Abstract
The superhelicity of the chromosome, which is controlled by DNA topoisomerases, modulates global gene expression. Investigations of transcriptional responses to the modulation of gyrase function have identified two types of topoisomerase-mediated transcriptional responses: (i) steady-state changes elicited by a mutation in gyrase, such as the D82G mutation in GyrA, and (ii) dynamic changes elicited by the inhibition of gyrase. We hypothesize that the steady-state effects are due to the changes in biochemical properties of gyrase, whereas the dynamic effects are due to an imbalance between supercoiling and relaxation activities, which appears to be influenced by the RecA activity. Herein, we present biochemical evidence for hypothesized mechanisms. GyrA D82G gyrase exhibits a reduced supercoiling activity. The RecA protein can influence the balance between supercoiling and relaxation activities either by interfering with the activity of DNA gyrase or by facilitating the relaxation reaction. RecA has no effect on the supercoiling activity of gyrase but stimulates the relaxation activity of topoisomerase I. This stimulation is specific and requires formation of an active RecA filament. These results suggest that the functional interaction between RecA and topoisomerase I is responsible for RecA-mediated modulation of the relaxation-dependent transcriptional activity of the Escherichia coli chromosome.
INTRODUCTION
In prokaryotes, DNA is maintained in negatively supercoiled state, which is essential for various cellular processes. DNA topoisomerases are responsible for controlling the superhelicity of DNA (1,2). Thus, topoisomerases play critical roles in many aspects of DNA transaction, as well as in the maintenance of chromosome structure. There are four topoisomerases in Escherichia coli. Topoisomerase I (Topo I) (3) and topoisomerase III (Topo III) (4) are type IA enzymes; DNA gyrase (5) and topoisomerase IV (Topo IV) (6) are type IIA enzymes.
Type IA enzymes are monomeric proteins that can relax negative supercoils, and catenate and decatenate nicked or gapped, double-stranded circular DNA molecules (1,2). These enzymes require a single-stranded region to bind to DNA. E.coli Topo I is the prototype of type IA protein family (1,2). The N-terminal 582 amino acid residues correspond to the catalytic domain containing the active site Tyr at position 319. The catalytic domain is followed by a non-homologous carboxyl-terminal domain. The non-homologous carboxyl-terminal domains are involved in determining the distinct substrate specificities and catalytic properties of these enzymes (7,8).
Type IIA topoisomerases alter the linking number in steps of two by breaking both strands, passing another segment of the helix through the break and then resealing the broken strands (1,2). DNA gyrase and Topo IV consist of GyrA and GyrB subunits and ParC and ParE subunits, respectively. GyrA and ParC subunits catalyze strand-breakage and reunion reactions, whereas GyrB and ParE subunits hydrolyze ATP. The active forms of gyrase and Topo IV are an α2β2 tetramer; these topoisomerases bind double-stranded DNA. Despite the high degree of similarity between gyrase and Topo IV, these two enzymes display distinct cellular functions. Gyrase is the only topoisomerase that introduces negative supercoils into DNA, whereas Topo IV is responsible for decatenation of replicating DNA molecules (1,2,9).
E.coli RecA has DNA-dependent ATPase activity and ATP-dependent DNA binding activity (10,11). RecA proteins from various bacteria are highly conserved and homologues have been identified in eukaryotes as well. RecA catalyzes strand-exchange reactions and thus plays a central role in the homologous recombination process (11). In addition, RecA, when activated, causes proteolytic cleavage of the LexA repressor to trigger the induction of the SOS response (11). RecA is also required for mutagenic lesion bypass synthesis during the SOS response and is involved in replication restart (11,12). RecA monomers polymerize on DNA to form a nucleoprotein filament. RecA can bind to and form a nucleoprotein filament on either single- or double-stranded DNA, although nucleation on single-stranded DNA is much faster than that on double-stranded DNA (13–17). Both assembly and disassembly of RecA filaments take place in the 5′ to 3′ direction (14,15,18). In the presence of ATP, dATP or an ATP analogue, such as ATPγS, RecA forms an ‘active’ filament (19–21). The active RecA filament is a right-handed helical filament with six RecA monomers per turn and 3 nt per RecA monomer. When it is formed on double-stranded DNA, the DNA is underwound relative to the B-form helix (22,23). In contrast, RecA forms an ‘inactive’ filament in the absence of cofactor or presence of ADP (21,24).
The ability of DNA microarrays to monitor transcriptional activity of entire genomes has allowed an assessment of transcriptional and replication states of the E.coli chromosome following inhibition of DNA gyrase (25–27). These studies confirmed that treating cells with norfloxacin, a fluoroquinolone inhibitor of gyrase, affects transcription of a largenumber of genes in the genome. Quinolone treatment also causes the replication fork arrest and generation of double-strand breaks (28–30). A systematic analysis of transcriptional effects would not be possible, however, without accounting for a various cellular responses, such as the SOS response, DNA relaxation and replication inhibition (26,31–33). We modeled transcriptional responses to the quinolone-induced inhibition of gyrase in E.coli as a function of the downstream processes, including DNA repair, supercoiling and DNA replication (34). We found that relaxation by Topo I was the dominant factor behind the transcriptional response followed by the effects of DNA replication and RecA. We also found the correlation between the effect of relaxation and that of RecA.
Decrease in the extent of negative supercoiling of the chromosomal DNA is accompanied by characteristic changes in gene expression, exemplified by a compensatory upregulation of the gyrA and gyrB genes (35) and downregulation of the topA gene (36). We observed such characteristic changes under two conditions: steady-state changes during the normal exponential growth of a strain carrying the D82G mutation in the gyrA allele (37) and dynamic changes following the inhibition of DNA gyrase with a quinolone drug (34). Despite the differences between the two conditions: one representing normal growth, another—cell killing, transcriptional states of the cells were strikingly similar. Depending on growth medium, we found that the overlap between significantly differentially expressed genes in those conditions is 15–25%, and the probability of such an overlap occurring by chance is less than one in a million. We hypothesized that the steady-state effects were due to the changes in biochemical properties of the gyrase enzyme itself, while the dynamic effects were largely due the imbalance between supercoiling and relaxation activities in the cell. Interestingly, the imbalance between supercoiling and relaxation activities appeared to be modulated by the RecA activity (34).
In the present work, we conducted biochemical studies to determine the molecular basis of two types of topoisomerase-mediated genomewide transcriptional responses, as well as the effect of RecA on the balance between supercoiling and relaxation activities. We found that GyrA D82G gyrase exhibited a reduced activity to catalyze the supercoiling reaction and support DNA replication. We also found that RecA was capable of stimulating Topo I-catalyzed relaxation reaction. The stimulatory effect of E.coli RecA was specific to E.coli Topo I and required the formation of an active RecA filament. Thus, the functional interaction between RecA and Topo I is likely to be responsible for the RecA-mediated modulation of transcriptional responses in quinolone treated E.coli cells.
MATERIALS AND METHODS
DNAs and proteins
The recA gene was generated by PCR using E.coli MG1655 genomic DNA as a template and cloned into the pET-11c vector (38). DNA sequence of the open reading frame was confirmed by dideoxy DNA sequencing (data not shown). The RecA protein was expressed and purified according to protocols described previously (39,40). Purified RecA protein purchased from Roche Molecular Biochemicals (Indianapolis, IN) was also used.
Four E.coli topoisomerases were purified as described previously (41,42). Purified Staphylococcus aureus Topo I and Bacillus cereus Topo I were provided by Imogen Wildin and Michael Gwynn (GlaxoSmithKline), and Zhiyu Li and Russell DiGate (Philadelphia College of Pharmacy), respectively.
Mutant gyrA genes, gyrA D82G and gyrA S83W, were constructed using the overlap extension PCR technique (43) and cloned into the pET-11c vector (38). DNA sequences of open reading frames were confirmed by dideoxy DNA sequencing (data not shown). The GyrA D82G and GyrA S83W proteins were overexpressed in E.coli BL21(DE3) (38) and purified by the protocol used to purify the wild-type GyrA protein (41,42). Purified GyrA D82G and GyrA S83W were mixed with the wild-type GyrB to reconstitute GyrA D82G gyrase and GyrA S83W gyrase, respectively.
Supercoiling reaction
Covalently closed, negatively supercoiled double-stranded circular (form I) DNA of plasmid pBR322 (4361 bp) was purchased from New England Biolabs (Beverly, MA). Covalently closed, relaxed double-stranded circular (form I′) DNA was prepared by incubating pBR322 form I DNA with E.coli Topo I and used as a substrate in the supercoiling reaction.
Standard supercoiling reaction mixtures (12.5 μl) contained 50 mM Tris–HCl (pH8.0), 10 mM magnesium chloride, 100 mM potassium glutamate, 10 mM DTT, 50 μg/ml BSA, 1 mM ATP, 0.29 μg (100 fmol as molecule) pBR322 form I′ DNA, the indicated amounts (as tetramer) of either the wild-type or a mutant gyrase, and, when present, the indicated concentrations of norfloxacin, a quinolone drug, and reaction mixtures were incubated for 15 min at 37°C. Reactions were terminated by the addition of EDTA to 25 mM and the DNA products were analyzed by electrophoresis through vertical 1.2% agarose (Seakem; BioWhittaker Molecular Applications, Rockland, ME) gels (14 × 10 × 0.3 cm) at 2 V/cm for 15 h in a running buffer of 50 mM Tris–HCl (pH 7.9 at 23°C), 40 mM sodium acetate and 1 mM EDTA (TAE buffer). Gels were stained with ethidium bromide and photographed using an Eagle Eye II system (Stratagene, La Jolla, CA).
The following modifications were made in the supercoiling reaction mixtures to assess the effect of the RecA protein on the supercoiling activity of gyrase. Standard reaction mixtures (20 μl) contained 50 mM Tris–HCl (pH 8.0), 10 mM magnesium chloride, 100 mM potassium glutamate, 10 mM DTT, 50 μg/ml BSA, 5 mM ATP and 0.29 μg (100 fmol as molecule) pBR322 form I′ DNA. The indicated amounts of RecA were first bound to the DNA in a first stage incubation of 5 min at 37°C. The indicated amounts (as tetramer) of the wild-type gyrase were then added and reaction mixtures were incubated during the second stage for 15 min at 37°C. Reactions were terminated by adding EDTA to 25 mM and incubating at 37°C for 5 min. SDS and proteinase K were then added to 10% and 50 μg/ml, respectively, and the incubation was continued for an additional 15 min. The DNA products were purified by extraction of the reaction mixtures with phenol-chloroform (1:1, v/v) and analyzed by agarose gel electrophoresis as described above.
oriC DNA replication reaction
An oriC plasmid, pBROTB535 type I (44), was prepared as described previously (45). E.coli replication proteins, generous gifts of Kenneth Marians (Memorial Sloan-Kettering Cancer Center), were as described previously (44–48).
The replication of an oriC plasmid in vitro was performed as described previously (45). Briefly, a standard reaction mixture (12.5 μl) contained 40 mM Hepes–KOH (pH7.6), 10 mM magnesium acetate, 10 mM DTT, 100 μg/ml BSA, 2 mM ATP, 0.4 mM each GTP, CTP and UTP, 40 μM [α-32P]dATP (2000–3000 counts/min/pmol), 40 μM each dGTP, dCTP and TTP, 140 ng (35 fmol as molecule) of pBROTB plasmid DNA, 300 nM DnaA, 40 nM DnaB, 160 nM DnaC, 80 nM DnaG, 16 nM DNA polymerase III holoenzyme, 270 nM SSB, 40 nM HU, and the indicated amounts (as tetramer) of either the wild-type or a mutant gyrase. Reaction mixtures were assembled on ice and the reaction was started by adding DnaA. Incubation was for 10 min at 30°C and the reaction was terminated by the addition of EDTA to a final concentration of 25 mM. Gel electrophoretic analysis of replication products was performed as described previously (45).
Relaxation of negatively supercoiled plasmid DNA
The relaxation assay was performed in a manner similar to the supercoiling assay described in the previous section. Briefly, standard relaxation reaction mixtures (20 μl) contained 50 mM Tris–HCl (pH 8.0), 5 mM (1 mM for E.coli Topo III) magnesium chloride, 100 mM (400 mM for S.aureus Topo I) potassium glutamate, 10 mM DTT, 50 μg/ml BSA, 5 mM ATP (or other cofactors when indicated) and 0.29 μg (100 fmol as molecule) pBR322 form I DNA. The indicated amounts of the RecA protein were first bound to the DNA in a first stage incubation of 5 min at 37°C. The indicated amounts of E.coli Topo I (or other type IA topoisomerases when indicated) were then added and reaction mixtures were incubated during the second stage for 30 min at 37°C. Reaction products were processed, purified and analyzed as described in the previous section.
RESULTS
The D82G mutation in the GyrA subunit affects the supercoiling activity of DNA gyrase
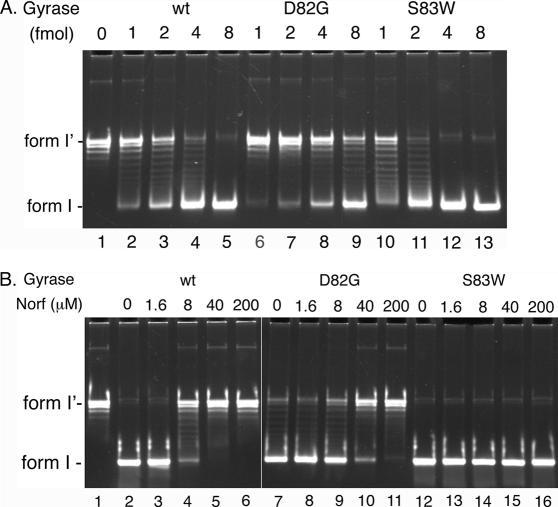
The D82G mutation in GyrA, which was previously identified as a part of a double mutation that confers quinolone resistance (49), caused the steady-state effects on the genomewide gene expression (37). To examine our hypothesis that the steady-state effects are due to the changes in biochemical properties of gyrase, we determined the biochemical properties of GyrA D82G gyrase. First, the effect of the D82G mutation on the supercoiling activity of gyrase was assessed (Figure 1A). Based on the amounts of form I DNA produced under these conditions, the supercoiling activity of GyrA D82G gyrase was 2–3 fold lower than that of the wild-type gyrase. In contrast, the S83W mutation, a commonly found quinolone resistance-conferring mutation at the conserved Ser in the GyrA subunit, did not affect the supercoiling activity of gyrase. In fact, the specific activity of GyrA S83W gyrase was slightly higher (2-fold or less) than that of the wild-type gyrase. We also measured the quinolone sensitivity of GyrA D82G gyrase in the supercoiling assay (Figure 1B). The D82G mutation conferred a low level of quinolone resistance to gyrase, whereas the S83W mutation conferred a high level of quinolone resistance to gyrase. These results suggested that a reduction of the supercoiling activity caused by the D82G mutation altered genomewide gene expression in E.coli.
Figure 1.
The D82G mutation in the GyrA subunit reduces the supercoiling activity of DNA gyrase. (A) The standard supercoiling reaction mixtures containing 100 fmol (as molecule) pBR322 form I′ DNA and the indicated amounts (as tetramer) of either the wild-type or a mutant gyrase were incubated and the DNA products were analyzed as described in Materials and Methods. (B) The standard supercoiling reaction mixtures containing 100 fmol (as molecule) pBR322 form I′ DNA, 10 fmol (as tetramer) of either the wild-type or a mutant gyrase, and the indicated concentrations of norfloxacin (Norf) were incubated and the DNA products were analyzed as described in Materials and Methods. Both experiments were repeated three times and virtually identical results were obtained. Representative results are shown. Lane 1 contains the substrate alone. wt, the wild-type gyrase; D82G, GyrA D82G gyrase; S83W, GyrA S83W gyrase.
Another transcriptional property of a strain carrying the D82G mutation is the constitutive induction of the dinP gene, encoding a Y-family DNA polymerase mediating translesion DNA synthesis (50). Since, despite the compensatory transcriptional changes in the mutant strain, the overall transcriptional state of the cells retained the signature of a suboptimal supercoiling activity, it seemed plausible that the constitutive activation of dinP might be reflecting an elevated steady-state level of DNA damage in the strain. One possible source of such damage could be the DNA replication itself, which may have a higher level of spontaneous arrests than in the wild type when elongation swivel, provided by DNA gyrase, is not fully effective (51). We examined the ability of the mutant gyrase to support DNA replication in vitro (Figure 2). GyrA D82G gyrase could not support oriC DNA replication reaction as efficiently as the wild-type gyrase could. These results indicated that the D82G mutation might affect the swivel activity of gyrase during chromosomal replication, which could elevate the steady-state level of DNA damage.
Figure 2.
GyrA D82G gyrase supports DNA replication less efficiently than the wild-type gyrase. The standard oriC replication reactions contained an oriC plasmid pBROTB535 type I (35 fmol as molecule = 420 pmol nt) as a template and the reaction was incubated at 30°C for 10 min. Incorporations of nt into acid-insoluble products were: 35 (lane 1), 43 (lane 1), 68 (lane 3), 115 (lane 4), 142 (lane 5), 44 (lane 6), 59 (lane 7), 88 (lane 8), 120 (lane 9), 51 (lane 10), 77 (lane 11), 132 (lane 12) and 154 (lane 13) pmol nt. LRI and ERI are late and early replication intermediates, respectively. Abbreviations were as in the legend to Figure 1.
RecA does not affect the supercoiling activity of gyrase
We observed that the average kinetics of transcriptional responses triggered by quinolone treatment was affected in a RecA-dependent manner (34). There seemed to be three possible mechanisms of the RecA effect. First, RecA can directly affect transcription of individual genes. Second, RecA can directly interfere with the supercoiling activity of gyrase. Third, RecA can directly facilitate the relaxation activity of Topo I. Since the RecA effect contributed to both accelerated induction and repression of transcription, we considered the first possibility to be unlikely, and we focused on a possible functional interaction(s) between RecA and a DNA topoisomerase(s).
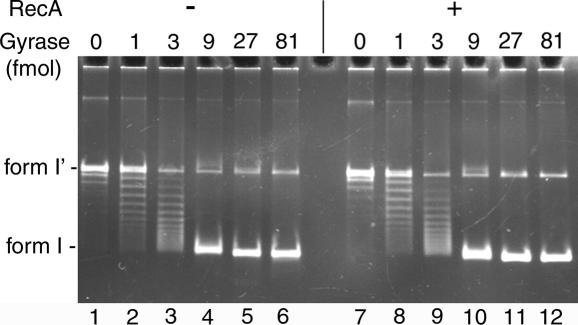
To examine a possibility that RecA could interfere with gyrase activity, we assessed the effect of RecA on gyrase-catalyzed supercoiling reaction in vitro. The supercoiling activity of gyrase was measured in the absence or presence of a saturated amount (3 nt per RecA monomer) of RecA (Figure 3). The presence of RecA did not affect the supercoiling activity of gyrase. We also varied the amounts of RecA and measured kinetics of the supercoiling reaction in the absence or presence of RecA. However, no effect of RecA on the supercoiling activity of gyrase was detected (data not shown). Thus, we could not find any evidence for the direct effect of gyrase on the RecA-mediated modulation of transcription in response to the quinolone treatment.
Figure 3.
RecA has no effect on the supercoiling activity of gyrase. The standard supercoiling reaction mixtures containing 100 fmol (as molecule) pBR322 form I′ DNA and the indicated amounts (as tetramer) of the wild-type gyrase were incubated in the absence (−) or presence (+) of 5.6 μg (3 nt per RecA monomer) of RecA and the DNA products were analyzed as described in Materials and Methods. Two independent experiments exhibited essentially identical results and representative results are shown.
RecA can stimulate the relaxation activity of Topo I
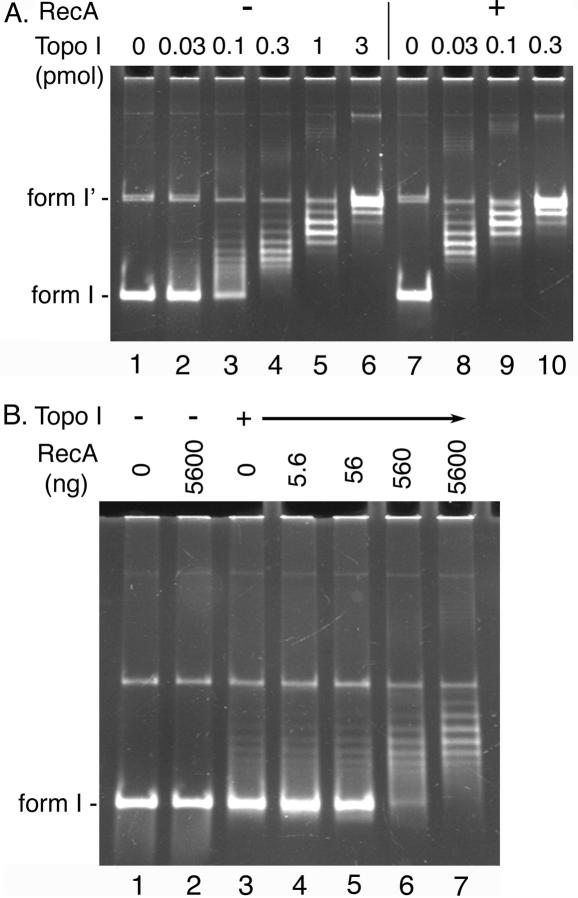
Next, we examined a possibility that RecA could facilitate the relaxation activity of Topo I. As shown in Figure 4, RecA was capable of stimulating the relaxation activity of Topo I. Based on the amount of Topo I required to completely relax 100 fmol of form I DNA, the relaxation activity of Topo I in the presence of RecA was about 10-fold higher than that in the absence of RecA (Figure 4A). The maximum level of the stimulation of the relaxation activity of Topo I by RecA required a saturated amount (3 nt per RecA monomer) of RecA (Figure 4B). Addition of higher amounts (up to 1 nt per RecA monomer) of RecA did not increase the level of stimulation any further (data not shown). These results suggested that RecA could modulate the balance, or imbalance, between supercoiling and relaxation activities in the cell by directly facilitating the relaxation activity of Topo I.
Figure 4.
RecA stimulates Topo I-catalyzed relaxation reaction. (A) The standard relaxation reaction mixtures containing 100 fmol (as molecule) pBR322 form I DNA and the indicated amounts of Topo I were incubated in the absence (−) or presence (+) of 5.6 μg (3 nt per RecA monomer) of RecA and the DNA products were analyzed as described in Materials and Methods. The assay was repeated three times and virtually identical results were obtained. Representative results are shown. (B) The standard relaxation reaction mixtures containing 100 fmol (as molecule) pBR322 form I DNA and either 0 fmol (−) or 33 fmol (+) of Topo I were incubated in the presence of the indicated amounts of RecA and the DNA products were analyzed as described in Materials and Methods. Two independent experiments showed essentially identical results and representative results are shown.
Active RecA filament formation is required for the stimulation of Topo I-catalyzed relaxation reaction
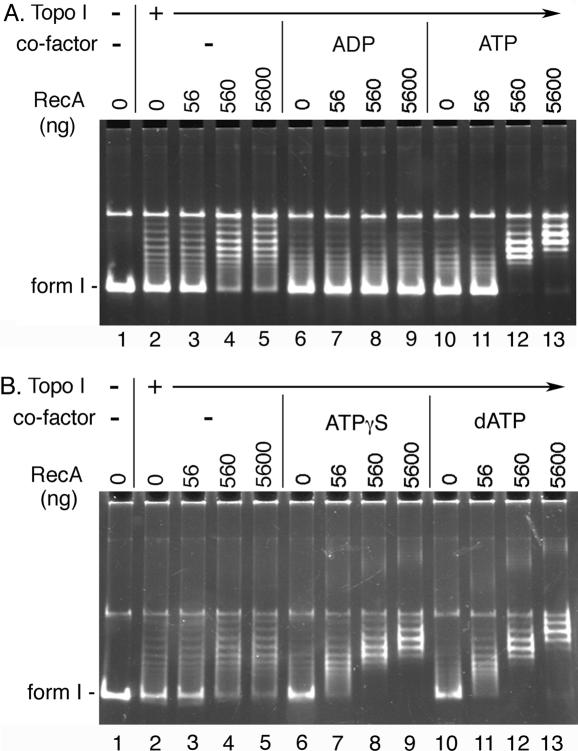
The RecA protein forms an active filament in the presence of ATP, dATP or ATPγS, whereas RecA forms an inactive filament in the absence of cofactor or in the presence of ADP (19–21,24). To determine the requirements for the stimulation of the relaxation activity of Topo I by RecA, we examine the effects of various cofactors on the functional interaction between RecA and Topo I.
A limited level of Topo I-catalyzed relaxation activity was detected in the absence of any cofactor (Figure 5). The presence of ATP was required for the RecA-mediated stimulation of the relaxation activity of Topo I (Figure 5A). In contrast, the presence of ADP exhibited an inhibitory effect (Figure 5A). RecA was also able to stimulate Topo I-catalyzed relaxation activity in the presence of either dATP or ATPγS (Figure 5B). These results suggested that active RecA filament formation was required for the stimulation of the relaxation activity of Topo I by RecA.
Figure 5.
Formation of active RecA filaments is required for the stimulation of Topo I-catalyzed relaxation activity by RecA. The standard relaxation reaction mixtures containing 100 fmol (as molecule) pBR322 form I DNA, 5 mM ADP, ATP (A), dATP, or ATPγS (B), as indicated, and either 0 fmol (−) or 100 fmol (+) of Topo I were incubated in the presence of the indicated amounts of RecA and the DNA products were analyzed as described in Materials and Methods.
E.coli RecA fails to stimulate the relaxation reaction catalyzed by either E.coli Topo III or S.aureus Topo I
Type IA topoisomerases, including E.coli Topo I, require a single-stranded region to bind to DNA (1,2). When an active RecA filament is formed on double-stranded DNA, the DNA becomes underwound (22,23). Thus, it seemed reasonable to argue that the RecA protein could stimulate the relaxation activity of Topo I by changing the conformation of DNA and generating single-stranded regions, binding sites for Topo I, in the negatively supercoiled plasmid DNA. In this case, RecA would have a general stimulatory effect on the relaxation activities of various type IA topoisomerases.
To directly examine this possibility, effects of the E.coli RecA protein on the relaxation activities of E.coli Topo III, S.aureus Topo I and B.cereus Topo I were assessed (Figure 6). RecA failed to stimulate either E.coli Topo III- or S.aureus Topo I-catalyzed relaxation reaction, although RecA was capable of slightly (about 2-fold) stimulating the relaxation activity of B.cereus Topo I. E.coli RecA did not exhibit a general stimulatory effect on type IA topoisomerases and its effect appeared to be specific to E.coli Topo I. Thus, RecA-induced conformational change of DNA alone could not explain the stimulatory effect of E.coli RecA on the relaxation activity of E.coli Topo I.
Figure 6.
E.coli RecA fails to stimulate the relaxation activity of either E.coli Topo III or S.aureus Topo I. The standard relaxation reaction mixtures containing 100 fmol (as molecule) pBR322 form I DNA, 5 mM ATP and the indicated amounts of E.coli Topo III (A), S.aureus (Sa) Topo I (B), or B.cereus (Bc) Topo I (C) were incubated in the absence (−) or presence (+) of 5.6 μg (3 nt per RecA monomer) of RecA and the DNA products were analyzed as described in Materials and Methods.
DISCUSSION
Investigations on the effects of the modulation of gyrase function on genomewide gene expression in E.coli have identified two types of topoisomerase-mediated genomewide transcriptional responses: steady-state changes elicited by a mutation in the GyrA subunit (37), and dynamic changes elicited by the inhibition of gyrase by a quinolone drug (34).We hypothesized that the steady-state effects were due to the changes in biochemical properties of gyrase, while the dynamic effects were due the imbalance between supercoiling and relaxation activities, which appeared to be modulated by the RecA activity. We conducted biochemical studies and provided evidence for these hypothesized mechanisms.
The D82G mutation in the GyrA subunit was selected by the screening of mutations, which increased levels of the gyrA and gyrB transcripts in E.coli (37). This mutation has been previously identified as a part of a double mutation in the E.coli gyrA gene that confers quinolone resistance (49). As expected, GyrA D82G gyrase exhibited a low level of resistance to norfloxacin, a quinolone drug, in the supercoiling (Figure 1B) and DNA cleavage (data not shown) assays. This mutation does not affect the growth rate in rich medium but alters the steady-state transcriptional activity of more than 800 genes in E.coli (37). We found that GyrA D82G gyrase had a reduced activity (2–3 fold lower than the wild-type enzyme) to catalyze the supercoiling reaction (Figure 1A) and support oriC DNA replication in vitro (Figure 2). Thus, the steady-state changes in the genomewide gene expression elicited by the D82G mutation in the GyrA subunit is due to its effect on the catalytic activity of DNA gyrase. The previous observation that small changes in the supercoiling activity of gyrase have widespread effects on the relative abundance of 88 proteins (52) supports our conclusion.
Our investigation on transcriptional responses to the inhibition of DNA gyrase in E.coli revealed that the average kinetics of transcriptional responses triggered by the quinolone treatment was affected in a RecA-dependent manner (34). The RecA protein could modulate the balance between supercoiling and relaxation activities either by interfering with the supercoiling activity of gyrase or by facilitating the relaxation activity of Topo I. To determine the molecular basis of the effect of RecA on the balance of supercoiling and relaxation activities in the cell, we assessed the effects of RecA on gyrase-catalyzed supercoiling reaction and Topo I-catalyzed relaxation reaction in vitro. The presence of RecA did not affect the supercoiling activity of gyrase (Figure 3). Furthermore, no effect of RecA on either the decatenation or relaxation activity of Topo IV was detected (data not shown). Thus, RecA does not affect the catalytic activities of bacterial type IIA topoisomerases.
RecA was capable of stimulating the relaxation activity of Topo I in vitro (Figure 4). The stimulation of Topo I-catalyzed relaxation reaction by RecA required formation of an active RecA filament (Figure 5). It is likely that RecA-mediated stimulation of Topo I activity accounts for the effect of a recA mutation on the balance between supercoiling and relaxation activities in E.coli. Topo I, in the presence of RecA, can relax the chromosomal DNA at its maximum rate upon the inhibition of gyrase in the wild-type strain, whereas, due to the loss of the stimulatory effect of RecA on Topo I, the relaxation of the chromosome is significantly slower in a recA− strain. A strong correlation between changes in transcript levels of a group of genes in a recA− strain and those in a topA− strain (34) also supports the conclusion that the functional interaction between RecA and Topo I is responsible for RecA-mediated modulation of the balance between supercoiling and relaxation activities in vivo.
What is the mechanism of the functional interaction between E.coli RecA and E.coli Topo I? One possibility is that the formation of an active RecA filament could alter the conformation of the DNA and generate single-stranded regions to provide binding sites for Topo I. An alternative possibility is that RecA might physically interact with Topo I and recruit Topo I to the DNA. It seemed more likely that RecA could stimulate Topo I by changing the conformation of DNA and generating single-stranded regions. In this case, it was reasonable to assume that RecA could have a general stimulatory effect on the activities of various type IA topoisomerases, which require a single-stranded region to bind to DNA (1,2). We found, however, that the stimulatory effect of E.coli RecA was somewhat specific to E.coli Topo I (Figure 6). Thus, conformational change of the DNA substrate alone may not explain the stimulation of the relaxation activity of E.coli Topo I by E.coli RecA.
The observed specificity of the effect of RecA on type IA topoisomerases supports a possible involvement of a protein–protein interaction between E.coli RecA and E.coli Topo I in RecA-mediated stimulation of Topo I activity. So far, we failed to detect any direct interaction between purified RecA and Topo I proteins in solution (data not shown) and the RecA–Topo I interaction was not detected in a systematic identification of protein–protein interactions in E.coli (53). Further studies are necessary to determine the molecular mechanism of the functional interaction between E.coli RecA and E.coli Topo I.
Acknowledgments
We thank Drs Imogen Wildin and Michael Gwynn for their gift of S.aureus Topo I, Drs Zhiyu Li and Russell DiGate for their gift of B.cereus Topo I, Dr Kenneth Marians for his gifts of replication proteins and comments on these studies, and Dr Lisa Oppegard for her critical reading of the manuscript. This work was supported in part by National Institutes of Health grants GM59465 (to H.H.) and GM66098 (to A.B.K.). Funding to pay the Open Access publication charges for this article was provided by the University of Minnesota Medical School.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Wang J.C. Interaction between DNA and an Escherichia coli protein ω. J. Mol. Biol. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 4.DiGate R.J., Marians K.J. Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 1989;263:13366–13373. [PubMed] [Google Scholar]
- 5.Gellert M., Mizuuchi K., O'Dea M.H., Nash H.A. DNA Gyrase: An Enzyme that Introduces Superhelical Turns into DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. New topoisomerase essential for chromosome segregation in E.coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H.L., DiGate R.J. The carboxyl-terminal residues of Escherichia coli DNA topoisomerase III are involved in substrate binding. J. Biol. Chem. 1994;269:9052–9059. [PubMed] [Google Scholar]
- 8.Zhang H.L., Malpure S., Li Z., Hiasa H., DiGate R.J. The role of the carboxyl-terminal amino acid residues in Escherichia coli DNA topoisomerase III-mediated catalysis. J. Biol. Chem. 1996;271:9039–9045. doi: 10.1074/jbc.271.15.9039. [DOI] [PubMed] [Google Scholar]
- 9.Adams D.E., Shekhtman E.M., Zechiedrich E.L., Schmid M.B., Cozzarelli N.R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 10.Roca A.I., Cox M.M. RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nucleic Acid Res. Mol. Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 11.Cox M.M. The bacterial RecA protein as a motor protein. Annu. Rev. Microbial. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- 12.Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 13.Register J.C., III, Griffith J. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem. 1985;260:12308–12312. [PubMed] [Google Scholar]
- 14.Lindsley J.E., Cox M.M. Assembly and disassembly of RecA protein filaments occurs at opposite filament ends: relationship to DNA strand exchange. J. Biol. Chem. 1990;265:9043–9054. [PubMed] [Google Scholar]
- 15.Shan Q., Bork J.M., Webb B.L., Inman R.B., Cox M.M. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- 16.Kowalczykowski S.C., Clow J., Krupp R.A. Properties of the duplex DNA-dependent ATPase activity of Escherichia coli RecA protein and its role in branch migration. Proc. Natl Acad. Sci. USA. 1987;84:3127–3131. doi: 10.1073/pnas.84.10.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pugh B.F., Cox M.M. General mechanism for RecA protein binding to duplex DNA. J. Mol. Biol. 1988;203:479–493. doi: 10.1016/0022-2836(88)90014-9. [DOI] [PubMed] [Google Scholar]
- 18.Bork J.M., Cox M.M., Inman R.B. RecA protein filaments disassemble in the 5′ to 3′ direction on single-stranded DNA. J. Biol. Chem. 2001;276:45740–45743. doi: 10.1074/jbc.M109247200. [DOI] [PubMed] [Google Scholar]
- 19.Di Capua E., Engel A., Stasiak A., Koller T. Characterization of complexes between RecA protein and duplex DNA by electron microscopy. J. Mol. Biol. 1982;157:87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- 20.Egelman E.H., Stasiak A. Structure of helical RecA–DNA complexes. Complexes formed in the presence of ATP-γ-S or ATP. J. Mol. Biol. 1986;191:677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- 21.Yu X., Egelman E.H. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J. Mol. Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- 22.Stasiak A., Di Capua E. The helicity of DNA in complexes with RecA protein. Nature. 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 23.Pugh B.F., Schutte B.C., Cox M.M. Extent of duplex DNA underwinding induced by RecA protein binding in the presence of ATP. J. Mol. Biol. 1989;205:487–492. doi: 10.1016/0022-2836(89)90219-2. [DOI] [PubMed] [Google Scholar]
- 24.Heuser J., Griffith J. Visualization of RecA protein and its complexes with DNA by quick-freeze/deep-etch electron microscopy. J. Mol. Biol. 1989;210:473–484. doi: 10.1016/0022-2836(89)90124-1. [DOI] [PubMed] [Google Scholar]
- 25.Peter B.J., Arsuaga J., Breier A.M., Khodursky A.B., Brown P.O., Cozzarelli N.R. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khodursky A.B., Peter B.J., Schmid M.B., DeRisi J., Botstein D., Brown P.O., Cozzarelli N.R. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc. Natl Acad. Sci. USA. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung K.J., Badarinarayana V., Selinger D.W., Janse D., Church G.M. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 2003;13:206–215. doi: 10.1101/gr.401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deitz W.H., Cook T.M., Goss W.A. Mechanism of action of nalidixic acid on Escherichia coli. III. Conditions required for lethality. J. Bacteriol. 1966;91:768–773. doi: 10.1128/jb.91.2.768-773.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snyder M., Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J. Mol. Biol. 1979;131:287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- 30.Pohlhaus J.R., Kreuzer K.N. Norfloxacin-induced DNA gyrase cleavage complexes block Escherichia coli replication forks, causing double-stranded breaks in vivo. Mol. Microbiol. 2005;56:1416–29. doi: 10.1111/j.1365-2958.2005.04638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiasa H., Yousef D.O., Marians K.J. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase–quinolone–DNA ternary complex. J. Biol. Chem. 1996;271:26424–26429. doi: 10.1074/jbc.271.42.26424. [DOI] [PubMed] [Google Scholar]
- 32.Zechiedrich E.L., Khodursky A.B., Bachellier S., Schneider R., Chen D., Lilley D.M.J., Cozzarelli N.R. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X., Malik M., Chan N., Drlica-Wagner A., Wang J.Y., Li X., Drlica K. Lethal action of quinolones against a temperature-sensitive dnaB replication mutant of Escherichia coli. Antimicrob. Agents Chemother. 2006;50:362–364. doi: 10.1128/AAC.50.1.362-364.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong K.S., Xie Y., Hiasa H., Khodursky A.B. Analysis of Pleiotropic Transcriptional Profiles: a Case Study of DNA Gyrase Inhibition. PLoS Genet. 2006;2:e152. doi: 10.1371/journal.pgen.0020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menzel R., Gellert M. Fusions of the Escherichia coli gyrA and gyrB control regions to the galactokinase gene are inducible by coumermycin treatment. J. Bacteriol. 1987;169:1272–1278. doi: 10.1128/jb.169.3.1272-1278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tse-Dinh Y.C., Beran R.K. Multiple promoters for transcription of the Escherichia coli DNA topoisomerase I gene and their regulation by DNA supercoiling. J. Mol. Biol. 1988;202:735–742. doi: 10.1016/0022-2836(88)90554-2. [DOI] [PubMed] [Google Scholar]
- 37.Jeong K.S., Ahn J., Khodursky A.B. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome Biol. 2004;5:R86. doi: 10.1186/gb-2004-5-11-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Studier W.F., Rosenberg A.H., Dunn J.J. Use of T7 RNA polymerase to direct the expression of cloned genes. Meth. Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 39.Lusetti S.L., Shaw J.J., Cox M.M. Magnesium Ion-dependent activation of the RecA protein involves the C-terminus. J. Biol. Chem. 2003;278:16381–16388. doi: 10.1074/jbc.M212916200. [DOI] [PubMed] [Google Scholar]
- 40.Lusetti S.L., Wood E.A., Fleming C.D., Modica M.J., Korth J., Abbott L., Dwyer D.W., Roca A.I., Inman R.B., Cox M.M. C-terminal deletions of the Escherichia coli RecA protein: characterization of in vivo and in vitro effects. J. Biol. Chem. 2003;278:16372–16380. doi: 10.1074/jbc.M212917200. [DOI] [PubMed] [Google Scholar]
- 41.Hiasa H., DiGate R.J., Marians K.J. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 1994;269:2093–2099. [PubMed] [Google Scholar]
- 42.Hiasa H., Shea M.E. DNA gyrase-mediated wrapping of the DNA strand is required for the replication fork arrest by the DNA gyrase–quinolone–DNA ternary complex. J. Biol. Chem. 2000;275:34780–34786. doi: 10.1074/jbc.M001608200. [DOI] [PubMed] [Google Scholar]
- 43.Morton R.M., Hunt H.D., Ho S.N., Pullen J.K., Pease L.R. Gene splicing by overlap extension. Meth. Enzymol. 1993;217:270–279. doi: 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- 44.Hiasa H., Marians K.J. Tus prevents over-replication of oriC plasmid DNA. J. Biol. Chem. 1994;269:26959–26968. [PubMed] [Google Scholar]
- 45.Hiasa H., Marians K.J. Primase couples leading- and lagging-strand DNA synthesis from oriC. J. Biol. Chem. 1994;269:6058–6063. [PubMed] [Google Scholar]
- 46.Minden J.S., Marians K.J. Replication of pBR322 DNA in vitro with purified proteins. Requirement for topoisomerase I in the maintenance of template specificity. J. Biol. Chem. 1985;260:9316–9325. [PubMed] [Google Scholar]
- 47.Parada C.A., Marians K.J. Mechanism of DnaA protein-dependent pBR322 DNA replication: DnaA protein-mediated trans-strand loading of the DnaB protein at the origin of pBR322 DNA. J. Biol. Chem. 1991;266:18895–18906. [PubMed] [Google Scholar]
- 48.Wu C.A., Zechner E.L., Marians K.J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork: I. Multiple effectors act to modulate Okazaki fragment size. J. Biol. Chem. 1992;267:4030–4044. [PubMed] [Google Scholar]
- 49.Truong Q.C., Van J.-C.N., Shlaes D., Gutmann L., Moreau N.J. A novel, double mutation in DNA gyrase A of Escherichia coli conferring resistance to quinolone antibiotics. Antimicrob. Agents Chemother. 1997;41:85–90. doi: 10.1128/aac.41.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohmori H., Friedberg E.C., Fuchs R.P.P., Goodman M.F., Hanaoka F., Hinkle D., Kunkel T.A., Lawrence C.W., Livneh Z., Nohmi T., Prakash L., Prakash S., Todo T., Walker G.C., Wang Z., Woodgate R. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 51.Grompone G., Ehrlich S.D., Michel B. Replication restart in gyrB Escherichia coli mutants. Mol. Microbiol. 2003;48:845–54. doi: 10.1046/j.1365-2958.2003.03480.x. [DOI] [PubMed] [Google Scholar]
- 52.Steck T.R., Franco R.J., Wang J.Y., Drlica K. Topoisomerase mutations affect the relative abundance of many Escherichia coli proteins. Mol. Microbiol. 1993;10:473–81. doi: 10.1111/j.1365-2958.1993.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 53.Butland G., Peregrín-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]