Abstract
In animals, most small nuclear RNAs (snRNAs) are synthesized by RNA polymerase II (Pol II), but U6 snRNA is synthesized by RNA polymerase III (Pol III). In Drosophila melanogaster, the promoters for the Pol II-transcribed snRNA genes consist of ∼21 bp PSEA and ∼8 bp PSEB. U6 genes utilize a PSEA but have a TATA box instead of the PSEB. The PSEAs of the two classes of genes bind the same protein complex, DmSNAPc. However, the PSEAs that recruit Pol II and Pol III differ in sequence at a few nucleotide positions that play an important role in determining RNA polymerase specificity. We have now performed a bioinformatic analysis to examine the conservation and divergence of the snRNA gene promoter elements in other species of insects. The 5′ half of the PSEA is well-conserved, but the 3′ half is divergent. Moreover, within each species positions exist where the PSEAs of the Pol III-transcribed genes differ from those of the Pol II-transcribed genes. Interestingly, the specific positions vary among species. Nevertheless, we speculate that these nucleotide differences within the 3′ half of the PSEA act similarly to induce conformational alterations in DNA-bound SNAPc that result in RNA polymerase specificity.
INTRODUCTION
The small nuclear RNAs (snRNAs) are involved in essential cellular functions such as pre-mRNA splicing, rRNA processing and histone mRNA 3′ end-formation (1–7). The genes that code for the snRNAs are unusual transcription units. Most of the snRNAs are synthesized by RNA polymerase II (Pol II), but some, such as U6 snRNA, are synthesized by RNA polymerase III (Pol III) (8–11). Despite this difference in RNA polymerase specificity, transcription of all the major snRNA genes in animals depends upon a unique promoter sequence termed the proximal sequence element (PSE) (8,9,12–27). The PSE is located in the 5′-flanking DNA with its center located ∼50–55 bp upstream of the transcription start site.
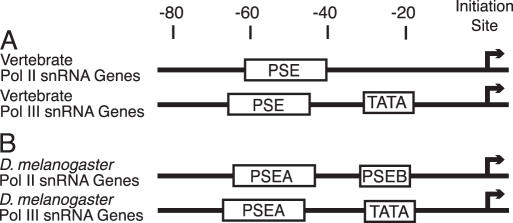
The transcription of animal snRNA genes has been studied most thoroughly in vertebrates (particularly the human system) and in the insect Drosophila melanogaster. A comparison of the organization of the vertebrate and D.melanogaster snRNA gene promoter elements is shown in Figure 1. The vertebrate genes (Figure 1A) that are transcribed by Pol II contain only a PSE and no other recognizably conserved promoter sequence. U6 genes, which are transcribed by Pol III, contain a PSE and a TATA box (8–11). In vertebrates, the TATA box acts as a dominant element for selection of Pol III and the PSEs of the Pol II- and Pol III-transcribed snRNA genes can be exchanged without affecting RNA polymerase specificity (11,28–30).
Figure 1.
Basal promoter elements of the snRNA genes of vertebrates (A) and of D.melanogaster (B). The approximate locations of the cis-acting elements conserved in the basal promoters of the snRNA genes of vertebrates and fruit flies are diagrammed. See the text for further details.
In the fruit fly D.melanogaster, both the Pol II (U1–U5) and Pol III (U6) snRNA gene promoters are bipartite (Figure 1B). The upstream PSEA is analogous to the vertebrate PSE. The fruit fly snRNA genes that are transcribed by Pol II contain also a conserved PSEB (25,31). Whereas mutation of the PSEA reduced U1 or U6 transcription to nearly negligible levels, mutation of the PSEB had only a 2- to 8-fold effect on transcription levels depending upon the method of measurement (25,32). The three D.melanogaster U6 genes each contain a TATA box, a feature that appears to be universally conserved among metazoan U6 genes. Mutation of the fruit fly TATA box reduced U6 transcription >30-fold (32).
Experiments have shown that the PSEA plays a predominant role in determining the RNA polymerase specificity of D.melanogaster snRNA genes. Fly U1 and U6 PSEAs, which differ at only a few nucleotide positions, are not functionally interchangeable. Each can support transcription only by Pol II or Pol III, respectively (33,34). The most important nucleotides involved in determining RNA polymerase specificity are those in the PSEA at positions 19 and 20. At these positions, different nucleotides are conserved in the PSEAs that recruit Pol III versus Pol II (33,34).
The protein that recognizes the PSE or PSEA has been best characterized in humans, fruit flies and trypanosomes. It is a multi-subunit transcription factor most often termed the snRNA activating protein complex (SNAPc) (35,36) but also known as PBP (37,38) or PTF (39,40). The human protein contains five distinct subunits (36,41–45), whereas only three subunits have been identified in fruit fly and trypanosome SNAPc (46–52). In accord with the names given to the human orthologs, the three subunits in D.melanogaster SNAPc have been named DmSNAP190, DmSNAP50 and DmSNAP43. High resolution protein–DNA photo-cross-linking experiments revealed that DmSNAP190 interacts with the entire length of the PSEA, but DmSNAP50 and DmSNAP43 interact only with the 3′ half of the PSEA (46,47). Moreover, the photo-cross-linking data indicate that the DmSNAPc binds in different conformations to the U1 versus U6 PSEAs. We have proposed a working model in which the U1 and U6 PSEAs act as differential allosteric effectors of DmSNAPc. We believe that when DmSNAPc binds in different conformations to Pol II or Pol III snRNA promoters, this leads to the differential recruitment of either the Pol II or Pol III general transcription factors and of the polymerases themselves (46,47).
To investigate the validity of this biochemical model within other species of insects across deep evolutionary time, we have carried out a comparative survey of the promoter sequences of the major spliceosomal Pol II- and Pol III-transcribed snRNA genes of six insect species for which complete or nearly complete genomic sequence is available. The species include two fruit flies (D.melanogaster and Drosophila virilis), two mosquitoes (Anopholes gambiae and Aedes aegypti), silkworm moth (Bombyx mori) and honeybee (Apis mellifera). The estimated divergence time of these organisms range from 40 to 60 million years ago for the two most closely related (D.melanogaster–D.virilis) to ∼400 million years ago for the most distantly related (honeybee versus the other insects).
The promoter comparisons have revealed interesting patterns of evolutionary conservation and divergence among the transcriptional regulatory sequences. Throughout the insecta, the 5′ half of the PSEA has remained highly conserved in sequence. The 3′ half of the PSEA is more free to evolve, but in all cases the data suggest that RNA polymerase specificity is determined by nucleotides in the 3′ half of the PSEA that are ‘conserved-to-be-different’ at specific positions in the Pol II and Pol III PSEAs. Interestingly, the specific nucleotide positions that are most important for making the determination of RNA polymerase specificity appear to vary from one species to another.
MATERIALS AND METHODS
Database searches and sequence alignments
D.melanogaster U1, U2, U4, U5, U6 and U6atac snRNA gene sequences were retrieved from the NCBI gene database. The D.melanogaster snRNA gene coding regions were used to retrieve similar sequences from the sequenced genomes of five other insect species (D.virilis, A.gambiae, A.aegypti, B.mori and A.mellifera). This was done by employing blastn to search FlyBase (53) (http://flybase.org/) for fly sequences, or the NCBI database for other insects. For each snRNA gene sequence, the hit with the highest sequence identity and lowest E-value was used as an inquiry sequence to re-BLAST the genome of the same organism. Potential genomic snRNA coding sequences were retrieved from the resulting BLAST ‘contig’ files using an in-house python script. The script extracted the sequences of interest (coding sequences and/or flanking sequences) using the coordinates produced for each BLAST hit.
Extracted sequences were aligned using MultAlin available at http://protein.toulouse.inra.fr/multalin/multalin.html. For each set of gene sequences, an alignment of the 5′-flanking DNA by MultAlin permitted the identification of the PSEA sequence of each of the genes. A second python script extracted a 57 or 61 bp sequence (for Pol II or Pol III-transcribed genes, respectively) that contained the PSEA and the PSEB or TATA box for each gene. These sequences were then aligned to maximize similarity among the promoter regions without gaps. Color-coding of the sequence alignments is explained in the figure legends.
Criteria for selection of snRNA genes
In many organisms, a large number of unexpressed pseudogenes exist that share partial sequence identity with the true snRNA genes. Such pseudogene sequences, if not eliminated from the analysis, would interfere with the interpretation of the data. Judgments were made manually based upon the following criteria. When sequence similarities were incomplete or truncated in the RNA-coding region, they were immediately eliminated from further consideration. Similarly, sequence identities that contained substantial internal insertions or deletions within the coding region were excluded from the analyses. If the coding region contained an excessive number of apparent point mutations, this led to a preliminary judgment that it was not likely to be a true gene. In those cases, examination of the 5′-flanking DNA usually failed to reveal an identifiable PSEA. Such sequences were judged to be pseudogenes and were eliminated from consideration.
In more rare instances, snRNA genomic similarities were identified that had several point mutations in the coding region and contained sequences in the 5′-flanking DNA that bore some, but less exact, resemblance to the PSEA and other promoter sequences of the true genes. In the absence of further data, these were considered to be homologs that were at one time true genes, but have evolved into pseudogenes or are becoming pseudogenes as a result of accumulating point mutations. It is possible that these sequences continue to be expressed at a low level, but even if that is true, they may have relatively ineffective promoter sequences. They were thus eliminated from consideration in order not to bias the results by including highly divergent sequences. Moreover, if a U6 homolog did not have a good TATA box sequence, it was considered to be a pseudogene. To broaden the number of Pol III snRNA gene promoters available for analysis, genes coding for U6atac, a U6-related snRNA present in a minor class of spliceosomes (54,55), were also included in the analyses. Although U6 and U6atac genes diverged early in metazoan evolution, they utilize similar promoter structures for the recruitment of Pol III.
In other rare cases, two apparently distinct copies of a true gene were identified that were identical or nearly identical in the entire 5′-proximal DNA. That is, similarity of 5′-flanking sequence was not restricted to the PSEA and other promoter elements. The identity or near identity of these sequences most likely arises from an evolutionarily recent gene duplication (or perhaps gene conversion) event. Therefore, only one copy was included in the promoter analysis in order to avoid bias from multiple representations of the same (or nearly the same) sequence. The primary goal of our analyses was to compare the sequences of snRNA gene promoter regions that had accumulated changes in non-essential nucleotides over a period of evolutionary time, whereas nucleotides important for promoter activity had been conserved.
The D.melanogaster snRNA genes have been annotated in FlyBase (53). The snRNA genes from the remaining insects were named numerically by us in the order that they appear in the figures. Accession information for the sequences shown in the figures is provided in the additional on-line material associated with this communication.
RESULTS AND DISCUSSION
D.melanogaster snRNA gene promoters
Pol II promoters
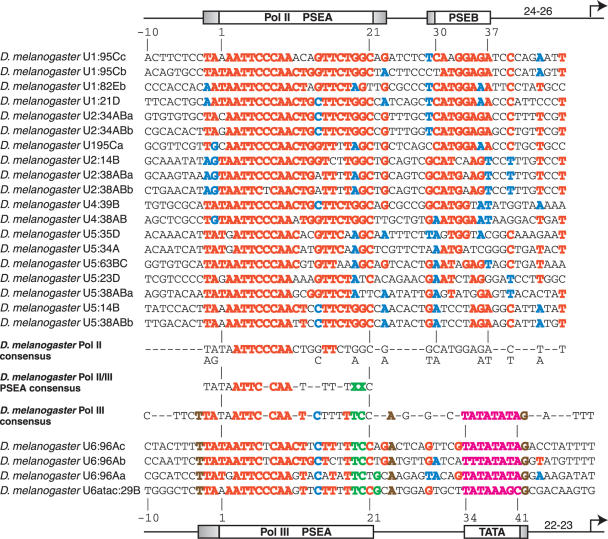
Nineteen U1, U2, U4 and U5 snRNA genes, evaluated to be true genes, were identified in the genome of the fruit fly D.melanogaster. An alignment of the promoter sequences of these 19 genes is shown in the upper section of Figure 2. From previous comparisons (25,31,33; S. Mount, personal communication) we have generally considered the D.melanogaster PSEA to be a 21 bp sequence extending between the nucleotides labeled in the figure as 1 and 21. Without altering the numbering system, the more comprehensive analysis in the current work reveals significant conservation beginning at position −2 (the second nucleotide 5′ of PSEA position 1). Position 23 is also fairly well conserved. This suggests that DmSNAPc may sequence-specifically recognize 23–25 bp of DNA in D.melanogaster snRNA gene Pol II promoters. The PSEA of these genes may therefore more properly be a 23–25 bp sequence, rather than a 21 bp sequence.
Figure 2.
Alignment and analysis of snRNA gene promoters of D.melanogaster. The sequences of the promoter regions of 19 D.melanogaster snRNA genes transcribed by RNA Pol II are shown in the upper section of the figure. The positions indicated as 1, 21, 30 and 37 indicate the extent of the PSEA and of the PSEB as identified in earlier work (25,31,33). The diagram at the top of the figure also indicates the boundaries of the PSEA and PSEB, but with the shaded areas indicating possible extensions identified as a result of the current work. Negative numbers are used to indicate nucleotides upstream of position 1 of the PSEA. The numbers on the diagram between the PSEB and the arrow at the right indicate the number of base pairs between the PSEB and the transcription start site. Red and blue colors indicate predominant nucleotides at a given position. A nucleotide is colored red if it is present at a given position in at least 50% of the sequences analyzed. Blue color is used to indicate the second most common nucleotide at a position but only if the following two conditions are met: (i) the nucleotide must be present in at least 25% of the sequences and (ii) a more common nucleotide (shown in red) must be present at the same position in at least 50% of the sequences. In the lower section of the figure, the promoters of three D.melanogaster U6 sequences and one U6-atac sequence are aligned and compared. In these Pol III promoter sequences, nucleotides are colored red if they are the same as a red-colored nucleotide in the Pol II sequences above. Similarly, nucleotides are colored blue if they are identical to a nucleotide colored blue in the Pol II sequences. Green color indicates nucleotides that are never found at the corresponding position in the Pol II sequences above. Brown colored nucleotides are 100% conserved in the Pol III sequences but are not well-conserved in the Pol II sequences. All nucleotides of the TATA box are in magenta color. The positions designated 1, 21, 34 and 41 indicate the extent of the PSEA and TATA box as identified in earlier work (33), as does the diagram at the bottom of the figure. The shaded areas represent possible extensions of these promoter elements based upon conservation of sequence. The numbers on the diagram between the TATA box and the arrow at the right indicate the number of base pairs between the TATA sequence and the transcription start site. Immediately below the Pol II gene sequences and above the Pol III gene sequences are shown consensus D.melanogaster snRNA promoter sequences for the Pol II- and Pol III-transcribed genes, respectively. In the Pol II sequence, consensus nucleotides are shown wherever there was color in the individual sequences above. In the consensus sequence, red color represents a very highly conserved nucleotide present in at least 90% of the individual Pol II promoter sequences. In the Pol III consensus promoter sequence, a consensus nucleotide is shown if present in at least three of the four individual sequences and is in color if it is conserved in all four individual sequences. Between the Pol II and Pol III consensus promoter sequences, a Pol II/III consensus PSEA is shown. A Pol II/III consensus nucleotide is shown only if it is present in both individual consensus promoter sequences. It is colored red only if red in both the Pol II and Pol III consensus sequences. The green upper case X's indicate positions where differences are always conserved in the Pol II and Pol III snRNA gene promoters.
In previous work, the PSEB was identified as a conserved 8 bp sequence located at positions 30–37 as numbered from the first base of the PSEA (25,32). The comparison shown in Figure 2 indicates there is significant conservation of 9 bp located between positions 29 and 37. Also, there are 3 nt positions downstream of the PSEB (positions 40, 44 and 47) that are conserved in most of the Pol II promoter sequences. In an earlier study (25), a cluster of point mutations encompassing these three nucleotides decreased in vitro transcription levels modestly (by 27%), so they probably represent nucleotides preferred for interaction with components of the Pol II general transcription machinery.
Pol III promoters
Three U6 genes and one U6-atac gene exist in the D.melanogaster genome, and an alignment of these four Pol III promoters is shown at the bottom of Figure 2. It should be noted that the color-coding in the Pol III promoter sequences is designed to emphasize both the similarities and the differences in these sequences as compared to the Pol II promoter sequences. Among these four genes, in addition to the conservation in the previously defined PSEA (positions 1–21) there is strong conservation of the sequence TTA immediately upstream of the PSEA (i.e. at positions −3 to −1 relative to position 1 of the canonical PSEA). The TATA box of each gene extends from position 34 to 41. In each case the TATA box is followed by a G nucleotide at position 42, whereas Gs are not abundant in the Pol II genes at this position. Thus, it is possible that this G may interact with components of the Pol III basal transcription machinery.
PSEA sequences and RNA polymerase specificity
Previous functional analyses indicated that nucleotides at positions 19 and 20 in D.melanogaster U1 and U6 PSEAs play an important role in determining the RNA polymerase specificity of these promoters. Figure 2 shows that T and C, respectively (shown in green) are absolutely conserved at these two positions in the four snRNA Pol III promoters analyzed. In stark contrast, these nucleotides are never found at these two positions in the 19 Pol II PSEAs; in these promoters position 19 is always a G or A, and position 20 is always a G or T. Other positions where specific nucleotides may favor Pol III specificity include G at position 22 (shown in green), T at position −3, A at position 24 (both shown in brown), and C at position 14 (shown in blue). In fact, experimental evidence indicates that a C at position 14 favors Pol III recruitment over Pol II in vitro (33). However, any effects of the nucleotides shown in blue and brown cannot be absolute since the Pol II promoters sometimes contain the same nucleotide at the corresponding position. Moreover, due to the relatively small number of Pol III snRNA promoters available for comparison, 100% identity at a position need not necessarily reflect a functional conservation within these more rare promoters.
Consensus sequences derived from the D.melanogaster Pol II and Pol III snRNA promoters are shown in the central region of Figure 2. Also shown in the center is a PSEA consensus sequence derived from the consensus Pol II and Pol III PSEAs. The 5′ half of the PSEA is highly conserved between the Pol II and Pol III promoters. The nucleotides shown in red (ATTC-CAA) are 100% conserved in all 23 D.melanogaster PSEAs and undoubtedly play an essential role in D.melanogaster snRNA transcription; most likely they are required for the high affinity binding of DmSNAPc. However, the Pol II and Pol III PSEAs diverge significantly in their 3′ halves. At least some of these nucleotides in the 3′ half of the PSEAs are known to play a role in determining RNA polymerase specificity, most importantly positions 19 and 20 denoted by green X's. Also, a C at position 14, although present in some Pol II PSEAs, favors Pol III specificity (33).
D.virilis snRNA gene promoters
Pol II promoters
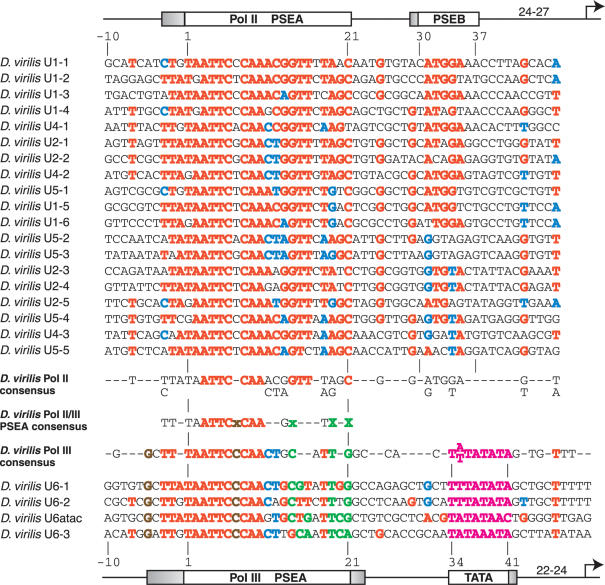
To examine which features of snRNA promoters are conserved at the nucleotide sequence level in other species of Drosophila, we next used sequence analysis tools to identify snRNA genes from the distantly related D.virilis. Nineteen D.virilis U1, U2, U4 and U5 snRNA gene promoters are compared in the upper section of Figure 3. When positions in the alignment were numbered to reflect the similarity to D.melanogaster snRNA gene promoters, the D.virilis PSEA exhibits strong conservation from position −3 to position 21, suggesting that a 24 bp sequence may comprise the Pol II PSEA in this organism. There is also a 7 bp cluster of sequence (G-ATGGA) that shares sequence similarity and location with the PSEB of D.melanogaster Pol II-transcribed snRNA genes, although the D.virilis PSEB appears to be less conserved than that of D.melanogaster.
Figure 3.
Alignment and analysis of snRNA gene promoters of D.virilis. Color-coding, symbolism and derivation of consensus sequences are as explained in the legend to Figure 2. Furthermore, a lower case green x indicates a position where the Pol III promoters most often contain a nucleotide that is never found at the corresponding position in the Pol II promoters. A lower case brown x indicates a fully conserved position in the Pol III PSEAs that is not conserved at the corresponding position in the Pol II PSEAs.
Pol III promoters
The lower part of Figure 3 shows an alignment of three U6 gene promoters and one U6atac gene promoter identified in the D.virilis genome. In these genes, there is extended conservation of PSEA sequence over a region of 26–28 bp. As in D.melanogaster, the TATA boxes of the genes extend between positions 34 and 41. A G is present immediately following the TATA box of three of the genes, similar to the conserved G found at this position in the four D.melanogaster genes.
PSEA sequences and RNA polymerase specificity
As in D.melanogaster, the D.virilis PSEAs that recruit Pol II and those that recruit Pol III have different nucleotides conserved at certain positions. The four PSEAs of the D.virilis U6 and U6-atac snRNA genes each contain a T at position 19, whereas the Pol II PSEAs always contain an A or G at this position. This is identical to the situation observed in D.melanogaster and strongly argues that this conserved difference at position 19 is involved in determining RNA polymerase specificity in D.virilis as well as in D.melanogaster.
Interestingly, it appears that position 21 in D.virilis has assumed the role played by position 20 in D.melanogaster. In the D.virilis PSEAs that recruit Pol II, position 21 is always a C (or in one case a T); in contrast, the Pol III PSEAs always have either an A or G at position 21. Based upon the conserved differences that exist at positions 19 and 21, it is likely that these two nucleotides play a critical role in the determination of RNA polymerase specificity in D.virilis. Furthermore, similar to D.melanogaster, a C at position 14 is the most common nucleotide at position 14 in D.virilis PSEAs that recruit Pol III, whereas a G is always present at this position in the PSEAs of genes transcribed by Pol II. Thus, a C at position 14 likely favors Pol III recruitment in both species.
In Figure 3, a Pol II consensus sequence is shown below the Pol II gene promoters and a Pol III consensus sequence is shown above the Pol III gene promoters. In the center of the figure is a consensus D.virilis PSEA derived from the two polymerase-specific PSEA sequences. The conservation of sequence is highest in the 5′ half of the PSEAs, while there is less conservation in the 3′ half, especially when cross-comparing the Pol II and Pol III PSEAs. The green X's represent positions where differences are conserved between the Pol II and Pol III PSEAs and therefore represent positions most likely involved in determining RNA polymerase specificity.
A.gambiae snRNA gene promoters
Pol II promoters
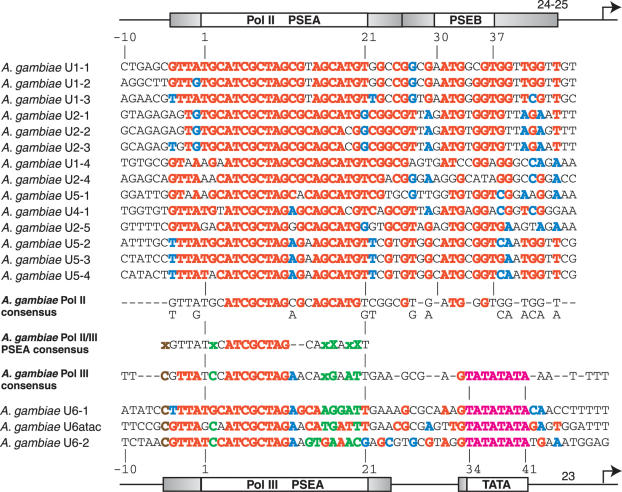
We wished to determine which features of snRNA gene promoters noted in Drosophila might be conserved in other insects, and which aspects were more free to evolve. Therefore, we examined the 5′-flanking sequences of the snRNA genes of two genera of mosquitoes, A.gambiae and A.aegypti. A comparison of the A.gambiae sequences from the Pol II-transcribed snRNA genes (Figure 4, upper section) reveals substantial conservation of sequence (as indicated by red and blue colored nucleotides) extending throughout the region from nucleotide −4 through position 45. (The numbering system employed aligns these sequences optimally with the Drosophila sequences in Figures 2 and 3). The most strongly conserved region extends from positions 4 through 20 as revealed by the red nucleotides in the Pol II consensus sequence shown below the individual sequences. Due to the significant conservation downstream of the PSEA, there is not a recognizable distinct block of nucleotides analogous to the Drosophila PSEB. However, there are three nucleotides (TG—G shown in red in the consensus sequence at positions 32, 33 and 36) that are well conserved at exactly the same positions as in the D.melanogaster promoter (Figure 2). This suggests a functional conservation of these nucleotides.
Figure 4.
Alignment and analysis of snRNA gene promoters of A.gambiae. Color-coding, symbolism and derivation of consensus sequences are as explained in the legends of Figures 2 and 3.
Pol III promoters
Only three A.gambiae Pol III promoters were available for alignment (Figure 4, bottom). As a result, care needs to be taken not to over-interpret the significance of nucleotides conserved in 2 of 3 sequences. Nonetheless, the Pol III PSEA exhibits strong conservation among the genes from position −5 to −1, as well as in the previously defined 21 bp PSEA. Weaker similarity extends beyond position 21. The spacing between the core PSEA sequence and the TATA box is precisely conserved with that found in Drosophila Pol III snRNA promoters. In all three A.gambiae sequences, the TATA box is preceded by a G nucleotide; a G is also highly conserved at the same position in the Pol II promoters. Whether this G has functional significance is an open question.
PSEA sequences and RNA polymerase specificity
In a pattern reminiscent of the findings with the Drosophila promoters, there are two positions in the A.gambiae Pol III PSEAs where nucleotides are always different from those found at the same positions in the Pol II PSEAs. At position 17, all A.gambiae Pol II PSEAs contain exclusively a C nucleotide, but the Pol III PSEAs have either a G or an A. At position 20, all Pol II PSEAs contain exclusively a G, but the Pol III PSEAs have either a T or a C. These positions are represented by the green capital X's in the A.gambiae consensus PSEA. Based upon our knowledge of D.melanogaster, these nucleotides very likely play a role in determining RNA polymerase specificity in A.gambiae. A conserved C nucleotide at position −5, an A nucleotide at position 12, as well as other nucleotides in the 3′ half of the PSEA (shown in green) may also favor Pol III recruitment. As noted in Drosophila, the Pol II and Pol III PSEAs are nearly identical in the 5′ half but diverge considerably in the 3′ half.
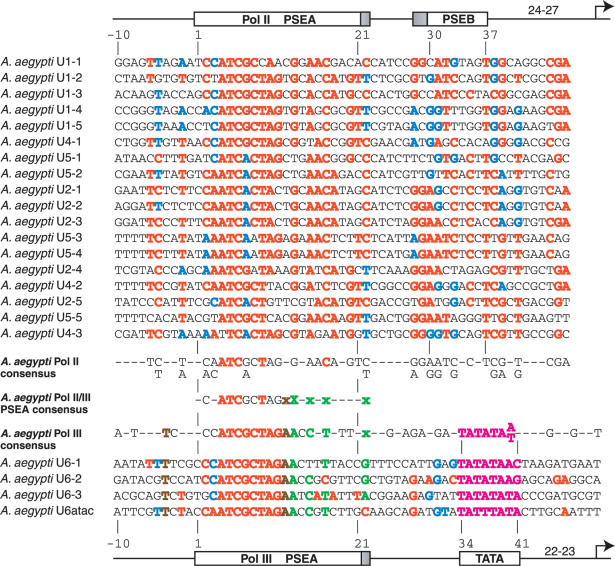
A.aegypti snRNA gene promoters
Pol II promoters
Eighteen Pol II snRNA promoters from A.aegypti are shown and compared in the upper section of Figure 5. The pattern of sequence conservation is quite different from that of A.gambiae. The 5′ half of the PSEA exhibits good conservation, but the 3′ half exhibits the most variation of any of the insects analyzed. Overall, the conserved area of the PSEA extends from position 2 to 22. Downstream of the PSEA, there is a block of conservation in a region that overlaps the PSEB of D.melanogaster.
Figure 5.
Alignment and analysis of snRNA gene promoters of A.aegypti. Color-coding, symbolism and derivation of consensus sequences are as explained in the legends of Figures 2 and 3.
Pol III promoters
The PSEAs of the Pol III genes are primarily conserved over a length of 19 bp from position 2 to position 20. As previously seen, the 3′ half of the PSEA is considerably less conserved than the 5′ half. The downstream TATA box is conserved at the same position it occupies relative to the PSEA in the A.gambiae and fly genes shown in Figures 2–4.
PSEA sequences and RNA polymerase specificity
At position 13, the A.aegypti PSEAs that recruit Pol II all contain a G (or rarely a C or T), but the PSEAs that recruit Pol III all contain an A nucleotide at position 13. Perfectly conserved differences are not found at any other position in this organism, but based upon the sequence patterns, three or four additional positions are strong candidates for contributing to polymerase specificity. Conserved differences predominate at positions 15, 17 and 22, as well as at position 12. These positions are denoted by the lower case green and brown x's in the Pol II/III consensus sequence shown in Figure 5.
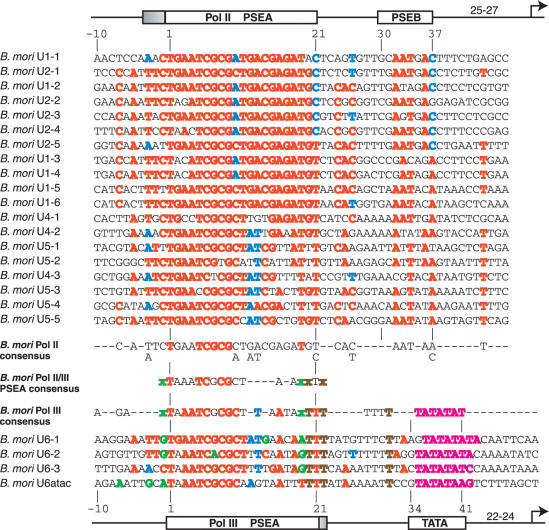
B.mori snRNA gene promoters
Pol II promoters
The B.mori PSEAs from the Pol II-transcribed snRNA genes are well conserved over a region of at least 24 bp from position −3 to position 21 (upper section of Figure 6). Also well-conserved are five nucleotides that lie at positions that correspond to the location of the D.melanogaster PSEB.
Figure 6.
Alignment and analysis of snRNA gene promoters of B.mori. Color-coding, symbolism and derivation of consensus sequences are as explained in the legends of Figures 2 and 3.
Pol III promoters. The PSEAs of the B.mori snRNA genes transcribed by Pol III are conserved from position 1 to position 22 (Figure 6, lower section). Interestingly, the location of the B.mori TATA box is different from those of the other insects. In three of the genes, the TATA box is displaced by one additional nucleotide farther away from the PSEA. In the other gene, a still additional nucleotide is present between the PSEA and the TATA box. This suggests that the interaction between SNAPc and TBP or TRF1 is slightly skewed in B.mori relative to the other insects.
PSEA sequences and RNA polymerase specificity
There are no positions that exhibit perfectly conserved differences between the Pol II and Pol III PSEAs in B.mori. However, several positions where differences predominate are likely to contribute to RNA polymerase specificity in this organism. These are positions 19, 20 and 22, and possibly position −1. These positions are denoted by lower case green and brown x's in the B.mori Pol II/III PSEA consensus sequence. It is also possible that a conserved T in the Pol III snRNA promoters at position 31 may contribute to Pol III specificity since a T at this position is extremely rare (1/19) in the Pol II PSEAs.
A.mellifera snRNA gene promoters
Pol II promoters
Conserved PSEA sequences of the honeybee snRNA genes transcribed by Pol II extend from position −1 to position 22 (Figure 7). There is also a block of well-conserved sequence (positions 32–38) that significantly overlaps the location occupied by the PSEB in D.melanogaster. Outside of these two regions, there is little conservation of sequence.
Figure 7.
Alignment and analysis of snRNA gene promoters of A.mellifera. Color-coding, symbolism and derivation of consensus sequences are as explained in the legends of Figures 2 and 3.
Pol III promoters
The PSEAs of the Pol III-transcribed snRNA genes of A.mellifera are highly conserved over only a 17 bp region from position 1 to 17 (Figure 7). In this insect, the separation of the TATA box from the PSEA is conserved at the same distance as observed for the two fruit flies and the two mosquitoes.
PSEA sequences and RNA polymerase specificity
In A.mellifera, position 16 of the PSEAs that recruit Pol II is nearly always conserved as an A (very rarely a G or T is present). In contrast, the same position in the A.mellifera Pol III gene PSEAs is always a C. This is the only position in the Pol II and Pol III PSEAs that exhibits perfectly conserved differences. We therefore expect that position 16 performs an important role in determining RNA polymerase specificity in the honeybee.
However, other positions are likely to contribute to the determination of RNA polymerase specificity in A.mellifera. For example, positions 14 and 17 are always G's in the PSEAs that recruit Pol III, whereas these positions are rarely occupied by G's in the Pol II gene PSEAs (Figure 7). A T at position 15 may also favor Pol III specificity whereas an A at this position likely favors Pol II recruitment. Finally, a pair of T's is conserved just upstream of the TATA box at positions 30 and 31 in the Pol III promoters, but not at these positions in the Pol II promoters. One of these, position 31, is the same position where a T was conserved in the B.mori Pol III-transcribed snRNA genes (Figure 6). This strengthens the possibility that this region just upstream of the TATA box may contribute to Pol III specificity in these two insects.
Inter-species comparison of insect snRNA gene promoters for Pol II
An alignment of the snRNA gene consensus Pol II promoter sequences for the six different insect species is shown in the upper part of Figure 8. A derived consensus sequence for insect Pol II snRNA promoters is shown below the sequences for the individual species. Overall, consensus nucleotides or pairs of nucleotides can be identified at 16 of 23 positions from −2 to 21 when the six species are compared. In the 5′ half of the PSEA, the consensus sequence is fairly rigid, but the 3′ half of the PSEA exhibits considerably more divergence.
Figure 8.
Interspecies comparison of insect snRNA gene Pol II and Pol III consensus promoter sequences. The consensus Pol II and Pol III promoter sequences for the individual species are repeated from Figures 2 to 7 with the color coding unchanged from those figures. Below the Pol II promoters and above the Pol III promoters, there are shown insect consensus Pol II and Pol III promoter sequences, respectively. These insect Pol II and Pol III consensus sequences were derived by the following criteria: a consensus nucleotide is shown if it is present in at least four of the six consensus sequences from the individual species. If a given position contains the same nucleotide in all six of the individual consensus sequences, it is colored red. Two nucleotides together were considered to form a consensus if together they are present in at least five of the six species. In the center of the figure, an overall insect PSEA consensus sequence is shown. The 5′ half of the PSEA is strongly conserved among the Pol II- and Pol III-transcribed snRNA genes of the insects. However, in the 3′ half of the PSEA, no positions of overall consensus are readily apparent. Superscripted x's indicate positions that are more conserved in the Pol II-transcribed snRNA genes and subscripted x's indicate positions that are more conserved in the Pol III-transcribed snRNA genes.
It is notable that the three nucleotides colored in red at positions 5, 6 and 8 of the Pol II consensus sequence are very stringently conserved. The T at position 5 is conserved in 100 of 101 Pol II PSEAs examined in this study. (Indeed, we cannot rule out the possibility that the single B.mori gene that contains a substitution may not be expressed or that a sequencing error may have occurred). Amazingly, the C at position 6 is conserved in all 101 Pol II PSEAs without exception. This strongly suggests that any mutation at position 6 is detrimental to gene activity. At position 8 in the PSEA, the C is conserved in 95 of 101 Pol II PSEAs.
Downstream of the PSEA in the Pol II promoters, nucleotides conserved across species occur at positions 29, 31, 32 and 37. These are within or near the area first identified as the PSEB in D.melanogaster. The conservation of sequence in this region argues that these nucleotides play a role in Pol II transcription of snRNA genes in other insects besides fruit flies.
Inter-species comparison of insect snRNA gene promoters for Pol III
The lower section of Figure 8 shows an alignment of the six consensus sequences for snRNA genes transcribed by Pol III, and a consensus sequence for insect Pol III snRNA promoters is shown above those six. The TATA box and the 5′ half of the PSEA is well conserved among the six species. Moreover, the 5′ half of the PSEA is highly conserved with the 5′ half of the PSEA of the Pol II snRNA genes. The most stringently conserved nucleotides are those at positions 5, 6 and 8 just as seen in the Pol II snRNA gene promoters. Each of these nucleotide positions is conserved in all 23 of the Pol III promoters analyzed in this study. Most likely they are essential (although not sufficient) for the high-affinity binding of insect SNAPc to both the Pol II and Pol III snRNA promoters.
As mentioned earlier, the spacing between the PSEA and TATA box is increased by 1 (or 2) bp in the B.mori promoters compared to the other five species.
Contribution of PSEA sequences to the RNA polymerase specificity of insect snRNA gene promoters
We have previously shown in D.melanogaster that the sequence of the PSEA plays an important role in determining the RNA polymerase specificity of the snRNA genes. This is primarily due to a few conserved sequence differences in the 3′ half of the PSEAs (33,34). Data from the five other insects is entirely consistent with this mechanism. In each species, the Pol III PSEAs commonly contain nucleotides that are never (or are very rarely) found at the corresponding positions in Pol II PSEAs. These positions are indicated by the green and brown colored letters in the six Pol III consensus sequences from the different species. Although conserved differences between the Pol II and Pol III PSEAs exist in every species, the exact positions of the conserved differences have changed during insect diversification.
When the insect Pol II and Pol III consensus PSEA sequences were compared and used to formulate a generalized insect PSEA, an interesting pattern emerged (center of Figure 8). As expected, nucleotide positions 1–10 are strongly conserved. However, from positions 12 to 21, each position that is more conserved in the Pol II PSEAs is less conserved in the Pol III PSEAs and vice versa. This is represented by the pattern of up-and-down x's in the 3′ half of the insect PSEA consensus sequence (center of Figure 8). Notably, positions 13, 15, 16, 18, 20 and 21 are more conserved in the Pol II PSEAs, whereas positions 12, 14, 17 and 19 are more conserved in the Pol III PSEAs. This suggests that within the insecta certain positions may be more important for Pol II transcription, and others could play a stronger role for Pol III transcription. Interestingly, these positions are interlaced with each other throughout the 3′ half of the PSEA.
Concluding remarks
Figure 9 shows an overall schematic summary of the results of this comparative study combined with previous empirical studies in D.melanogaster that examined the differential binding of DmSNAPc to U1 and U6 snRNA gene promoters. The comparative analysis of snRNA gene promoters indicates that the 5′ half of the PSEA (shaded in dark red in Figure 9) is highly conserved within the insecta phylogenetic class. The 3′ half of the PSEA is much less conserved among species as well as among Pol II- versus Pol III-transcribed snRNA genes. The light pink shading indicates nucleotide positions that are relatively more conserved across species in the Pol II-transcribed snRNA genes, and the light blue shading indicates positions that are more conserved in the Pol III-transcribed snRNA genes.
Figure 9.
Schematic working model of SNAPc–DNA interactions that lead to RNA polymerase specificity at insect snRNA gene promoters. The model is based upon the results of this study and previous work (32–34,46,47). The dark red shading indicates the 5′ half of the PSEA that is highly conserved in both the Pol II- and Pol III-transcribed snRNA gene promoters as well as across insect species. The pink and light blue boxes indicate nucleotide positions that are preferentially conserved in the Pol II and Pol III promoters, respectively. These exist in the 3′ half of the PSEAs as well as in the PSEB and TATA regions. The ovals represent the three conserved subunits of SNAPc drawn to indicate the regions of the DNA that are differentially contacted in the Pol II and Pol III snRNA gene promoters.
In previous work (46), site-specific protein-DNA photo-cross-linking studies revealed that the 5′ half of the U1 or U6 PSEA was contacted exclusively by DmSNAP190. In contrast, the 3′ half of the PSEA contacted all three subunits: DmSNAP43, DmSNAP50 and DmSNAP190 (46). Furthermore, the cross-linking pattern of DmSNAPc to the 5′ half of the Pol II and Pol III PSEAs was very similar, but the patterns of cross-linking to the 3′ halves of the Pol II and Pol III PSEAs exhibited significant differences (46). Those data suggested that DmSNAPc assumes different conformations when bound to U1 and U6 PSEAs. In Figure 9, the three subunits are drawn to reflect the region of DNA to which each subunit can be cross-linked when DmSNAPc binds to Pol II vs. Pol III snRNA gene promoters.
Overall, the data suggest that the DNA/SNAPc interactions in the 5′ half of the PSEA are highly conserved and have been relatively fixed at the molecular level during insect evolution. In contrast, those in the 3′ half are more pliable and are apparently involved in determining the RNA polymerase specificity of snRNA gene promoters by inducing alternative conformations in SNAPc (Figure 9). More than likely, differences in the 3′ half may also lead to species specificity for snRNA gene promoter activity.
The Pol II-transcribed genes of each species also possess a second conserved region of sequence near to where the PSEB was originally identified in D.melanogaster. However, the extent and the degree of conservation of the ‘PSEB’ can vary significantly among the different insects (Figures 2–8). Nevertheless, some of the nucleotides in this region are well-conserved across species (Figures 8 and 9). In D.melanogaster, this region of the U1 promoter is contacted by DmSNAP43 (32,47) and quite possibly by the Pol II general transcription machinery. Our analysis suggests the interaction of DmSNAP43 with the PSEB region is likely to be a general phenomenon among the insecta.
Empirical results from earlier studies indicated that the U1 and U6 PSEAs in D.melanogaster had different properties and that they were not interchangeable even though they were recognized by the same protein complex, DmSNAPc (33,34,46). Conserved nucleotide differences were noted between the PSEAs that recruit Pol II and those that recruit Pol III, and those differences were shown empirically to affect RNA polymerase specificity. The bioinformatic analyses reported here support the notion that similar mechanisms of determining RNA polymerase specificity are widely utilized throughout the insecta phylogenetic class of organisms. The pattern of conservation in the 3′ half of the PSEAs further suggests that some of the nucleotides in this region may play a stronger role for Pol II transcription and other nucleotides for Pol III transcription.
Supplementary Material
Acknowledgments
We thank Abraham Motola and Mun kyoung Kim for assistance with preparing the figures and Elizabeth Waters for a critical reading of the manuscript. This work was supported by National Science Foundation Grant MCB-0131151 (including an REU supplement to support G.H.) and in part by the California Metabolic Research Foundation. Funding to pay the Open Access publication charges for this article was provided by the California Metabolic Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sharp P.A. Split genes and RNA splicing. Cell. 1994;77:805–816. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 2.Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991;253:157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 3.Steitz J.A., Black D.L., Gerke V., Parker K.A., Kramer A., Frendewey D., Keller W., Birnsteil M.L. Functions of the Abundant U-snRNPs. Heidelberg, Federal Republic of Germany: Springer-Verlag KG; 1988. pp. 115–154. [Google Scholar]
- 4.Kass S., Tyc K., Steitz J.A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 5.Peculis B.A., Steitz J.A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 6.Birnstiel M.L., Schaufele F.J., Birnstiel M.L. Structure and Function of Minor snRNPs. Heidelberg, Federal Republic of Germany: Springer-Verlag KG; 1988. pp. 155–182. [Google Scholar]
- 7.Bond U.M., Yario T.A., Steitz J.A. Multiple processing-defective mutations in a mammalian histone pre-mRNA are supressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- 8.Dahlberg J.E., Lund E., Birnstiel M.L. The Genes and Transcription of the Major Small Nuclear RNAs. Heidelberg, Federal Republic of Germany: Springer-Verlag KG; 1988. pp. 38–70. [Google Scholar]
- 9.Parry H.D., Scherly D., Mattaj I.W. Snurpogenesis: the transcription and assembly of U snRNP components. Trends Biochem. Sci. 1989;14:15–19. [Google Scholar]
- 10.Hernandez N., McKnight S.L., Yamamoto K.R. Transcription of Vertebrate snRNA Genes and Related Genes. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 281–313. [Google Scholar]
- 11.Lobo S.M., Hernandez N.T., Conaway R.C., Conaway J.W. Transcription of snRNA Genes by RNA Polymerases II and III. New York: Raven Press; 1994. pp. 127–159. [Google Scholar]
- 12.Parry H.D., Tebb G., Mattaj I.W. The Xenopus U2 gene PSE is a single, compact, element required for transcription initiation and 3′ end formation. Nucleic Acids Res. 1989;17:3633–3644. doi: 10.1093/nar/17.10.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earley J.M., III, Roebuck K.A., Stumph W.E. Three linked chicken U1 RNA genes have limited flanking DNA sequence homologies that reveal potential regulatory signals. Nucleic Acids Res. 1984;12:7411–7421. doi: 10.1093/nar/12.19.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman M.L., Korf G.M., McNamara K.J., Stumph W.E. Structural and functional analysis of chicken U4 small nuclear RNA genes. Mol. Cell. Biol. 1986;6:3910–3919. doi: 10.1128/mcb.6.11.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris G.F., Price D.H., Marzluff W.F. Synthesis of U1 RNA in a DNA-dependent system from sea urchin embryos. Proc. Natl Acad. Sci. USA. 1986;83:3674–3678. doi: 10.1073/pnas.83.11.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazmaier M., Tebb G., Mattaj I.W. Functional characterization of X. laevis U5 snRNA genes. EMBO J. 1987;6:3071–3078. doi: 10.1002/j.1460-2075.1987.tb02614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNamara K.J., Walker R.J., Roebuck K.A., Stumph W.E. Transcription signals of a U4 small nuclear RNA gene. Nucleic Acids Res. 1987;15:9239–9254. doi: 10.1093/nar/15.22.9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Southgate C., Busslinger M. In vivo and in vitro expression of U7 snRNA genes: cis- and trans-acting elements required for RNA polymerase II-directed transcription. EMBO J. 1989;8:539–549. doi: 10.1002/j.1460-2075.1989.tb03408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNamara K.J., Stumph W.E. Site-directed mutational analysis of a U4 small nuclear RNA gene proximal sequence element: Localization and identification of functional nucleotides. J. Biol. Chem. 1990;265:9728–9731. [PubMed] [Google Scholar]
- 20.Das G., Henning D., Wright D., Reddy R. Upstream regulatory elements are necessary and sufficient for transcription of a U6 RNA gene by RNA polymerase III. EMBO J. 1988;7:503–512. doi: 10.1002/j.1460-2075.1988.tb02838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das G., Henning D., Reddy R. Structure, organization, and transcription of Drosophila U6 small nuclear RNA genes. J. Biol. Chem. 1987;262:1187–1193. [PubMed] [Google Scholar]
- 22.Gruber A., Soldati D., Burri M., Schümperli D. Isolation of an active gene and of two pseudogenes for mouse U7 small nuclear RNA. Biochim. Biophys. Acta. 1991;1088:151–154. doi: 10.1016/0167-4781(91)90167-k. [DOI] [PubMed] [Google Scholar]
- 23.Lescure A., Carbon P., Krol A. The different positioning of the proximal sequence element in the Xenopus RNA polymerase II and III snRNA promoters is a key determinant which confers RNA polymerase III specificity. Nucleic Acids Res. 1991;19:435–441. doi: 10.1093/nar/19.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wendelburg B.J., Marzluff W.F. Two promoter elements are necessary and sufficient for expression of the sea urchin U1 snRNA gene. Nucleic Acids Res. 1992;20:3743–3751. doi: 10.1093/nar/20.14.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zamrod Z., Tyree C.M., Song Y., Stumph W.E. In vitro transcription of a Drosophila U1 small nuclear RNA gene requires TATA box-binding protein and two proximal cis-acting elements with stringent spacing requirements. Mol. Cell. Biol. 1993;13:5918–5927. doi: 10.1128/mcb.13.9.5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J.M., Parsons R.A., Marzluff W.F. Transcription of the sea urchin U6 gene in vitro requires a TATA-like box, a proximal sequence element, and sea urchin USF, which binds an essential E box. Mol. Cell. Biol. 1994;14:2191–2200. doi: 10.1128/mcb.14.3.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J.M., Haberman R.P., Marzluff W.F. Common factors direct transcription through the proximal sequence elements (PSEs) of the embryonic sea urchin U1, U2, and U6 genes despite minimal sequence similarity among the PSEs. Mol. Cell. Biol. 1996;16:1275–1281. doi: 10.1128/mcb.16.3.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattaj I.W., Dathan N.A., Parry H.D., Carbon P., Krol A. Changing the RNA polymerase specificity of U snRNA gene promoters. Cell. 1988;55:435–442. doi: 10.1016/0092-8674(88)90029-3. [DOI] [PubMed] [Google Scholar]
- 29.Lobo S.M., Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez N. snRNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- 31.Lo P.C.H., Mount S.M. Drosophila melanogaster genes for U1 snRNA variants and their expression during development. Nucleic Acids Res. 1990;18:6971–6979. doi: 10.1093/nar/18.23.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai H.T., Chen H., Li C., McNamara-Schroeder K.J., Stumph W.E. The PSEA promoter element of the Drosophila U1 snRNA gene is sufficient to bring DmSNAPc into contact with 20 base pairs of downstream DNA. Nucleic Acids Res. 2005;33:6579–6586. doi: 10.1093/nar/gki972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen R.C., Wang Y., Hardin S.B., Stumph W.E. The proximal sequence element (PSE) plays a major role in establishing the RNA polymerase specificity of Drosophila U-snRNA genes. Nucleic Acids Res. 1998;26:616–622. doi: 10.1093/nar/26.2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNamara-Schroeder K.J., Hennessey R.F., Harding G.A., Jensen R.C., Stumph W.E. The Drosophila U1 and U6 gene proximal sequence elements act as important determinants of the RNA polymerase specificity of snRNA gene promoters in vitro and in vivo. J. Biol. Chem. 2001;276:31786–31792. doi: 10.1074/jbc.M101273200. [DOI] [PubMed] [Google Scholar]
- 35.Sadowski C.L., Henry R.W., Lobo S.M., Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 1993;7:1535–1548. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- 36.Henry R.W., Sadowski C.L., Kobayashi R., Hernandez N. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerases II and III. Nature. 1995;374:653–656. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- 37.Waldschmidt R., Wanandi I., Seifart K.H. Identification of transcription factors required for the expression of mammalian U6 genes in vitro. EMBO J. 1991;10:2595–2603. doi: 10.1002/j.1460-2075.1991.tb07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wanandi I., Waldschmidt R., Seifart K.H. Mammalian transcription factor PBP. Characterization of its binding properties to the proximal sequence element of U6 genes. J. Biol. Chem. 1993;268:6629–6640. [PubMed] [Google Scholar]
- 39.Murphy S., Yoon J.B., Gerster T., Roeder R.G. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol. Cell. Biol. 1992;12:3247–3261. doi: 10.1128/mcb.12.7.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon J.B., Murphy S., Bai L., Wang Z., Roeder R.G. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol. Cell. Biol. 1995;15:2019–2027. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai L., Wang Z.X., Yoon J.B., Roeder R.G. Cloning and characterization of the β subunit of human proximal sequence element-binding transcription factor and its involvement in transcription of small nuclear RNA genes by RNA polymerases II and III. Mol. Cell. Biol. 1996;16:5419–5426. doi: 10.1128/mcb.16.10.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon J.B., Roeder R.G. Cloning of two proximal sequence element-binding transcription factor subunits (γ and δ) that are required for transcription of small nuclear RNA genes by RNA polymerases II and III and interact with the TATA-binding protein. Mol. Cell. Biol. 1996;16:1–9. doi: 10.1128/mcb.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henry R.W., Ma B.C., Sadowski C.L., Kobayashi R., Hernandez N. Cloning and characterization of SNAP50, a subunit of the snRNA-activating protein complex SNAPc. EMBO J. 1996;15:7129–7136. [PMC free article] [PubMed] [Google Scholar]
- 44.Wong M.W., Henry R.W., Ma B.C., Kobayashi R., Klages N., Matthias P., Strubin M., Hernandez N. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol. Cell. Biol. 1998;18:368–377. doi: 10.1128/mcb.18.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henry R.W., Mittal V., Ma B.C., Kobayashi R., Hernandez N. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev. 1998;12:2664–2672. doi: 10.1101/gad.12.17.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Stumph W.E. Identification and topological arrangement of Drosophila proximal sequence element (PSE)-binding protein subunits that contact the PSEs of U1 and U6 snRNA genes. Mol. Cell. Biol. 1998;18:1570–1579. doi: 10.1128/mcb.18.3.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li C., Harding G.A., Parise J., McNamara-Schroeder K.J., Stumph W.E. Architectural arrangement of cloned proximal sequence element-binding protein subunits on Drosophila U1 and U6 snRNA gene promoters. Mol. Cell. Biol. 2004;24:1897–1906. doi: 10.1128/MCB.24.5.1897-1906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das A., Bellofatto V. RNA polymerase II-dependent transcription in trypanosomes is associated with a SNAP complex-like transcription factor. Proc. Natl Acad. Sci. USA. 2003;100:80–85. doi: 10.1073/pnas.262609399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das A., Zhang Q., Palenchar J.B., Chatterjee B., Cross G.A., Bellofatto V. Trypanosomal TBP functions with the multisubunit transcription factor tSNAP to direct spliced-leader RNA gene expression. Mol. Cell. Biol. 2005;25:7314–7322. doi: 10.1128/MCB.25.16.7314-7322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schimanski B., Laufer G., Gontcharova L., Gunzl A. The Trypanosoma brucei spliced leader RNA and rRNA gene promoters have interchangeable TbSNAP50-binding elements. Nucleic Acids Res. 2004;32:700–709. doi: 10.1093/nar/gkh231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schimanski B., Nguyen T.N., Gunzl A. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei. Mol. Cell. Biol. 2005;25:7303–7313. doi: 10.1128/MCB.25.16.7303-7313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schimanski B., Nguyen T.N., Gunzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell. 2005;4:1942–1950. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grumbling G., Strelets V., The Flybase Consortium FlyBase: anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarn W.Y., Steitz J.A. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 55.Mount S.M., Salz H.K. Pre-messenger RNA processing factors in the Drosophila genome. J. Cell. Biol. 2000;150:F37–F44. doi: 10.1083/jcb.150.2.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.