Abstract
Entrez Gene (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene) is NCBI's database for gene-specific information. Entrez Gene includes records from genomes that have been completely sequenced, that have an active research community to contribute gene-specific information or that are scheduled for intense sequence analysis. The content of Entrez Gene represents the result of both curation and automated integration of data from NCBI's Reference Sequence project (RefSeq), from collaborating model organism databases and from other databases within NCBI. Records in Entrez Gene are assigned unique, stable and tracked integers as identifiers. The content (nomenclature, map location, gene products and their attributes, markers, phenotypes and links to citations, sequences, variation details, maps, expression, homologs, protein domains and external databases) is provided via interactive browsing through NCBI's Entrez system, via NCBI's Entrez programing utilities (E-Utilities), and for bulk transfer by ftp.
INTRODUCTION
Entrez Gene is the gene-specific database at the National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine (NLM), located on the campus of the US National Institutes of Health (NIH) in Bethesda, MD, USA. Entrez Gene provides unique integer identifiers for genes and other loci (such as officially named mapped markers) for a subset of model organisms. It tracks those identifiers, and is integrated with the Entrez system for interactive query, LinkOut and access by E-Utilities (1). The information that is maintained includes nomenclature, defining sequence, chromosomal localization, gene products and their attributes (e.g. protein interactions), associated markers, phenotypes, interactions and a wealth of links to citations, related sequences, variation, maps, expression, homologs, protein domain content and external databases.
Data in Entrez Gene result from a mixture of curation by RefSeq staff and automated analyses. Annotation in sequences from NCBI's Reference sequence project (2) or the International Nucleotide Sequence Database Collaboration (DDBJ, EMBL, GenBank) (3) is integrated with information from collaborating model organism databases, public users and literature review (especially the Gene References into Function or GeneRIFs).
Entrez Gene is an integral part of representation of gene-specific information at NCBI. The information conveyed by establishing the relationship between sequence and a GeneID is used by other NCBI resources (1) such as BLAST, dbSNP, GEO, HomoloGene, Map Viewer, Probe, UniGene, UniSTS and NCBI's genome annotation pipeline. For example, the names associated with GeneIDs are used in HomoloGene, UniGene and the Mammalian Gene Collection (4). Inconsistencies in representation of genes and their sequences are investigated, and resolved by NCBI RefSeq staff in consultation with multiple authorities (2). Although providing a stable interface is a goal of Entrez Gene, the content, display or methods for bulk transfer may change. One method to receive advanced notification of changes is via subscription to gene-announce@ncbi.nlm.nih.gov.
FUNCTION OF THE DATABASE
The primary goals of Entrez Gene are to provide tracked, unique identifiers for genes of multiple genomes and to report information associated with those identifiers for unrestricted public use. The identifier that is assigned (GeneID) is an integer, and is species-specific. In other words, the integer assigned to dystrophin in human is different from that in any other species. The GeneID is reported in RefSeq records as a ‘db_xref’ (e.g. /db_xref=‘GeneID:856646’, in GenBank format).
Entrez Gene provides multiple reports. For the interactive user, the defaults are the HTML summary display resulting from an Entrez query (Figure 1) or a gene-specific report accessed by clicking on the symbol in the summary page (Figure 2). The Gene Table display option is useful to obtain a report of the intron/exon organization of the gene as annotated on a RefSeq genomic sequence, and to navigate quickly to the sequence of any of those gene features. In addition to the standard views from Entrez, Gene provides a complete database extraction as well as several special reports for ftp transfer (ftp://ftp.ncbi.nih.gov/gene/README). The data are also available from the programatic interface to Entrez, namely E-Utilities (1).
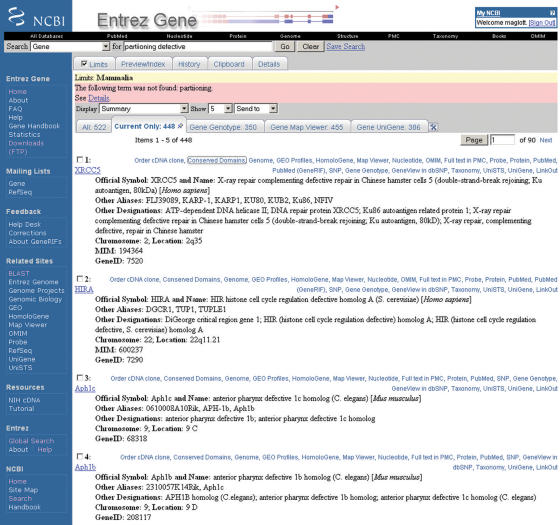
Figure 1.
Representative ‘Summary’ report of query results. Result of a query to retrieve information about partitioning-defective genes in mammals. This figure illustrates several points: (i) the display when limits is invoked to restrict result sets; (ii) spell checking; (iii) use of My NCBI to customize tabs to highlight subcategories of records in the result set; (iv) use of My NCBI to alter the display of the links menu. Limits:mammalia indicates that mammalia was selected from the page accessed via the limits tab to restrict results to genes in mammals. The term partitioning had no matches in the database; the ‘details’ page explains that only the term ‘defective’ was processed. Entrez identifies possible misspellings and suggests an alternate query (Did you mean:partitioning defective?). Of the 522 results that were returned, the tabs indicate that 448 are current (current only), 350 have genotype information available in dbSNP (Gene Genotype), 455 can be viewed in Map Viewer (Gene Map Viewer) and 386 have expression data in UniGene (Gene UniGene). Because, My NCBI environment replaces the default links menu with text, the databases connected to each record are displayed directly on the results page. The summary display includes the species of origin, preferred and alternate symbols, preferred and other descriptive names, chromosome localization, the GeneID and the MIM number when appropriate. Click on any symbol to link to the full report (Figure 2). The top black navigation bar and the blue side-bar at the left provide general links to other sites, including genome-specific resource guides (Genomic Biology), the FTP site, forms to submit feedback (Feedback) and forms to subscribe to mail lists (Mailing Lists).
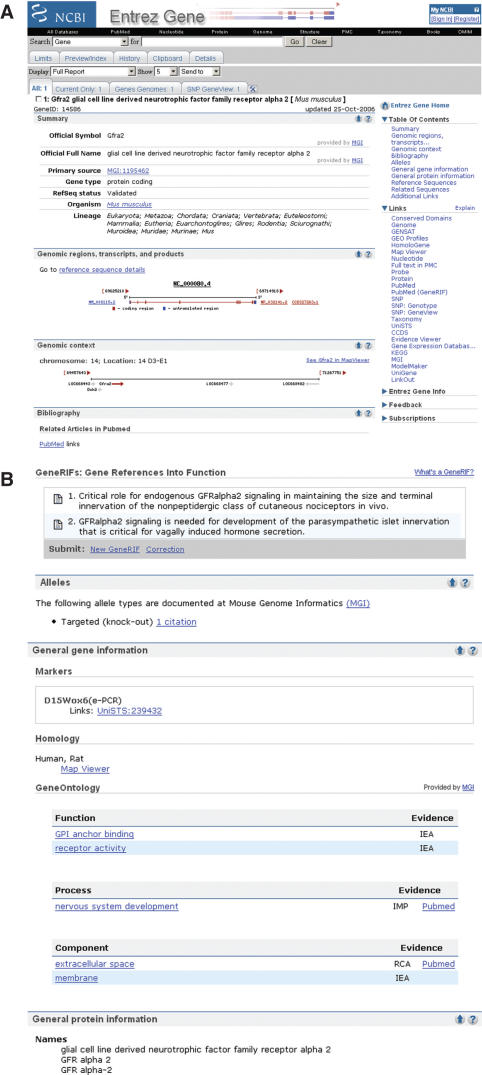
Figure 2.
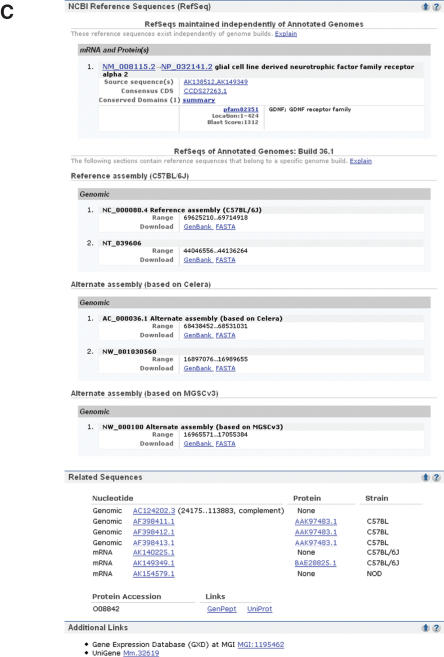
(a) Representative Entrez gene full-report page, part 1. The full-report display. The standard gene-specific report page starts with summary information about the gene, a table of contents and a links menu. The summary section includes names and symbol aliases. If the gene has official names provided by a nomenclature authority, those names are reported as official symbol and official full name, with the named source anchoring a link. The database identifier provided by that source is displayed, anchoring a link to that source's specific record. The review status of all RefSeq RNAs for the gene is reported as RefSeq status. If the gene has been annotated on a RefSeq genomic sequence, a graphic is provided diagramming the intron/exon organization of the gene (genomic regions, transcripts and products) with the accessions for the genomic, mRNA and protein RefSeqs anchoring links to the sequence records in NCBI's Entrez system and, in the case of proteins, to BLink (1). If a RefSeq protein is a member of a CCDS group (2), the CCDS identifier to the right of the RefSeq protein accession anchors a link to the CCDS database. The genomic context section diagrams the placement of the gene and its neighbors. Each symbol anchors a link to another record in Entrez Gene. A link to NCBI's map viewer is in this section, identical to the one in the links menu. All citations in PubMed associated with a record in gene can be accessed by clicking on PubMed in the bibliography section or in the links menu. Navigation to PubMed for citations associated with specific information, such as GeneRIFs or gene ontology terms, is repeated explicitly with those elements (b). The links menu should be used to determine the types and sources of additional information that may be available about a gene. In this example, information about expression is available from GENSAT, GEO, UniGene and MGI; homology from HomoloGene, variation from SNP; cDNAs supporting genomic annotation from evidence viewer and ModelMaker, pathways from KEGG, etc. (b) Representative Entrez gene full report page, part 2. This portion of a full report display includes the sections of the record indicated in the table of contents (a) as bibliography, alleles, general gene information and general protein information. In the GeneRIFs section, the icons to the right of the text anchor a link to the PubMed that supports the GeneRIF. The data in the alleles and gene ontology sections were imported from MGI, as indicated in the links anchored by MGI. Alternate names for the gene, and the protein it encodes, are listed under general protein information/names. If this gene encoded an enzyme, the E.C. designation would be in this section as well. (c) Representative Entrez gene full report page, part 3. This portion of a full report display includes the sections of the record indicated in the table of contents (a) as reference sequences, related sequences and additional links. The reference sequences section is subdivided into subsections based on the type of RefSeq being reported. The first section (RefSeqs maintained independently of annotated genomes) reports the RefSeq genomic, RNA and protein accessions that can be updated at any time, and thus may differ in version or number from what was included in a genomic annotation (2). The sequences reported under RefSeqs of annotated genomes are the genomic RefSeqs for the chromosomes and contigs of reference and alternate assemblies. Each of these RefSeq sections, and the related sequences sections below, anchors links to records in NCBI's Entrez system, where standard tools are provided to process the sequence (e.g. altering the range, displaying annotated SNPs or downloading in multiple formats). The related sequences section lists the accessions and strains of public sequences of this gene or its encoded protein. The items in the additional links section are included in the Links menu (a), but are selected to be repeated here to enhance access, for example to display UniGene cluster number.
SCOPE OF THE DATABASE
When are GeneIDs assigned?
Identifiers are always assigned to what is annotated as a gene feature on a RefSeq record. Identifiers may also be assigned when no RefSeq exists. This may occur when an authoritative source for a genome, such as a model organism-specific database, assigns an identifier to what is termed a gene, mapped locus or trait, even though that entity is not completely defined by sequence. When a Gene record is established, it is assigned a category (e.g. protein-coding, pseudogene, rRNA, unknown). The term ‘unknown’ is used when the category is under review, as when some of the sequences defining the gene are annotated with coding regions, but the support for that annotation is inconclusive. The assigned category can change without changing the GeneID.
Some current statistics
As of September, 2006, there were >2 million current records in Gene, distributed among >3500 taxa (Table 1). Not all the taxa are completely represented in Gene; most of the eukaryotes, for example, have Gene records only for their mitochondrial genomes. The Gene Statistics site (http://www.ncbi.nlm.nih.gov/projects/Gene/gentrez_stats.cgi) reports both current and historical counts of records by taxonomic node and species.
Table 1.
Representative Statistics
| Category | Taxa | GeneIDs |
|---|---|---|
| Records with GO terms | 30 | 194446 |
| Records with GeneRIFs | 631 | 30726 |
| From Eukaryota | 1077 | 777108 |
| From Fungi | 66 | 135771 |
| From Archea | 68 | 71805 |
| From Bacteria | 785 | 1151407 |
| From Viruses | 1569 | 46484 |
Record content
Figure 2 displays representative gene-specific information that can be retrieved through Entrez Gene. For example, GeneRIFs, contributed by the general public and the Index Section of the National Library of Medicine, provide an annotated bibliography of the function, discovery and mapping of genes from the current literature. Not all categories of information are displayed completely in the Gene Report; many details may be retrieved by links (Links menu, Figure 2a) provided to other databases such as Nucleotide and Protein for sequence, HomoloGene for integration of information about homologs, Map Viewer for extended genomic context and comparative maps, GENSAT, UniGene and GEO for expression data, Conserved Domain Database for domain content of proteins, OMIM for human Mendelian disorders, PubMed and Books for publications, species-specific databases and LinkOut link for navigation to external databases that have reported they have more information related to a GeneID. Links are also provided to tools such as BLink (1), which supports many views of related proteins determined by BLAST alignments. The goal is to integrate sufficient text, keywords and links to make Entrez Gene an effective starting place to retrieve information of interest.
ACCESS TO ENTREZ GENE
The information in Entrez Gene can be accessed in multiple ways at NCBI (Table 2). The most direct is to submit a query to Entrez from the NCBI home page and display the results in Gene, or enter a query in any Entrez query bar and restrict the database search to Gene. Another way is to take advantage of the Links computed by the Entrez system. For example, you might find a PubMed record of interest and from PubMed's Links menu discover that there is a record in Entrez Gene connected to the publication. The BLAST group uses the GeneID<->sequence relationship maintained by Entrez Gene to help you navigate from protein or mRNA accessions matching your query to Entrez Gene via the blue G icon. Map Viewer provides links from annotated genes to Entrez Gene. And RefSeq records include the GeneID as a db_xref in the gene feature. Thus you can navigate to Gene not only by text but by genomic position (Map Viewer), RefSeq annotation and sequence data (BLAST, Nucleotide, Protein).
Table 2.
Accessing Entrez Gene
| Direct query | |
|---|---|
| Enter search term(s) and select results shown in the Gene section | www.ncbi.nlm.nih.gov or http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi? |
| Enter search term(s) and query only Entrez Gene | www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene or select Gene as the search option from any Entrez query bar |
| E-Utilities: check the result interactively. (Hint: view source if your Browser does not display the XML.) | http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?db=gene&id=19,11303,313210,373945,378973,464631&retmode=xml |
| Record-specific connections in other NCBI databases | |
| Gene option in the Links menu at the upper right of a display in a non-Gene record | Click on Gene to find Gene records related to the record being displayed |
| Links called Gene or G | Map Viewer's annotation of Genes; BLAST retrieval of accessions connected to Gene records |
| More information | |
| Help documentation | http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=helpgene.TOC&depth=2 |
| How Entrez Links are computed | http://www.ncbi.nlm.nih.gov/entrez/query/static/entrezlinks.html |
If you register for MyNCBI (http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=helpmyncbi.chapter.MyNCBI), you can elect to receive e-mails when records satisfying your favorite search are created or updated. You can also customize your default display to identify what subset of records returned by a query has particular attributes (Figure 1).
LINKS TO EXTERNAL DATABASES FROM ENTREZ GENE
Entrez Gene can serve as a directory to gene-specific information for databases outside of NCBI. There are two major categories of connections. One comes from active collaborations with multiple data providers such as model organism databases, the GO consortium, KEGG and Reactome (http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=helpgene.table.EntrezGene.T1). The others are generated from data providers who register with the NCBI LinkOut (1) system. Any user of Entrez Gene retrieving a record with a LinkOut will then be able to connect to the registered database according to the specification of the data provider.
FEEDBACK
We welcome your feedback with respect to the Entrez Gene interface, or any data contained therein. Please select from the Feedback options on any Gene page (Figure 1).
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acid Res. 2007 doi: 10.1093/nar/gkl1031. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pruitt K.D., Tatusova T., Maglott D. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acid Res. 2007 doi: 10.1093/nar/gki025. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acid Res. 2007 doi: 10.1093/nar/28.1.15. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl Acad. Sci. USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]