Abstract
The Rat Genome Database (RGD, http://rgd.mcw.edu) is one of the core resources for rat genomics and recent developments have focused on providing support for disease-based research using the rat model. Recognizing the importance of the rat as a disease model we have employed targeted curation strategies to curate genes, QTL and strain data for neurological and cardiovascular disease areas. This work has centered on rat but also includes data for mouse and human to create ‘disease portals’ that provide a unified view of the genes, QTL and strain models for these diseases across the three species. The disease curation efforts combined with normal curation activities have served to greatly increase the content of the database, particularly for biological information, including gene ontology, disease, pathway and phenotype ontology annotations. In addition to improving the features and database content, community outreach has been expanded to demonstrate how investigators can leverage the resources at RGD to facilitate their research and to elicit suggestions and needs for future developments. We have published a number of papers that provide additional information on the ontology annotations and the tools at RGD for data mining and analysis to better enable researchers to fully utilize the database.
INTRODUCTION
The Rat Genome Database (RGD, http://rgd.mcw.edu) is the model organism database for the laboratory rat, Rattus norvegicus. The primary goal of the database is to provide convenient access to high quality data about the genes and genome of the Rat and to support the goals of researchers using the rat as a genetic and genomic model [see (1) for a review of the impact of genomics in rat research]. In pursuit of these goals we curate a variety of other relevant information such as mapping information (genetic maps, radiation hybrid maps and most recently the genome assembly), quantitative trait loci (regions of the rat genome believed to contain a gene or genes related to a particular phenotype or trait), rat strains used in genetic and genomic experiments and the microsatellite markers used to genotype inbred rat strains. This data is acquired from a variety of sources: the literature, other scientific databases and through bioinformatic analyses performed locally. The public website provides convenient access to this data through search tools, web page reports and more specialized bioinformatics tools that can be used to do novel analyses directly online. Data curated at RGD is also available through other global genomics resources such as NCBI/Entrez Genes (2) and on the Ensembl (3) and UCSC genome browsers (4). In addition, our data is freely available as FTP files to enable anyone to use it in their research or analysis.
A resource like RGD evolves in parallel with its community and for a model organism database, this community includes the researchers using that model organism and also the broader genomics and bioinformatics communities wishing to access and use this data. One of the most significant trends in recent years that has greatly influenced the database has been the use of biological ontologies, of which the Gene Ontology (5) is perhaps the most well known. By providing a set of standardized terms, definitions and relationships between the terms, ontologies are very convenient for researchers as a way to consistently annotate and categorize data. For the bioinformatics community they are emerging as a way to standardize data representations, enabling cross-organism data mining and analysis and have opened up avenues for novel data representation and integration (6,7). Another significant trend facing anyone using online resources and one that is particularly acute for modern biologists is the massive increase in information available (6). The problem ranges from not finding enough data to finding too much and being unable to efficiently comprehend what is available. Driven by the needs of our community we have been exploring ways to utilize the descriptive power of our ontology annotations combined with innovative visualization tools and web designs to begin to address this problem.
DATABASE CONTENTS
For a number of years we have been using a variety of biological ontologies as annotation tools to organize and classify the data we curate. These include the Gene Ontology (5), the Mammalian Phenotype ontology (8), a disease ontology that we developed from MeSH disease terms and a Pathway ontology created at RGD. These have provided a comprehensive platform with which to annotate genes, QTL and strains and provide a snapshot of the most relevant information from the molecular level to that of the whole organism. As of September 2006 RGD has entries for 23 599 genes, 746 rat strains, 1093 rat QTL and 535 human QTL. These in turn have been annotated with ontology terms, giving rise to 92 137 GO annotations, 2990 disease annotations, 1756 pathway annotations and 8242 phenotype annotations.
The increasing numbers of annotations and other data has necessitated improvements to the layout of our gene report page. Information has been organized into sections that provide brief summaries of the gene or QTL with links to more comprehensive information. The ontology annotation section was combined with the free text notes into a functional annotation block, providing a single location for this information. The technical details of each annotation such as evidence codes, ontology identifiers and reference links were moved to a new detail page to reduce the clutter on the main gene report. A similar approach was taken to move the lists of literature references and sequence identifiers from the main page to separate detail pages. Other improvements to the gene report included providing SNP and synteny tracks on the genome image for a gene and more comprehensive external database links.
DISEASE PORTALS
The major new addition to the RGD site in the past year was guided by three user-centric goals—(i) providing easy access to data related to diseases, (ii) allowing multiple perspectives of RGD data according to the needs of the user, and (iii) presenting a broader overview of data and allowing users to zoom in, filter the data and then drill down to the details as required.
Analysis of rat publications and trends in rat research demonstrate that much rat research is done in the context of disease-related studies. This is borne out by the types of grants that are funded and the types of searches that are undertaken on RGD. Figure 1 shows a tag cloud view of a typical month's top searches on RGD. The search keywords of interest to our users are primarily disease terms, centered on cardiovascular, autoimmune and neurological disease areas. Based on this demonstrated need, we have introduced a variety of disease-centric resources to the database.
Figure 1.
Top search terms from the RGD web logs (June 1–July 4, 2006). The individual search terms are shown with the number of times that term was searched shown in parentheses. Font sizes are proportional to search frequency.
A general disease portal page was released to enable ‘one-click’ access to some of the most popular disease areas (http://rgd.mcw.edu/tools/diseases/disease_search.cgi). For each disease this provides preconfigured links into our ontology annotation tools and the genome browser to enable users to quickly find gene lists and see genomic locations for genes and QTLs.
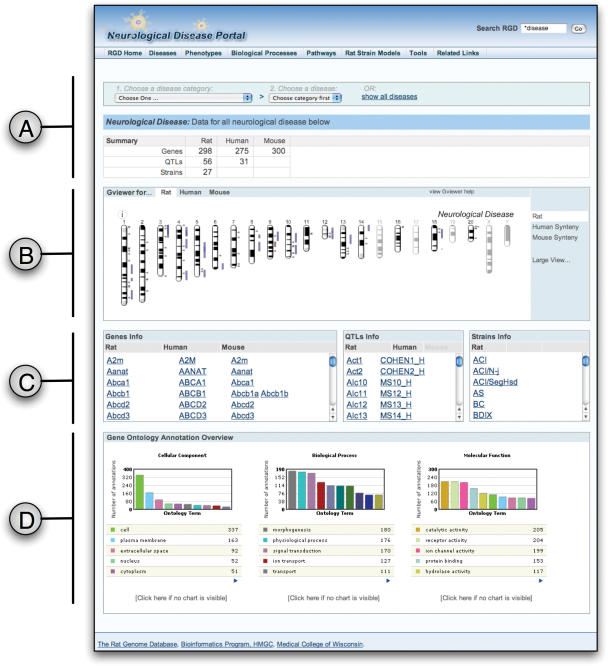
Building from this, we identified a subset of diseases that were clearly of high interest to the research community and developed a two-pronged approach to providing enhanced support for research in these areas. For each disease area, two methods were utilized to identify data for inclusion. Disease-related genes were identified from existing sources such as Online Mendelian Inheritance in Man (OMIM) (9), the Genetic Association Database (10), GeneCards (11) and NCBI's Genes and Disease database. In addition, genes at the Mouse Genome Database (MGD) (12) annotated with related phenotypes were included. These genes were prioritized and targeted searches of rat and human literature were undertaken to provide comprehensive annotations for functional, disease, phenotype and pathway information. In a complimentary approach, focused literature searches were conducted to identify additional genes, QTLs and strains related to the disease area. To facilitate translational studies Human and Mouse data was also included. As there is limited data on human QTL available electronically, a similar strategy was followed to identify relevant Human QTL papers for inclusion in the portal. Mouse and Human gene orthologs are already curated as part of the normal RGD gene curation process. By following this targeted approach, all rat genes, strains, QTL related to a disease area could be added to the database, along with their functional annotations (GO, pathway, phenotype, disease). Similarly, the Human and Mouse gene orthologs and Human QTL were also identified to provide comparative resources for the database. To complement the dedicated curation effort, an online portal was created to provide access to this information. To date, one disease portal has been released for neurological diseases; a second for Cardiovascular Diseases will be released in the autumn of 2006. The portal combines text data with visual elements to allow the user to quickly get an overview of knowledge in a disease area while providing hyperlinks to more details as desired. A screenshot of the Neurological portal is shown in Figure 2 and the main elements are described below:
Disease Category Selection—Based on the disease ontology structure, three levels of disease specificity are provided. When the page is first opened data is presented for All Neurological Diseases. Using the two dropdown menus a disease category and, optionally, a disease from this category can be selected to further narrow down the data displayed. A summary table lists the number of genes, QTL and strains for the three species, rat, mouse and human for the selected disease.
Genome view using Flash Gviewer—This provides a graphical overview of the chromosomal locations of all genes and QTL annotated to the selected disease category. Maps for rat, mouse and human genomes are provided and syntenic maps are also available. An enlarged map can also be selected showing the chromosomes spread over two rows. The Gviewer package is written using Adobe Flash and allows zooming to view individual chromosomes and their features. Hyperlinks are available to jump to gene and QTL reports. The zoomed view also allows chromosomal regions to be selected, providing a dynamic link out to visualize the selected region in a genome browser. GViewer is freely available via the GMOD project (http://www.gmod.org/flashgviewer/)
Gene, QTL and Strain lists—The symbols for the genes, QTL and strains associated with the selected disease are shown in tabular form in the center of the page. These allow easy browsing of the data related to the disease and each symbol is a hyperlink to the appropriate object report in RGD. The Strain table provides quick access to the rat models used to study the selected disease.
Gene Ontology Overview—Three bar charts provide an overview of the prominent gene ontology annotations available for the Rat genes annotated to the selected disease. The individual GO annotations for each rat gene are converted to the corresponding GO Slim annotation and graphed to provide a visual indication of popular GO categories relevant to the selected disease.
Figure 2.
Screenshot of the RGD Neurological Disease portal showing the combined data for all neurological diseases. The various subsections of the page (A–D) are described in more detail in the text.
Complementing the Disease section of the portal are similar views for Phenotype, Biological Process and Pathways. These list selected phenotype ontology terms, biological process terms or pathway ontology terms for genes that have been linked to Neurological or Cardiovascular disease. These allow the scientist to view the disease data from alternative perspectives, to quickly ask questions like ‘what pathways are involved in neurological disease, where are these genes on the genome, what cellular location do they typically occupy?’ The strain models section provides a comprehensive background on specific disease models and the strains used to study these diseases. It includes information on the experimental model, how it can be induced, the disease course, phenotypic indices and strains that are susceptible or resistant to the induction of the disease phenotypes.
The disease portal approach utilizes in-depth curation and dedicated web tools to provide detailed coverage for specific disease areas. Upcoming portals will cover Autoimmune Diseases, Cancer, Metabolic and Nutritional Diseases, Renal Diseases, Respiratory Diseases and other high priority research areas for the rat model. Until these become available the general disease portal page and search tool (http://rgd.mcw.edu/tools/diseases/disease_search.cgi) does provide a convenient way to find genes, orthologs and QTL associated with any disease that are curated by the regular RGD literature curation effort.
OUTREACH
RGD continues to explore different ways to provide outreach to the community. Two recent papers describe the tools and data available at RGD. The first describes strategies for using RGD in support of specific research areas including comparative genomics, positional cloning and microarray with a particular focus on using the RGD tools to access data useful in these types of studies (13). A chapter is also available in Current Protocols in Bioinformatics which provides a more step-by-step practical guide to using RGD and its various resources (14). As part of its role in supporting and informing the broader rat genomics community RGD hosts the Rat Community Forum (http://rgd.mcw.edu/RCF/), an online mailing list with almost 900 subscribers that is regularly used to discuss practical questions pertaining to rat genetics and biology. We also produce a quarterly newsletter, the Pied Piper (http://rgd.mcw.edu/newsletter/), which contains more in depth articles and announcements on current developments in rat genomics in both academia and the commercial fields. In addition to providing information to the community, RGD is also trying to solicit input from the community. A recent addition has been the Genome Discrepancy Form (http://rgd.mcw.edu/GenomeErrorReport/errEntry.jsp), developed as a way for RGD to begin to collect and publicize known or suspected errors in the current genome assembly. We will take information submitted through this form and display it on the genome browser to publicly identify potential problem regions across the genome. Ultimately these regions may be considered for further sequencing and analysis, as the rat genome assembly is refined at a future date. The importance of continuing to improve the assembly cannot be overstated as the quality of annotations and sequence analyses are directly proportional to the quality of the underlying sequence. Critical genomic reagents such as microarray chips are designed and annotated using the latest assemblies and the utility of these reagents are lagging behind those in human and mouse due to the difference in assembly coverage and quality.
DISCUSSION AND FUTURE DIRECTIONS
The focus of much rat research and of federally sponsored biomedical research as a whole is translational medicine (15), the application of basic research to address problems in human health. RGD has been supporting this trend through an emphasis on comparative mapping resources and tools (16) and most recently through the disease portals. This will continue as we develop and release future disease portals and refine the portal interface. We will be introducing improved data mining tools through the use of the Biomart package that will be integrated with the disease portals and other RGD tools and reports. This will greatly improve access to raw data within RGD and also allow RGD data to be more directly integrated with other external Biomart resources such as Ensembl's Ensmart (17) and rat proteomic and phenotypic datasets housed at the Medical College of Wisconsin. Biomedical ontologies have been and will continue to be a core part of our data curation process with an emphasis on incorporation into tools that benefit both the novice and power user. The disease portals demonstrate this commitment, utilizing the underlying power of the disease and other ontologies through a more user-centered, visual interface. We hope to continue this trend, building on the visual nature of much biological data (and many biologists) by developing graphical tools to present and summarize the ever more complicated biological datasets.
Acknowledgments
In addition to the listed authors RGD is built and maintained by the following group of dedicated people: Jiali Chen, Weihong Jin, Nataliya Nenasheva, Rajni Nigam, Andrew Patzer, Victoria Petri, Dorothy Reilly, Jennifer Smith, Renee White, Stacy Zacher, Angela Zuniga-Meyer. This work was funded by National Heart, Lung, and Blood Institute Grant HL-64541 and National Human Genome Research Institute Grant HG-002273. Funding to pay the Open Access publication charges for this article was provided by NHLBI, grant HL-64541.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lazar J., Moreno C., Jacob H.J., Kwitek A.E. Impact of genomics on research in the rat. Genome. Res. 2005;15:1717–1728. doi: 10.1101/gr.3744005. [DOI] [PubMed] [Google Scholar]
- 2.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blake J.A., Bult C.J. Beyond the data deluge: data integration and bio-ontologies. J. biomed. Inform. 2006;39:314–320. doi: 10.1016/j.jbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Bodenreider O., Stevens R. Bio-ontologies: current trends and future directions. Brief Bioinform. 2006;7:256–274. doi: 10.1093/bib/bbl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith C.L., Goldsmith C.A., Eppig J.T. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome. Boil. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker K.G., Barnes K.C., Bright T.J., Wang S.A. The genetic association database. Nature Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 11.Safran M., Solomon I., Shmueli O., Lapidot M., Shen-Orr S., Adato A., Ben-Dor U., Esterman N., Rosen N., Peter I., et al. GeneCards 2002: towards a complete, object-oriented, human gene compendium. Bioinformatics. 2002;18:1542–1543. doi: 10.1093/bioinformatics/18.11.1542. [DOI] [PubMed] [Google Scholar]
- 12.Blake J.A., Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E. The Mouse Genome Database (MGD): updates and enhancements. Nucleic Acids Res. 2006;34:D562–D567. doi: 10.1093/nar/gkj085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Twigger S.N., Pasko D., Nie J., Shimoyama M., Bromberg S., Campbell D., Chen J., dela Cruz N., Fan C., Foote C., et al. Tools and strategies for physiological genomics: the Rat Genome Database. Physiol. Genomics. 2005;23:246–256. doi: 10.1152/physiolgenomics.00040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Twigger S., Smith J., Zuniga-Meyer A., Bromberg S. Exploring phenotype data at the Rat Genome Database. In: Baxevanis A.D., editor. Current Protocols in BIoinformatics. Hoboken, NJ: Wiley; 2006. pp. 1.14.1–1.14.27. [DOI] [PubMed] [Google Scholar]
- 15.Zerhouni E.A. US biomedical research: basic, translational, and clinical sciences. JAMA. 2005;294:1352–1358. doi: 10.1001/jama.294.11.1352. [DOI] [PubMed] [Google Scholar]
- 16.Twigger S.N., Nie J., Ruotti V., Yu J., Chen D., Li D., Mathis J., Narayanasamy V., Gopinath G.R., Pasko D., et al. Integrative genomics: in silico coupling of rat physiology and complex traits with mouse and human data. Genome. Res. 2004;14:651–660. doi: 10.1101/gr.1974504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasprzyk A., Keefe D., Smedley D., London D., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome. Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]