partition the sequences into dense, non-overlapping, clusters using an iterative heuristic method. These non-overlapping clusters are then used as seeds to a more rigorous process of alignment coupled with searching for new members. This process does all versus all alignment of sequences within the cluster to find the maximal regions within each sequence relevant to alignment with any other member. These trimmed regions are aligned by MUSCLE (12). This alignment is used to build an HMM using the HMMER package (13) which is used to search the set of all sequences having any BLAST hit (at 1E-6) to any member of the seed cluster. All top scoring sequences are included in the growing set down to a score threshold as a proportion of the top scoring sequence (we have used 0.3 and 0.5). The set of included sequences, trimmed to the endpoints of the HMM hit, are then aligned using Muscle and this alignment is stored to the database with statistics on overall conservation. Prior to tree building, columns of the alignment are filtered to mask out poorly aligned regions. Briefly, a score is calculated for each column consisting of the product of the column occupancy (proportion not gaps), the occupancy of a 10-column window centered on the column and the window average of the mean squared frequencies of each amino acid frequency within each column (a measure of conservation). Columns with scores below the threshold (typically 0.3) are masked out for tree building and shown in grey in the alignment visualization.
An individual sequence may participate in more than one alignment with alignments containing overlapping sequence sets. This is an inherent requirement to describing the patterns observed. As an example, one sequence may contain a conserved domain that has a clear phylogenetic relationship to many other instances of the domain, while the remainder sequence may align to only a handful of close homologs. This partial redundancy is considered an advantage but needs to be handled appropriately. Each alignment display is accompanied by a table describing and linking to overlapping alignments when they exist.
Phylogenetic trees are then constructed using the program MrBayes version 3.1.2 (14) which uses Markov-chain Monte Carlo (MCMC) methods to infer the distribution of likely tree topologies and branch lengths, which are then summarized as a consensus tree. The advantage of MCMC methods is that they allow estimating unknown parameters such as substitution matrices and site-specific rate variation without having to specify these a priori. A typical MrBayes run consists of an initial hill-climbing phase where it finds successively better trees by random permutations, one each ‘generation’, then asymptotically reaching a plateau where it continues to sample tree space, but with no trend towards better trees. This MCMC strategy attempts to accurately sample the distribution of likely trees without overly emphasizing a single ‘best’ tree. Achieving this plateau stage for all alignments in the GeneTrees database is not possible in the short term. The correlation between tree likelihood and generation number is used in GeneTrees as an index of the thoroughness of the analysis of each alignment, with low correlation scores indicating the MCMC run has levelled off. The system applies a limited length initial analysis run of 10 000 generations to each alignment starting from a random tree. A second script will revisit alignments in descending order of this correlation score, worst ones first, starting a new MCMC search starting from the best tree previously found. This strategy allows us to move the entire population of trees gradually to better statistical status in a breadth-first manner. Some alignments require many rounds of MrBayes searching to reach the correlation threshold and therefore accumulate many tens of thousands of generations. Currently, of over 105 alignments with 250 sequences or fewer, over half have a best MrBayes run to date with a negative correlation of log likelihood versus generation, which we believe is strong evidence of having run to saturation. For alignments with 100 or fewer sequences, 12% have correlations worse than 0.3 and are being further refined. We intend to continue this process until trees of 100 or fewer sequences have correlations no >0.1 and trees between 100 and 250 have correlations <0.3, which is a concession to the high-throughput nature of this database.
DATABASE UPDATES
As new whole genomes are added to NCBI, the database needs to be updated. We currently have a semi-automated pipeline for this. We update our BLASTP table by searching all new sequences against all old and new sequences combined. Then for each alignment in the database we identify all new candidate member sequences by BLAST hit. An HMM is built and used to score all candidates plus original members. Again, all sequences with score >0.3 times the top score are used to build the derived alignment. The original alignment is retained in the database, leading to the frequent occurrence of many instances of strongly overlapping alignments, with one including only one or a few additional sequences. Handling this redundancy in a graceful manner is one of the development goals. Besides using the old alignments to interpret new sequences, the sequence partitioning program is run again, though rejecting any seed cluster which is a subset of sequences within any alignment. Surviving seeds are run through the alignment-HMMsearch-alignment routine described above. This update pipeline was last run on May 24, 2006, which brought the prokaryote database to 325 species, 890 000 sequences and over 100 000 alignments. We intend to run the update process at least every 2 months and more frequently as it becomes more automated. Statistics on the most recent update will be presented on the website.
WEBSITE AND INTERFACE
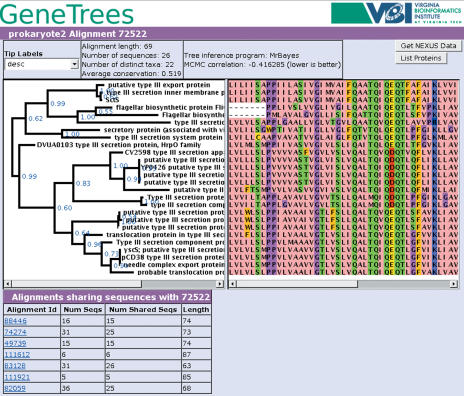
Homology groups can be queried by sequence identifiers such as GI number or annotation terms such as ‘potassium transporter’. These queries return a table of alignment identifiers with summary information. Selecting one of the rows in this table takes the user to a web page with an image of the consensus tree and an image of the color-coded alignment. These are juxtaposed so that each tree tip aligns with the sequence it describes, allowing visual perusal of the evidence for the tree (Figure 1). Regions of the alignment that were excluded from tree building due to low conservation are shaded in gray.
Figure 1.
An example of the central tree plus alignment visualization from GeneTrees. This alignment describes type III secretion proteins from a variety of bacteria.
Bayesian support numbers are shown for the interior branches of the tree. A drop-down menu allows the user to select which class of information is used to label the tree tips. Useful tip-labeling options include taxon name, annotation description, GI number, accession and COG/Pfam patterns. A textual description of some statistics for the alignment and tree are provided. Below the graphics is presented an optional table listing links to alignments with overlapping sequence members, if these exist. A button is provided to present the entire alignment, tree and current choice of tip labels in the NEXUS format (15).
Another browsing option is provided in the form of a gene map of each replicon (chromosome or plasmid) of each species. These maps show the location of each protein gene along the genome sequence and present simple glyphs for the locations corresponding to members of multiple sequence alignments. Clicking on such a glyph takes one to the tree and alignment display.
AVAILABILITY
The GeneTrees database is hosted at the Virginia Bioinformatics Institute at Virginia Tech and can be accessed at: http://genetrees.vbi.vt.edu. A collection of software used to build the database will be made available at the site. The alignment and tree visualization web pages utilize dynamic HTML and Javascript which is tested on Internet Explorer (IE) 6 and Firefox 1.5. This requires Javascript to be enabled (called ‘Active script’ in IE).
CONCLUSIONS AND FUTURE DIRECTIONS
With the growing number of completely sequenced bacterial genomes, the scientific value of a specific phylogenomics resource for this set of biological sequences is clear. We hope to increase the update frequency so that the database stays current as each new genome becomes available at NCBI. GeneTrees is an evolving system and should add significant functionality over time. One direction of ongoing development is to enable automated comparison of individual gene trees to a ‘species tree’ to highlight differences such as gene duplication, loss and horizontal transfer. This will also flag strongly supported orthologs for better functional inference (16). Doing this is somewhat challenging in the prokaryotes where the concept of a ‘species tree’ is less crisp than in higher groups (10). GeneTrees may also grow by adding divisions for other taxonomic groups where data density justifies a whole-genome phylogenomics approach. We plan to follow the recommended standards for minimum information about a phylogenetic analysis (MIAPA) as these evolve (17).
Acknowledgments
We thank Rebecca Wattam and Kelly Williams for constructive commentary. A.W.D is funded in part by NSF grant 0114866. This work was funded in part by DOD contract W911SR-04-C-0045 to Bruno W. Sobral and in part by the Virginia Bioinformatics Institute. Funding to pay the Open Access publication charges for this article was provided by Virginia Bioinformatics Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whelan S., de Bakker P.I., Quevillon E., Rodriguez N., Goldman N. PANDIT: an evolution-centric database of protein and associated nucleotide domains with inferred trees. Nucleic Acids Res. 2006;34:D327–D331. doi: 10.1093/nar/gkj087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roth C., Betts M.J., Steffansson P., Saelensminde G., Liberles D.A. The Adaptive Evolution Database (TAED): a phylogeny based tool for comparative genomics. Nucleic Acids Res. 2005;33:D495–D497. doi: 10.1093/nar/gki090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann S., Lu D., Phillips J., Vision T.J. Phytome: a platform for plant comparative genomics. Nucleic Acids Res. 2006;34:D724–D730. doi: 10.1093/nar/gkj045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H., Coghlan A., Ruan J., Coin L.J., Heriche J.K., Osmotherly L., Li R., Liu T., Zhang Z., Bolund L., et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitch W.M. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 8.Sonnhammer E.L., Koonin E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 9.Eisen J.A. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- 10.Bapteste E., Susko E., Leigh J., MacLeod D., Charlebois R.L., Doolittle W.F. Do orthologous gene phylogenies really support tree-thinking? BMC Evol. Biol. 2005;5:33. doi: 10.1186/1471-2148-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 12.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 14.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 15.Maddison D.R., Swofford D.L., Maddison W.P. NEXUS: an extensible file format for systematic information. Syst. Biol. 1997;46:590–621. doi: 10.1093/sysbio/46.4.590. [DOI] [PubMed] [Google Scholar]
- 16.Storm C.E., Sonnhammer E.L. Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics. 2002;18:92–99. doi: 10.1093/bioinformatics/18.1.92. [DOI] [PubMed] [Google Scholar]
- 17.Leebens-Mack J., Vision T., Brenner E., Bowers J.E., Cannon S., Clement M.J., Cunningham C.W., de Pamphilis C., deSalle R., Doyle J.J., et al. Taking the first steps towards a standard for reporting on phylogenies: Minimum Information About a Phylogenetic Analysis (MIAPA) OMICS. 2006;10:231–237. doi: 10.1089/omi.2006.10.231. [DOI] [PMC free article] [PubMed] [Google Scholar]