Abstract
We describe the synthesis of an abasic RNA phosphoramidite carrying a photocleavable 1-(2-nitrophenyl)ethyl (NPE) group at the anomeric center and a triisopropylsilyloxymethyl (TOM) group as 2′-O-protecting group together with the analogous DNA and the 2′-OMe RNA abasic building blocks. These units were incorporated into RNA-, 2′-OMe-RNA- and DNA for the purpose of studying their chemical stabilities towards backbone cleavage in a comparative way. Stability measurements were performed under basic conditions (0.1 M NaOH) and in the presence of aniline (pH 4.6) at 37°C. The kinetics and mechanisms of strand cleavage were followed by High pressure liquid chromotography and ESI-MS. Under basic conditions, strand cleavage at abasic RNA sites can occur via β,δ-elimination and 2′,3′-cyclophosphate formation. We found that β,δ-elimination was 154-fold slower compared to the same mechanism in abasic DNA. Overall strand cleavage of abasic RNA (including cyclophosphate formation) was still 16.8 times slower compared to abasic DNA. In the presence of aniline at pH 4.6, where only β,δ-elimination contributes to strand cleavage, a 15-fold reduced cleavage rate at the RNA abasic site was observed. Thus abasic RNA is significantly more stable than abasic DNA. The higher stability of abasic RNA is discussed in the context of its potential biological role.
INTRODUCTION
Abasic sites are well known DNA lesions that occur spontaneously via depurination at a frequency of ∼10 000 per genome per day in mammalian cells (1). Acidic or alkylating conditions as well as oxidative stress greatly enhance this number. Abasic DNA is unstable and undergoes strand cleavage 3′ to the abasic site with an average lifetime of 8 days at 37°C, pH 7.4 and physiological ionic strength (2). Due to the degradation of the genetic material and to missing coding information such sites are highly mutagenic and a major threat to living cells (3,4). Living organisms have therefore evolved a highly efficient and complex DNA repair machinery that maintains genome integrity (5). One component of this machinery is the base excision repair pathway in which abasic sites are produced as intermediates by DNA glycosylases and cleaved either by AP-endonucleases or by AP-lyases. It is only recently that cellular repair processes were also found in RNA. For example, the human oxidative demethylases hABH3 that reverse alkylation of bases in DNA were found to repair also RNA (6). When expressed in Escherichia coli they were found to reactivate methylated RNA bacteriophage MS2 in vivo. This illustrates the biological relevance of such a repair activity and points towards RNA repair being a potentially important defense mechanism in living cells.
Spontaneous or induced abasic lesions are a priori not restricted to DNA but can also occur in RNA. Given the fact that not only short lived mRNA but long-lived tRNAs and ribosomal RNAs are present at all times in a cell in much higher concentrations than DNA, and given also the fact that RNA viruses as well as retroviruses store their genetic information in form of RNA, it is not excluded that abasic RNA plays a significant biological role. Literature about the chemistry and chemical biology of RNA abasic sites is, however, scarce and their biological impact is largely unexplored with a few exceptions. For example, RNA abasic sites are known to be the result of the action of RNA N-ribohydrolases (7). A famous example is the peptide toxin ricin which depurinates a specific adenosine residue on 28S rRNA, one of the RNA chains of eukaryotic ribosomes. This leads to poor binding of elongation factors and knockout of protein synthesis. A single molecule of ricin suffices to kill a whole cell. Besides this there is evidence for a class of RNA specific lyases that act on rRNA abasic sites, leading to complete inactivation of ribosomes (8,9).
Data on the mechanism and kinetics of strand scission of abasic RNA in comparison to abasic DNA is virtually inexistent. In vitro generation and cleavage of abasic RNA has been of some interest in the context of RNA sequencing via base alkylation and depurination followed by strand scission induced by aniline under neutral to slightly acidic conditions. It is believed that under these conditions only β-elimination of the phosphate unit on the 3′-side occurs, leaving behind a 5′-oligonucleotide with a pendent 4,5-dihydroxy-2-oxovaleraldehyde moiety. In another example similar chemistry was used to excise one specific abasic nucleotide out of yeast tRNAPhe. Here the intrinsic lability of the glycosidic bond of wybutosine was used to selectively produce one abasic site, followed by treatment with 2-aminopyridine to effect β,δ-elimination, leaving behind 3′ and 5′-phosphorylated ends that were religated in three consecutive enzymatic steps (10).
Given the lack of knowledge on the chemistry and chemical biology of RNA abasic sites we set out to investigate this in more detail. More precisely we synthesized an abasic RNA precursor phosporamidite that carries a photocleavable protecting group at the anomeric center (11). This unit can be introduced into oligoribonucleotides via automated RNA synthesis and the abasic site can be revealed after detachment from solid support and high pressure liquid chromotagraphy (HPLC) purification by photolysis. In preliminary work we reported on the trans-lesion synthesis of HIV-1 reverse transcriptase on a RNA-template/DNA-primer system with an abasic site in the RNA template, and on the degradation of the damaged template by the RNase H activity of the polymerase in comparison to a non-damaged template (12). We found efficient incorporation of deoxynucleotides opposite the lesion following the ‘A-rule’, previously described for DNA-templated trans-lesion synthesis (13). Here we present now a detailed analysis of the kinetics and mechanisms of strand cleavage of RNA abasic sites at various conditions in comparison to abasic DNA.
MATERIALS AND METHODS
Synthesis, deprotection and purification of oligonucleotides
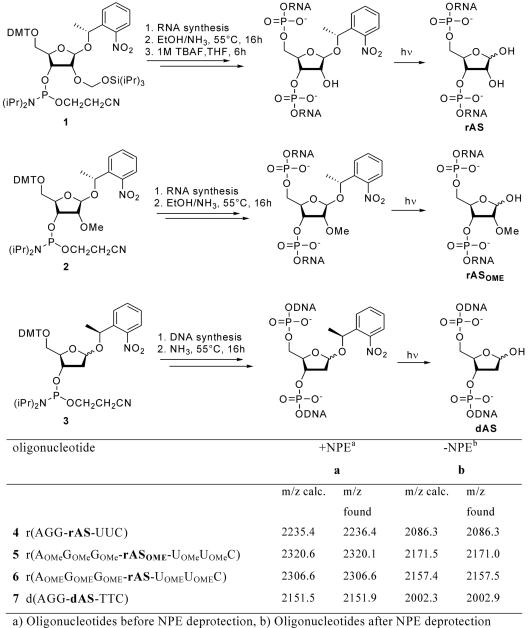
Full experimental details on the synthesis of the RNA and DNA abasic site phosphoramidites 1–3 (Scheme 1) containing the photocleavable 1-(2-metyhl)nitrophenethyl (NPE) group is given in the supplementary data section. All oligonucleotides were prepared by automated oligonucleotide synthesis with an Expedite 8909 nucleic-acid synthesis system (PerSepitve Biosystems Inc., Framingham, MA) by using the cyanoethyl-phosphoramidite approach. RNA synthesis was performed with 2′-O-TBDMS protected PAC-phosphoramidites (Glen Research Corp., Sterling, VA) on polystyrene solid supports (Amersham Biotech). 2′-OMe-RNA oligoribonucleotides were prepared with benzoyl (A, C) and dimethylformamidino (G) protected 2′-OMe RNA phosphoramidites (Glen Research Corp., Sterling, VA). All 2′-OMe-RNA sequences were synthesized on normal RNA solid supports thus leading to oligonucleotides with an unmodified 3′-terminus. For DNA synthesis benzoyl (dA, dC) and isobutyryl (dG) protected phosphoramidites and controlled pore-glass solid supports (Glen Research Corp., Sterling, VA) were used. The syntheses were performed using the standard coupling protocols for DNA and RNA synthesis, with 5-(ethylthio)-1H-tetrazole (Aldrich) as activator and a coupling time of 90 s for DNA and 6 min for RNA and 2′OMe-RNA building blocks. The modified phosphoramidites 1–3 were allowed to couple for 6 min and 12 min for DNA and RNA, respectively. Solid supports were treated with EtOH/NH4OH (1:4) at 55°C, overnight. The DNA and RNA heptamers were purified by using RP-HPLC on an ÄKTA 900 HPLC system (Amersham Pharmacia Biotech). A SOURCE™ 15RPC ST 4.6/100 (polystyrene/divinyl benzene, 15 μm, 100/4.6 mm) column (Amersham Biosciences, Uppsala, Sweden) was used with a linear gradient of solvent A (0.1 M Et3NOAc in H2O) and solvent B [0.1 M Et3NOAc, CH3CN/H2O (4:1)]. RNA sequences were further silyl deprotected by treatment with Bu4NF (1 M in THF) for 16 h at RT. Deprotected oligoribonucleotides were desalted by using Sep-Pak® C18 columns (Waters Corp., Milford, MA) before purification by RP-HPLC. Purified oligonucleotides were dissolved in DEPC-treated water and the concentrations of these stock solutions (O.D. 260 nm) were determined with a NanoDrop® ND-100 UV/Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE). All abasic oligonucleotides as well as their NPE-protected precursors were analyzed by ESI-MS.
Scheme 1.
Synthesis and sequences of rAS-, rASOMe- and dAS-containing oligonucleotide heptamers.
Strand cleavage kinetics
Sample preparation
Aliquots (0.3 or 0.4 ml of a 1 OD260 ml−1 solution) of NPE-protected RNA heptamers in H2O in a quartz cuvette were deprotected at room temperature by using a UV immersion lamp TQ 150 (UV-RS-2, Heraeus) for 2 min. Alternatively, deprotection was achieved using a slide projector with a 250 W tungsten halogen lamp for 6 min. For both methods a conventional glass plate was used as a filter to remove light with wavelengths <300 nm to avoid damage to the oligonucleotides.
Alkaline strand cleavage
Deprotected oligonucleotides were treated with 1 M NaOH to a final concentration of 0.1 M NaOH and incubated at 37°C. Samples were quenched with a stoichiometric amount of 1 M HCl and the reaction mixtures were immediately analyzed by RP-HPLC. The ratio of the remaining abasic heptamer to reaction products was determined by peak integration. The resulting values were fitted with a first order exponential decay using OriginPro 7.5 software (OriginLab Corp., Northhampton, MA). Resulting half-life times T1/2 were calculated from the fitted curves that were corrected, where necessary, for the observed amount of products already present at t = 0. Alternatively, the ln([oligo]/[oligo]0) was plotted against the time t. In all cases a linear relationship was observed, indicating first order kinetics. The first order rate constants for strand scission ks (s−1) were obtained from the slopes of the respective linear fits.
Strand cleavage in the presence of aniline at pH 4.6
Deprotected oligonucleotides were diluted to a final concentration of 1 OD260 ml−1 in 0.5 M aniline–HCl, pH 4.6 (0.4 ml). The aliquots are incubated at 37°C for different time intervals. To quench the reactions, aniline was removed by quick DEAE HPLC (Nucleogen® 60-7 125/4 mm column, Macherey–Nagel) with the following gradient: 0% solvent A (0.25 mM Na2HPO4 + 0.25 mM NaH2PO4 in 20% acetonitrile) for 10 min, 100% solvent B (A + 1 M NaCl) for 5 min. The oligonucleotides eluted with solvent B are collected and carefully concentrated (Speedvac) to a volume of ∼0.4 ml and further analyzed by RP-HPLC as described above.
Fragment identification
Cleavage products of three independent cleavage reactions each were separated by RP-HPLC as described, pooled, lyophilized and analyzed by ESI− mass spectrometry. In the case of oligonucleotide 4b the pooled fragments after aniline removal were desalted over Sep-Pak® C18 columns (Waters Corp., Milford, MA) and measured directly by ESI-MS without RP-HPLC separation.
RESULTS
Synthesis of abasic building blocks and oligonucleotides
The synthesis of the RNA and DNA abasic site phosphoramidites 1–3 (Scheme 1) containing the photocleavable 1-(2-nitrophenyl)ethyl (NPE) group, which was used successfully before in oligonucleotide synthesis as protecting group for the 2′-OH function of ribonucleosides (14) or for cageing RNA and DNA bases (15–17), is described in the supplementary data section. Their synthesis is straight forward and resulted in overall yields of 21% for the rAS and 32% for the rASOMe phosphoramidite starting from tetraacetyl ribose, as well as 44% for the dAS building block starting from 1,3,5-tri-O-acetyl-2′-deoxy-d-ribose. Both configurations at the chiral center of the NPE protecting group were used for reasons of synthetic economy and in both cases photolytic removal was quantitative (see Supplementary Data). These building blocks were stable for more than 1 year when kept in the dark at −20°C. Direct contact to sunlight was avoided but otherwise no special light protection was used during synthesis or manipulation of these abasic site precursors. The synthesis of oligonucleotides was performed by standard phosphoramidite chemistry (Scheme 1) and resulted in coupling yields for building blocks 1–3 that are in the same range as for natural nucleoside phosphoramidites (>95%). Typically, oligoribonucleotides were HPLC purified before silyl deprotection. We found that this lead to purer oligonucleotide samples after desilylation.
In a first step we determined the deprotection kinetics of the NPE group in the ribonucleotide 4a at a concentration of 1 OD260 ml−1 with glass-filtererd light from a UV immersion lamp and followed the progress of deprotection by RP-HPLC at different time intervals. The HPLC traces (see Supplementary Data) showed clean conversion to the AS-containing oligonucleotide 4b following first order kinetics. The half-life time was calculated to be 12.5 s. Traces of 4b (<5%) resulting from deprotection during manipulations could already be seen at the beginning. An irradiation time of 2 min was found to be sufficient to achieve almost quantitative deprotection. In an alternative setup we used a slide projector with a halogen lamp for irradiation. Due to its lenses, UV light <365 nm was filtered out. Irradiation times of 6–10 min were found sufficient to promote full NPE-cleavage. This latter method is very mild and was generally preferred in further experiments.
We applied the same NPE-deprotection protocols also to the dAS percursor 7a. Again we found first order kinetics for NPE deprotection to 7b with a half-life time of 3.4 s. Long time (∼15 min) UV irradiation with the UV lamp lead to a side product that was not identified but that was clearly not the result of strand cleavage. Most likely it is a product resulting from T-T cyclobutane formation. Again irradiation with the slide projector for 5 min effected complete revelation of the abasic site without producing traces of this side product and was therefore considered superior.
Mechanisms of RNA abasic site cleavage
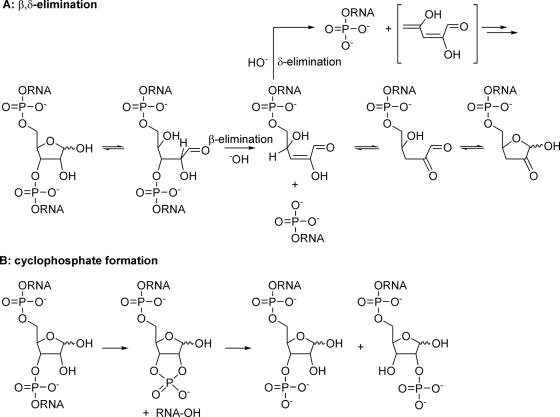
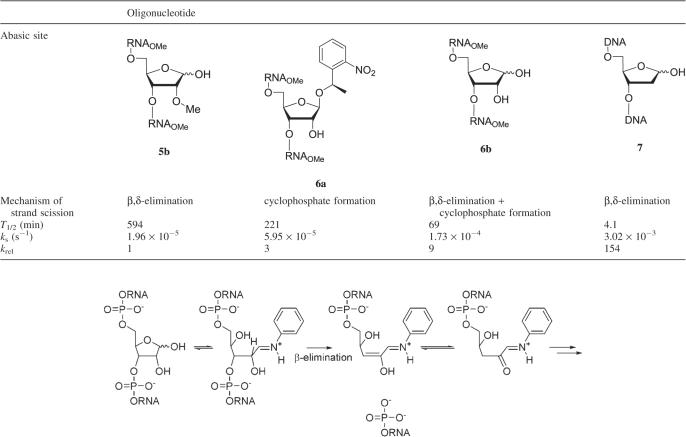
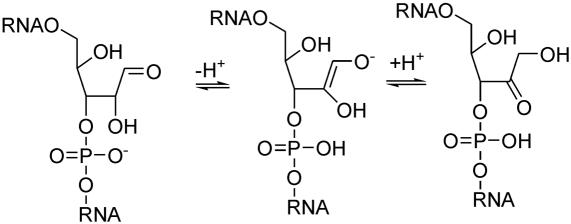
The presence of the 2′-OH group at an RNA- compared to a DNA abasic site substantially influences the mechanism of cleavage at high pH. The structurally and kinetically relevant intermediates for hydroxide induced strand scission are depicted in Scheme 2. As in the case of DNA, β-elimination of the oligonucleotide fragment 3′ to the abasic site can occur from the aldehyde form yielding a 5′-phosphorylated 3′-fragment and an enol intermediate which after enol-ketone tautomerization leads to a 4,5-dihydroxy-2-oxo-valeraldehyde unit attached to the 5′-fragment of the original RNA (Scheme 2A). Alternatively, the enol form is suited to a further δ-elimination step leading to complete excision of the abasic ribose unit and leaving behind a 3′-phosphorylated 5′ end. Besides this an alternative mechanism via cyclophosphate formation (Scheme 2B) does also apply under basic conditions producing a 5′-fragment with an abasic site that is phosphorylated in the 2′ or 3′ position and a 3′-fragment with a 5′-OH end. Cyclophosphate formation must not only occur in the hemiacetal form but can also happen in the alddehyde form of the abasic site (not shown in Scheme 2B).
Scheme 2.
(A and B) Mechanisms of strand cleavage of abasic RNA at high pH.
To determine the contribution of each of these mechanisms to overall strand scission we performed separate kinetic analyses with oligonucleotide 5b and 6a and compared the results to that of 6b in which both mechanisms are active simultaneously. In 5b, the 2′-OH positions are methylated (with the exception of the 3′ terminal C) and the abasic site is deprotected, thus excluding cyclophosphate formation, but admitting β-elimination. In 6a the abasic site is still NPE protected, while the 2′-position of the abasic ribose is free. Here β-elimination is excluded but cyclophosphate formation at the abasic ribose is allowed. Oligonucleotide 7b served as the DNA abasic site model that was used for comparison. The kinetics of strand scission was followed by reversed phase HPLC after incubating the oligonucleotides (Na+ form) with 0.1 M NaOH at 37°C. In regular time intervals samples were injected and the relative amount of residual starting material determined by peak integration. The half-life times (T1/2) were calculated from a first order exponential fit while the first order rate constants ks for strand scission were obtained from the slope of a linear plot of ln [S]/[S0] versus t where [S]/[S0] means the relative amount of unreacted abasic oligonucleotide at the time t.
β,δ-elimination at an RNA abasic site
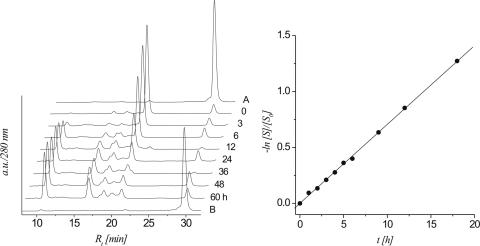
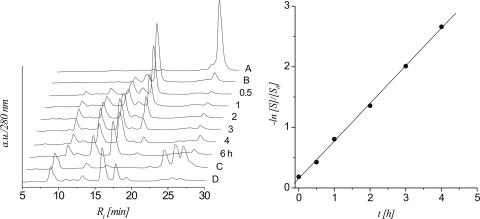
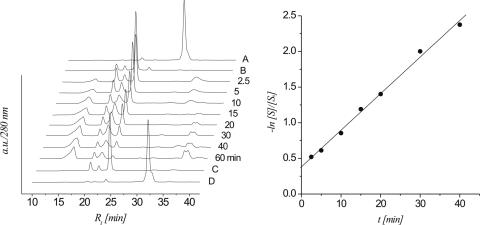
The time course of strand cleavage of oligonucleotide 5b is depicted in Figure 1. From the HPLC traces it becomes clear that the abasic heptamer 5b (peak 4) decreases while at the same time three major fragments with lower retention times appear (peaks 1–3). Lane B is a control with protected oligonucleotide 5a proving that no cleavage occurs if the anomeric center is still protected. The T1/2 and ks for 5b under these conditions were calculated to be 594 min and 1.96 × 10−5 s−1, respectively. Peaks 1–3 were isolated and analyzed by ESI-MS (Table 1). Peaks 2 and 3 contain the two 5′-RNA-fragments after β-elimination and β,δ-elimination, respectively, while peak 1 corresponds to the phosphorylated 3′-fragment. Thus it is clear that both β-and δ-elimination steps take place, however at much slower rates compared to DNA 7b (Table 4).
Figure 1.
left: HPLC traces (280 nm) of the β-elimination reaction of the 2′-OMe heptamer 5b at different time intervals; controls: trace A: protected heptamer 5a; trace B: protected 5a after treatment with 0.1 M NaOH for 48 h at 37°C; right: linear fit (R2 > 0.999) of ln[S]/[S0] versus t calculated from the HPLC traces.
Table 1.
Fragment analysis of cleavage of 5b
| peak | Rt [min] | Structure | Formula | m/z calc. | m/z found |
|---|---|---|---|---|---|
| 1 | 10.5 | r(pUOMeUOMeC) | C29H40N7O24P3 | 963.60 | 963.64 |
| 2 | 16.1 | r(AOMeGOMeGOMep) | C33H44N15O21P3 | 1079.73 | 1079.26 |
| 3 | 18.2 | r(AOMeGOMeGOMeprb)a | C39H52N15O24P3 | 1207.86 | 1207.34 |
arb = 4,5-dihydroxy-2-methoxy-2-pentenal.
Cyclophosphate formation at the RNA abasic site
To study strand scission via cyclophosphate formation independently from β,δ-elimination chemistry, oligonucleotide 6a was subjected to the same basic conditions and its decay followed again by HPLC (Figure 2).
Figure 2.
HPLC left: traces of strand cleavage via cyclophosphate formation of 6a (0.1 M NaOH, 37°C) at different time intervals; controls: trace A: oligomer 6b; right: linear fit (R2 > 0.994) of ln[S]/[S0] versus t calculated from the HPLC traces.
The HPLC traces show as expected a decrease of peak 4 corresponding to the heptamer 6a and the appearence of three novel fragments (peaks 1–3) with lower retention times. Trace A shows oligonucleotide 6b in which the abasic site was deprotected as a control. The absence of this peak in the other traces clearly shows that the 1NPE group is stable under the basic conditions and that no β,δ-elimination can be observed. We calculated again the T1/2 for the disappearance of 6a as described before and found a value of 221 min ks in this case was found to be 5.95 × 10−5 s−1.
To follow the mechanism of cleavage we isolated the newly arising peaks 1–3 and analyzed the RNA fragments by ESI-MS (Table 2). Peak 1 contains the 3′-RNA fragment with a free 5′-OH group which is in accord with hydrolysis via cyclophosphate formation. Peaks 2 and 3 both contain fragments with the same mass that correspond to the isomeric 5′-fragment in which the terminal phosphate group is located either at O2′ or O3′. The corresponding cyclophosphate intermediate could not be isolated in this case.
Table 2.
Fragment analysis of cleavage of 6a
| Peak | Rt [min] | Structure | Formula | m/z calc. | m/z found |
|---|---|---|---|---|---|
| 1 | 11.6 | r(UOMeUOMeC) | C29H39N7O21P2 | 883.62 | 883.20 |
| 2 | 19.5 | r(AOMeGOMeGOMeASp) | C33H44N15O21P3 | 1440.98 | 1440.35 |
| 3 | 20.6 | r(AOMeGOMeGOMeASp) | C39H52N15O24P3 | 1440.98 | 1440.34 |
Combination of β,δ-elimination and cyclophosphate formation in strand cleavage at the RNA abasic site
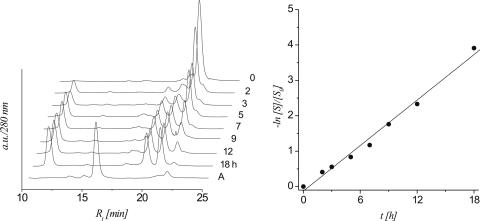
Since the two strand cleavage mechanisms studied separately so far act in concert in abasic RNA, it was necessary to study the effect of their combination on strand cleavage efficiency. For this purpose oligonucleotide 6b was subjected to 0.1 M NaOH and strand cleavage analyzed as described before (Figure 3).
Figure 3.
left: HPLC traces of strand cleavage via cyclophosphate formation and β,δ-elimination of 6b (0.1 M NaOH, 37°C) at different time intervals; controls: trace A: 6a, trace B: 6b, trace C: 6a (0.1 M NaOH, 6.5 h, 37°C), trace D: 6a (0.1 M NaOH, 6.5 h, 37°C, followed by photolysis); right: linear fit (R2 > 0.999) of ln[S]/[S0] versus t calculated from the HPLC traces.
Olignucleotide 6b disappears under standard basic conditions with a T1/2 of 69 min and a ks of 1.73 × 10−4 s−1 as can be seen from the fit. This is considerably faster compared to 6a and 6b and is due to the overlay of the two mechanisms for strand scission (Table 4). A calculated T1/2 for 6b from those of 5b and 6a resulted in a value of 165 min. The difference between calculated and measured value is in reasonable agreement and validates the 2′-OMe modification as a model for studying the β-elimination mechanism.
The overlay of the two modes for strand cleavage gives rise to an increased number of fragments. Fractions of the newly arising three main peaks (peaks 1–3) in the HPLC were again isolated and subjected to fragment analysis (Table 3). To have a first indication weather cyclophosphate formation would be dominant, protected heptamer 6a was also submitted to basic conditions (0.1 M NaOH at 37°C, 6.5 h, trace C) and deprotected afterwards (trace D). Fraction 1 contained the 5′-phosphorylated 3′-fragment of 6b arising from β,δ-elimination while fraction 2 contained the corresponding non-phophorylated fragment which is expected from strand scission via cyclophosphate formation. The MS spectrum of fraction 3 revealed four major signals from four different fragments which could be identified as the 3′-phosphorylated 5′-fragment of 6b together with that with the pending 4,5-dihydroxy-2-oxo-pentanal residue, arising from β,δ- and β-elimination, respectively. A second set of signals could be attributed to the same RNA fragment with the abasic ribose unit in its cyclophosphate and its hydrolyzed phosphate form. These products clearly arise from the hydrolytic strand scission pathway. Thus, fragment analysis clearly proves that both strand scission mechanisms are simultaneously active and contribute to the observed faster overall rate of strand cleavage as compared to 5b and 6a.
Table 3.
Fragment analysis of strand scission of 6b
| Nr | Rt (min) | Fragmenta | Formula | m/z calc. | m/z found |
|---|---|---|---|---|---|
| 1 | 10.3 | r(pUOMeUOMeC) | C29H40N7O24P3 | 963.60 | 963.26 |
| 2 | 13.6 | r(UOMeUOMeC) | C29H39N7O21P2 | 883.62 | 883.22 |
| 3 | 16.2 | r(AOMeGOMeGOMep) | C33H44N15O21P3 | 1079.73 | 1079.34 |
| r(AOMeGOMeGOMe-rb) | C38H52N15O25P3 | 1211.85 | 1211.51 | ||
| r(AOMeGOMeGOmeAScp) | C38H51N15O27P4 | 1273.81 | 1273.36 | ||
| r(AOMeGOMeGOMeASp) | C38H53N15O28P4 | 1291.83 | 1291.34 |
arb = 4,5-dihydroxy-2-oxo-pentanal, cp = 2′,3′-cyclophosphate, AS = abasic residue.
β,δ-elimination at a DNA abasic site
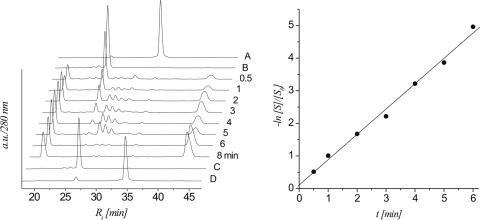
To compare the relative stability of the RNA abasic site with its DNA analogue, the abasic DNA heptamer 7b was prepared and subjected to basic degradation under the same conditions (0.1 M NaOH, 37°C). The mechanism and kinetics of decay were measured as described before (Figure 4).
Figure 4.
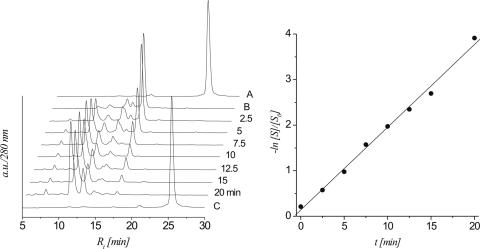
left: HPLC traces of strand cleavage via β,δ-elimination of 7b (0.1 M NaOH, 37°C) at different time intervals; controls: trace A: 7a; trace B: 7b; trace C: 7a after treatment with 0.1 M NaOH, 37°C for 20 min; right: linear fit (R2 > 0.996) of ln[S]/[S0] versus t calculated from the HPLC traces.
Oligodeoxynucleotide 7b rapidly degrades into at least one intermediate and two stable products with a T1/2 of only 4.1 min and a ks of 3.02 × 10−3 s−1. HPLC analysis indicated the d(pTTC) fragment (m/z = 915.15) to be the faster eluting and d(AGGp) (m/z = 989.38) to be the slower eluting fragment, both being the expected products of β,δ-elimination. We assume that the transiently formed intermediate corresponds to the 5′-DNA fragment after β-elimination with the pending 4,5-dihydroxypent-2-enal unit but we did not investigate this in further detail.
For an overview of abasic site decay under alkaline conditions, all measured first order rate constants and the corresponding half-life times were summarized in Table 4, together with krel, indicating relative differences in the rates of abasic site cleavage. It becomes immediately evident that β,δ-elimination at a DNA abasic site is 154-fold faster than at an RNA abasic site. This highlights the strong influence of the 2′-OH group on this mechanism for strand scission. Interestingly, strand cleavage via cyclophosphate formation is 3-fold faster compared to strand cleavage via β,δ-elimination in RNA. In the case of an unperturbed abasic site, where both mechanisms can contribute to strand clavage, the RNA abasic site shows ∼16-fold enhanced stability compared to a DNA abasic site.
Table 4.
Summary of the T1/2 and ks and krel data measured for alkaline induced strand scission of the abasic RNA and DNA heptamers
Aniline mediated strand cleavage at pH 4.6
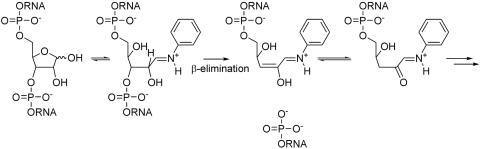
It is long known that strand cleavage of abasic DNA at physiological ionic strength and neutral to slightly acidic pH is greatly enhanced in the presence of amines (2). It was therefore of interest to investigate strand cleavage of abasic RNA in the presence of an amine and compare it to DNA. We have chosen aniline as a model amine because abasic RNA cleavage by this amine at pH 4.6 was used in the past for sequencing and site specific modification of RNA (10,18). A potentially simplified mechanism for the aniline mediated abasic RNA decay is given in Scheme 3.
Scheme 3.
Structurally and kinetically relevant intermediates in the aniline mediated strand scission at RNA abasic sites at slightly acidic pH.
It is beyond doubt that strand cleavage is initiated by immonium ion formation which renders the 2′-H base-labile and the 3′-phosphate unit prone to β-elimination. The intermediate enol strongly prefers the keto tautomeric form which renders δ-elimination that could in principle be initiated via vinylogous deprotonation of 4′-H slow. Given the fact that strand cleavage occurs in a slightly acidic medium, concomittant cyclophosphate formation, as observed under under alkaline conditions, is no longer obscuring β-elimination and for this reason the following experiments were performed with the 2′-unmodified abasic RNA oligonucleotide 4b. Again, abasic DNA 7b was used for comparison. The extent of cleavage was followed also in this case by HPLC (Figures 5 and 6). T1/2 and the apparent first order rate constant for strand scission ks were determined as described before. We found a T1/2 of 14 min and a ks of 8.56 × 10−4 s−1 for the abasic RNA 4b. The control experiments at neutral pH (Figure 5, trace B and C) clearly show that strand cleavage is much slower in the absence of aniline and essentially does not progress significantly over the time course of the experiment. The same analysis in the case of the DNA abasic oligomer 7b resulted in a T1/2 of 53 s and a ks of 1.29 × 10−2 s−1. Also, here strand cleavage in the absence of aniline was negligible over the time course of the experiment (Figure 6, trace B and C).
Figure 5.
left: HPLC traces of aniline-induced strand cleavage reaction of 4b at the given time intervals; controls: trace A: protected heptamer 4a; trace B: deprotected heptamer 4b; trace C: 4b after 60 min in 0.3 M TEAA buffer, pH 7.0 at 37°C in the absence of aniline; trace D: protected heptamer 4a after treatment with aniline for 60 min; right: linear fit (R2 > 0.997) of the ln[S]/[S0] versus t for 4b.
Figure 6.
left: HPLC traces of aniline-induced strand cleavage reaction of 7b at the given time intervals; controls: trace A: protected heptamer 7a; trace B: deprotected heptamer 7b; trace C: deprotected heptamer 7b after 10 min in 0.3 M TEAA buffer, pH 7.0 at 37°C in the absence of aniline; trace D: protected heptamer 7a after treatment with aniline for 10 min; right: linear fit (R2 > 0.996) of the ln[S]/[S0] versus t for 7b.
Fragment analysis in the case of aniline induced strand cleavage by ESI-MS proved more difficult than in the case of base induced cleavage for a yet unknown reason. We were unable to obtain masses for cleavage products in all isolated fractions. Those that could be identifed for 4b and 7b were listed in Table 5. In both cases, the trimers 3′ to the abasic sites, as expected from β-elimination, were identified. Interestingly, in the case of the deoxyoligonucleotide 7b, the 5′-fragment containing the absic residue bound to aniline as well as the aniline adduct of the intact abasic heptamer could be identified. This is a direct proof of the intermediacy of imines or hemiaminals in the cleavage mechanism.
Table 5.
Identified fragments of aniline induced cleavage at pH 4.6 of oligonucleotides 4b and 7b
| oligonucleotide | Rt (min) | Identified fragmentsa | Formula | m/z calc. | m/z found |
|---|---|---|---|---|---|
| 7b | 16.5 | r(UUC) | C27H35N7O21P2 | 855.6 | 855.1 |
| 20.3 | r(pUUC) | C27H36N7O24P3 | 953.5 | 935.1 | |
| 4b | 20.5 | d(pTTC) | C29H40N7O21P3 | 915.6 | 915.2 |
| 20.5 | d(TTC) | C29H39N7O18P2 | 835.6 | 835.2 | |
| 28.3–30.3 | d(AGG)-AS-NHC6H5 | C41H48N16O19P3 | 1161.9 | 1162.3 | |
| d(AGG-AS-NHC6H5-TTC) | C70H88N23O40P6 | 2077.4 | 2077.4 |
aAS-NHC6H5 = imine or hemiaminal between the abasic site and aniline.
From these experiments, it becomes clear that amines greatly accelerate strand cleavage near neutral pH also in abasic RNA. But again, abasic DNA is cleaved 15-fold faster. If one compares the ks values of cleavage via β-elimination in the presence of aniline with those obtained for alkaline cleavage it becomes clear that a DNA abasic site is only cleaved 4-fold faster while a RNA abasic site decays 42-fold faster. Thus the 2′-OH group seems to increase the β-elimination chemistry next to the immonium group.
DISCUSSION
We describe here the first direct comparison of the chemical stability of an RNA versus a DNA abasic site under basic conditions and under slightly acidic conditions in the presence of an amine. While under basic conditions no other mechanism than β-elimination is operative in DNA, the situation is more complex in the case of RNA, where cyclophosphate formation is acting in concert with β-elimination. In order to analyze the contribution of each mechanism to strand scission separately we used a 2′-OMe abasic site analogue, incapable of cyclophosphate formation, and a 1′-protected abasic RNA analogue incapable of β-elimination as well as a true abasic site within a 2′-OMe-oligoribonucleotide.
An initial concern was whether the 2′OMe-RNA abasic site analogue would be an adequate model reflecting correctly the β-elimination properties of a true RNA abasic site. This was a priori not clear as e.g. reversible, competitive deprotonation at 2′-OH or the shift of the keto function from C1′ to C2′ via a C1′–C2′dienolate could interfere with β-elimination kinetics (Figure 7).
Figure 7.
1′,2′-shift of keto function via a dienolate intermediate.
We compared therefore T1/2 of 6b with that calculated from the sum of the measured exponential decays of 6a and 5b. We found a theoretical T1/2 of 165 min which is 2.5-fold longer than that measured for 6b. From this we conclude that O2′-methylation slightly but not significantly retards β-elimination in abasic RNA which validates the O2′-methylated ribose unit as abasic site model. At the same time this comparison also validates the NPE protected abasic RNA unit with a free 2′-OH group to be a valid model. In a true RNA abasic site not only the hemiacetal form but also the open chain, aldehyde form can undergo strand cleavage via the cyclophosphate pathway. The above described comparison underlines that if this is the case there is either no significant difference in the velocity of cyclophosphate formation between the open chain and the furanose form, or the open chain form is kinetically not relevant. By comparing the relative rate of strand cleavage we noted that cleavage via cyclophosphate formation is ca 3-fold faster than cleavage by β-elimination. Extrapolated to longer RNA as, e.g. viral RNA this means that under basic conditions strand scission via cyclophosphate formation is clearly the dominant mechanism of decay even in the presence of an abasic site.
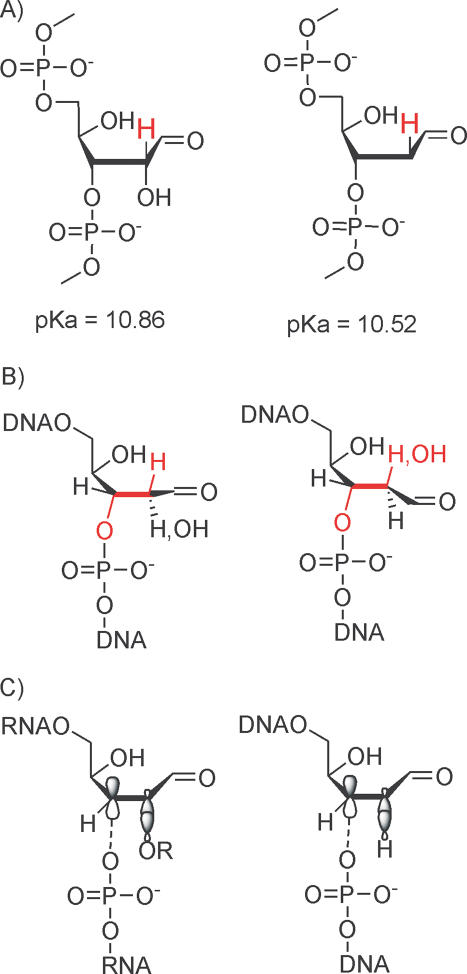
From the mechanistic point of view by far the most challenging fact is the roughly 150-fold higher stability of an RNA absic site towards β-elimination compared to DNA under alkaline conditions. Several factors could be responsible for this. A thermodynamic argument would invoke differences in the pKa of the C2′-H as a function of the presence or absence of an OH function. We calculated the C2′-H acidity of a DNA and RNA abasic site model using the SPARC calculator (19) and found only minor differences in pKa (Figure 8A). According to theory, the C2′-H of the RNA abasic site is slightly less acidic compared to DNA but the ΔpKa of only 0.34 can not explain the differences in reactivity. Also retardation of β-elimination via competitive, reversible deprotonation of the 2′-OH group can be excluded as the reason due to the fact that the corresponding 2′-OMe derivative shows slightly slower strand scission kinetics as compared to the non-methylated abasic residue. Also to be excluded are statistical effects resulting from 2 scissile C2′-H bonds in DNA compared to only one in RNA, as this certainly does not explain a more than 150-fold rate difference.
Figure 8.
(A) C2′–H acidities of an RNA and a DNA abasic site in the aldehyde form; (B) syn and anti conformers at the C2′–C3′–bond and implications on the concerted trans-β-elimination process for RNA and DNA; (C) Model of differential hyperconjugative assistance of C3′–O3′ bond dissociation by a C2′–H or C2′–OH σ-bond in a DNA and RNA abasic site.
The origin of the stability differences could in principle arise from differential stereoelectronic effects in a concerted trans-β-elimination mechanism (Figure 8B). The conformer for which less reactivity of RNA over DNA can be expected is that with the anti-arrangement of the C2′–C3′-bond (Figure 8B, right). Although steric arguments favor this conformation, stereoelectronic effects disfavor this arrangement in the case of RNA due to the anti-alignment of the C2′–O- and the C3′–O-bond, violating the gauche effect. Thus, there is no strong stereoelectronic argument in support of the higher reactivity of a DNA over RNA abasic site.
If one assumes that β-elimination follows a polar, non-concerted pathway, another hypothesis opens up. Under the basic conditions applied it seems likely that C2′–H dissociation is fast and reversible and therefore dissociation of the C3′–O3′ bond is rate determining. An important role has then to be attributed to the C2′–O2′ σ-bond in RNA which renders dissociation of the C3′–O3′ bond more difficult compared to the C2′–H bond in DNA due to less hyperconjugative stabilization of the transition state upon progressing C3′–O3′ bond dissociation (Figure 8C). We currently favor this hypothesis as it is in agreement with our experimental observations.
In the Schiff-base induced β-elimination at pH 4.6, the relative rates of cleavage between an RNA and a DNA abasic site are less different. Under these conditions, abasic RNA is still 15-fold more stable compared to abasic DNA. It may well be that under these conditions the deprotonation of the 2′C–H becomes kinetically more dominant compared to the C3′–O3′ bond dissociation in the overall β-elimination mechanism due to the increased difference of the pH of the medium compared to the pKa of the C2′–H. Thus the differences in kinetics between RNA and DNA decay are expected to correlate with the differences of the pKa of the respective C2′–H which is at least qualitatively the case. As a bottom line it appears that also under these more biorelevant conditions there is an advantage in stability of abasic RNA over abasic DNA.
The relative stability of abasic RNA may have interesting biological consequences for long lived RNAs. It is known that spontaneous generation of abasic RNA is slower compared to DNA (20), but once such lesions are formed, their potential biological effect is expected to be higher due to their relative longevity. Ribosome inactivating proteins (RIPs) are long known, but only recently it was found that class-I RIPs such as the pokeweed antiviral protein can not only depurinate the sarcin/ricin loop of the large rRNA but also specific adenine and guanine residues throughout the sequence of capped mRNAs (21) or other viral RNAs (22). This means that there exist not only spontaneous but also enzymatic pathways for abasic mRNA generation. It will therefore be of interest to study the effect of abasic RNA sites on the translation process in more detail.
Another open question associated with RNA abasic sites in long-lived RNA is whether they have any impact on the evolution of viral RNA. In preliminary experiments we studied the trans-lesion synthesis of an abasic RNA template/DNA-primer system by HIV-1 reverse transcriptase and found efficient insertion of purine bases opposite the lesion (12). Thus it is not excluded that abasic RNA also contributes to mutations in viral or retroviral RNA during replication or reversed transcription, in addition to the error rate of RNA dependent RNA polymerases (23) or reversed transcriptases (24).
Finally it will be of interest to find out whether there exists a primitive cellular repair system for abasic RNA. There is a growing list of genes involved in RNA damage control (25,26), and it is assumed that many of them are not yet discovered. There is also evidence that defects in the RNome stability might play a role in cancer formation. The study of mRNA biogenesis over the last years lead to the insight that there exists an elaborate RNA quality control mechanism involving, e.g. proteins that select aberrantly spliced mRNAs and mRNAs with premature termination codons (26). mRNAs are the working copies of the genomic information and a cell should therefore have a great investment in maintaining the integrity of its RNA. So, why should there not be a specific mechanism for dealing with abasic RNA? It could be as simple as just a mechanism for inactivating and metabolizing abasic RNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Financial support from the Swiss National Science Foundation (grant No. 200020-107692) is gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by Swiss National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T., Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972;11:3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- 3.Loeb L.A., Preston B.D. Mutagenesis by apurinic/apyrimidinic sites. Ann. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 4.Lhomme J., Constant J.F., Demeunynck M. Abasic DNA structure, reactivity and recognition. Biopolymers. 1999;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 5.Scharer O.D. Chemistry and biology of DNA repair. Angew. Chem. Int. Ed. Engl. 2003;42:2946–2974. doi: 10.1002/anie.200200523. [DOI] [PubMed] [Google Scholar]
- 6.Aas P.A., Otterlei M., Falnes P.O., Vagbo C.B., Skorpen F., Akbari M., Sundheim O., Bjoras M., Slupphaug G., Seeberg E., et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 7.Schramm V.L. Enzymatic N-riboside scission in RNA and RNA precursors. Curr. Opin. Chem. Biol. 1997;1:323–331. doi: 10.1016/s1367-5931(97)80069-5. [DOI] [PubMed] [Google Scholar]
- 8.Ogasawara T., Sawasaki T., Morishita R., Ozawa A., Madin K., Endo Y. A new class of enzyme acting on damaged ribosomes: ribosomal RNA apurinic site specific lyase found in wheat germ. EMBO J. 1999;18:6522–6531. doi: 10.1093/emboj/18.22.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozawa A., Sawasaki T., Takai K., Uchiumi T., Hori H., Endo Y. RALyase; a terminator of elongation function of depurinated ribosomes. FEBS Lett. 2003;555:455–458. doi: 10.1016/s0014-5793(03)01304-8. [DOI] [PubMed] [Google Scholar]
- 10.Nishikawa Y., Adams B.L., Hecht S.M. Chemical excision of apurinic acids from rna. a structurally modified yeast tRNAPhe. J. Am. Chem. Soc. 1982;104:326–328. [Google Scholar]
- 11.Trzupek J.D., Sheppard T.L. Photochemical generation of ribose abasic sites in RNA oligonucleotides. Org. Lett. 2005;7:1493–1496. doi: 10.1021/ol050120h. [DOI] [PubMed] [Google Scholar]
- 12.Kuepfer P.A., Leumann C.J. RNA abasic sites: preparation and trans-lesion synthesis by HIV-1 reverse transcriptase. Chem. Bio. Chem. 2005;6:1970–1973. doi: 10.1002/cbic.200500204. [DOI] [PubMed] [Google Scholar]
- 13.Strauss B.S. The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 14.Pitsch S., Weiss P.A., Wu X., Ackermann D., Honegger T. Fast and reliable automated synthesis of RNA and partially 2′-O protected precursors (‘caged RNA’) based on two novel orthogonal 2′-O protecting groups. Helv. Chim. Acta. 1999;82:1753–1761. [Google Scholar]
- 15.Wenter P., Furtig B., Hainard A., Schwalbe H., Pitsch S. Kinetics of photoinduced RNA refolding by real-time NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2005;44:2600–2603. doi: 10.1002/anie.200462724. [DOI] [PubMed] [Google Scholar]
- 16.Wenter P., Furtig B., Hainard A., Schwalbe H., Pitsch S. A caged uridine for the selective preparation of an RNA fold and determination of its refolding kinetics by real-time NMR. Chembiochem. 2006;7:417–420. doi: 10.1002/cbic.200500468. [DOI] [PubMed] [Google Scholar]
- 17.Heckel A., Mayer G. Light regulation of aptamer activity: an anti-thrombin aptamer with caged thymidine nucleobases. J. Am. Chem. Soc. 2005;127:822–823. doi: 10.1021/ja043285e. [DOI] [PubMed] [Google Scholar]
- 18.Rajbhandary U.L. Recent developments in methods for RNA sequencing using in vitro 32P-labeling. Fed. Proc. 1980;39:2815–2821. [PubMed] [Google Scholar]
- 19.Hillal S.H., Karickhoff S.W., Carreira L.A. A rigorous test for SPARC's chemical reactivity models: estimation of more than 4300 ionization pK(a)s. Quant. Struct. -Act. Relat. 1995;14:348–355. [Google Scholar]
- 20.Kochetkov N.K., Budovskii E.I. Hydrolysis of N-glycosidic bonds in nucleosides, nucleotides and their derivatives. In: Kochetkov N.K., Budovskii E.I., editors. Organic Chemistry of Nucleic Acids. New York: Plenum; 1972. pp. 425–448. [Google Scholar]
- 21.Hudak K.A., Bauman J.D., Tumer N.E. Pokeweed antiviral protein binds to the cap structure of eukaryotic mRNA and depurinates the mRNA downstream of the cap. RNA. 2002;8:1148–1159. doi: 10.1017/s1355838202026638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parikh B.A., Tumer N.E. Antiviral activity of ribosome inactivating proteins in medicine. Mini. Rev. Med. Chem. 2004;4:523–543. doi: 10.2174/1389557043403800. [DOI] [PubMed] [Google Scholar]
- 23.Castro C., Arnold J.J., Cameron C.E. Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res. 2005;107:141–149. doi: 10.1016/j.virusres.2004.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menendez-Arias L. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:91–147. doi: 10.1016/s0079-6603(02)71042-8. [DOI] [PubMed] [Google Scholar]
- 25.Bregeon D., Sarasin A. Hypothetical role of RNA damage avoidance in preventing human disease. Mutat. Res. 2005;577:293–302. doi: 10.1016/j.mrfmmm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Bellacosa A., Moss E.G. RNA repair: damage control. Curr. Biol. 2003;13:R482–R484. doi: 10.1016/s0960-9822(03)00408-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.