Abstract
The Ensembl (http://www.ensembl.org/) project provides a comprehensive and integrated source of annotation of chordate genome sequences. Over the past year the number of genomes available from Ensembl has increased from 15 to 33, with the addition of sites for the mammalian genomes of elephant, rabbit, armadillo, tenrec, platypus, pig, cat, bush baby, common shrew, microbat and european hedgehog; the fish genomes of stickleback and medaka and the second example of the genomes of the sea squirt (Ciona savignyi) and the mosquito (Aedes aegypti). Some of the major features added during the year include the first complete gene sets for genomes with low-sequence coverage, the introduction of new strain variation data and the introduction of new orthology/paralog annotations based on gene trees.
INTRODUCTION
The genome sequence of an organism provides a natural index for organizing and understanding biological data. Ensembl is a software system to store, analyze, use and display genomic information. Ensembl's primary focus is around providing gene annotation and comparative genome integration for chordate genomes, the vast majority of which are vertebrates. Ensembl concentrates particularly on mammalian genomes having developed initially around the human genome sequence. Some major distinguishing features of the project compared to other major sites providing access to these genomes at UCSC (1) and NCBI (2) are that Ensembl creates a gene set, using an automatic gene build pipeline, for each species for which no manually curated gene set exists and that Ensembl makes all its data and software source code available to all users to encourage its reuse and programatic access. As discussed below, for the relatively mature vertebrate genomes of human and mouse where there are active efforts to curate gene structures by the Havana group through Vega (3) the RefSeq group (2) and UniProt (4) Ensembl is actively collaborating with these groups to converge on the reference gene set.
The genomes of 28 chordates are currently available through Ensembl, from mammals such as human and mouse through to the ‘primitive’ chordates Ciona intestinalis and Ciona savignyi. Ensembl sites for the genomes of three key eukaryote model organisms, yeast (Saccharomyces cerevisiae), fruitfly (Drosophila melanogaster) and nematode (Caenorhabditis elegans), are provided to allow integration, primarily around predicted ortholog/paralog relationships, between these organisms and chordates within a common database environment. For these organisms, no automatic gene build is performed and instead gene annotation is imported from their respective model organism databases. Finally, a number of insect genomes are also available through Ensembl due to Ensembl's participation in the Vectorbase consortium (http://www.vectorbase.org/). Vectorbase is an NIH–NIAID Bioinformatics Resource Center for Invertebrate Vectors of Human Pathogens and is using the Ensembl platform to present key vector genomes. It is likely that in the future these vector genomes will only be available on the Vectorbase site, though Vectorbase will continue to use Ensembl software. This year's increase in the number of genomes provided by Ensembl is the largest so far (Figure 1). More than half the genomes in Ensembl are mammalian. In keeping with the focus on chordates, Ensembl has this year stopped providing an Ensembl site for the honeybee (Apis mellifera) genome, while being involved in the initial analysis and producing an initial geneset (5), just as the previous year it stopped providing a site for the second nematode Caenorhabditis briggsae. This is because it recognizes that access to these genomes is being provided by the dedicated Beebase (http://racerx00.tamu.edu/bee_resources.html) and Wormbase (6) model organism databases. In both cases the old Ensembl databases for these genomes remain accessible via Ensembl archive sites.
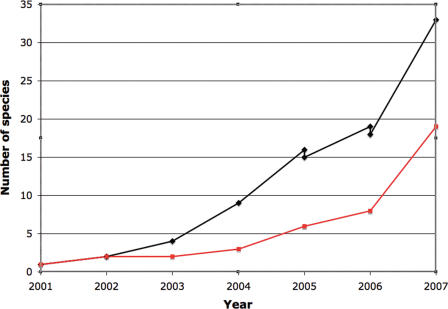
Figure 1.
Figure shows the growth in the number of genomes provided by Ensembl over the past 5 years. The discontinuities at the start of 2006 and 2005 represent the removal of the honeybee (Apis mellifera) and nematode (Caenorhabditis briggsae) Ensembl sites, respectively. The black line shows all genomes and the red line shows mammalian genomes.
Ensembl provides a variety of ways to access these data to suit different audiences and types of use. The majority of researchers using Ensembl use the website (http://www.ensembl.org/) and can rapidly locate individual items of interest either by entering keywords or from the built-in sequence similarity search interface. For cases where researchers are working with sets of items, such as a particular class of genes, Ensembl provides data mining tools via the BioMart system (7). For bioinformaticians Ensembl provides access to all the data behind the Ensembl website both as downloadable datasets and by allowing programatic access to databases hosted on the Ensembl site (ensembldb.ensembl.org). The later is growing in popularity as complete database dumps become large to download. Increasingly bioinformaticians are carrying out their own custom data analysis by accessing the databases remotely via the Perl language application programing interfaces (APIs) that Ensembl provides. Extensive documentation and tutorials are provided to help researchers get started programing using the APIs as well as describing the database schemas (http://www.ensembl.org/info/software). Ensembl runs courses and training around the world, has a full-time helpdesk and online tutorial materials (http://www.ensembl.org/info). Over the year courses have been held in the following countries: UK (16×), Austria (2×), Belgium (4×), Finland (2×), France (2×), Germany (2×), Hungary, Italy (6×), Spain (4×), USA (3×), Brazil (2×), South Africa (2×), Australia (2×) and Singapore. There are many practical details concerning data processing algorithms developed and used by Ensembl and the overall system's design and operation. For detailed descriptions, researchers are referred to the series of papers published in 2004 that describe both technical aspects of the software implementation and the scientific aspects of the genome annotation system (7–16). Whilst the system has evolved considerably since these articles were published, they provide a background to the technical documentation maintained on the Ensembl website and distributed with each software release. As an open data, open source software project Ensembl encourages participation and discussion on development issues mostly via the development email list (send ‘subscribe ensembl-dev’ to majordomo@ebi.ac.uk to join). We are seeing this email list being increasingly used to exchange advice on API usage.
Ensembl continues to improve both in terms of the analysis of genome information and its usability both via programatic means and for web-based browsers. This paper details only some of the major improvements since the last report (17). For more comprehensive information about new features and data contained in the bi-monthly updates of Ensembl, researchers are also recommended to read the ‘what's new’ pages accompanying every release (http://www.ensembl.org/Multi/newsview) and/or subscribe to the ‘announce’ email list by sending ‘subscribe ensembl-announce’ to majordomo@ebi.ac.uk.
RESULTS
Improvements to protein-coding genes
Providing expressed gene sets which are as accurate as possible is one of the major goals in Ensembl. Ensembl gene sets are all based on evidence from alignments of protein and cDNA sequences to genomic sequence. The completeness of each gene set depends on the amount of transcript data, either specific for the genome in question, or evolutionarily close enough to be reliably aligned. Gene set accuracy depends on alignment quality and being able to reconcile evidence, including detecting erroneous data from truncated and chimeric transcripts and identifying pseudogenes. This year's major improvements and changes to the Ensembl gene build systems and strategy cover the following three different situations for gene building: (i) the new class of low-sequence coverage mammalian genomes; (ii) high-coverage genomes which have little organism-specific transcript data; and (iii) the high-quality reference genomes of human and mouse.
New projection build pipeline for low-sequence coverage genomes
Ensembl has this year incorporated the first nine genomes which have been sequenced at low-coverage [2× whole genome shotgun (WGS)]. These are elephant (Loxodonta africana), rabbit (Oryctolagus cuniculus), armadillo (Dasypus novemcinctus), tenrec (Echinops telfairi), cat (Felis catus), bush baby (Otolemur garnettii), common shrew (Sorex araneus), microbat (Myotis Lucifugus) and european hedgehog (Erinaceus europaeus). In total 16 mammals will be sequenced at this coverage as part of the Mammalian Genome Project, funded by the National Institutes of Health (NIH). Currently, automatically generated gene set are provided for the first four of these genomes and gene builds are in progress for the remaining five. The low sequence coverage of these genomes means that all of the normal problems experienced when predicting gene structures in draft genome assemblies (missing sequence, fragmentation, misassemblies, misplacements, small insertions/deletions/substitutions) are exacerbated. In particular, many genes will be represented only partially (or not at all) in the assembly, and many others (particularly those with large genomic extent) will be found in pieces, distributed across more than one scaffold. The standard Ensembl gene build pipeline (11), which relies on aligning expressed transcript sequences onto the genome sequence, is unsuitable as the main approach for annotating such a low-coverage genome.
To address this, a new gene building methodology for low-coverage genomes has been developed that relies on a whole genome alignment (WGA) to an annotated, reference genome. The WGA underlying each annotated gene structure in the reference genome are used to infer ‘gene-scaffold’ assemblies of scaffolds in the target genome that contain complete gene structures. WGAs are generated in-house using BLASTz (18) with the resulting set of local alignments processed into a form suitable for the above method using the Axt tools (19). The protein-coding transcripts of the reference gene structures are then projected through the WGA onto the implied gene-scaffolds in the target genome. These projections frequently contain small insertions/deletions with respect to the protein-coding transcript of the reference gene structure. In some cases this raw projection would result in a frame shift in the translation. In the vast majority of such cases this apparent frame shift is most likely the result of a sequence/assembly error in the target genome, resulting from the low sequence coverage. To correct for these apparent errors, the gene build introduces an artificial intron, 1 or 2 bp long, into the predicted gene structure to correct the frame shift at each point where one is introduced. In this way the reading frame of the predicted transcripts are preserved. We refer to these introns as ‘frame-shift introns’. When the WGA implies that the sequence contains an internal exon missing from the assembly, and the location is consistent with an intra- or inter-scaffold gap, the exon is placed on the gap sequence. This results in a run of X's of the correct length in the translation. An example of this situation in the elephant (L.africana) genome is shown in Figure 2.
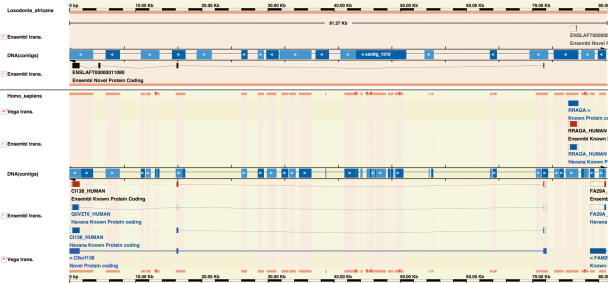
Figure 2.
Figure shows a screenshot of part of an AlignSliceView web page from the elephant (Loxodonta africana) genome as an example of the output from the Ensembl gene build system when applied to low-coverage shotgun genomes. The top panel shows elephant genome sequence and the bottom panel shows the region of human genome sequence that aligns to it. In the DNA(contigs) track blue regions indicate sequence and blank regions indicate gaps. The track for elephant gives an idea of fragmentation of the genome assembly (the gaps in the track for human do not indicate gaps in the genome but rather gaps in the alignment between elephant and human). Elephant DNA contigs have been organized into ‘gene-scaffolds’ based on whole genome alignments (WGA) to a reference genome, in this case human (see text). Elephant transcripts, such as the reverse strand transcript ENSLAFT00000011080 shown here, are built by projecting protein-coding part of human transcripts through the WGA. In this case the elephant transcript has been built by projecting the annotated transcript C9orf138; however, there is no WGA alignment for the third exon of this transcript [the third exon from the right is positioned against a gap between contigs in the DNA(contigs) track]. As a result this exon is missing from the view of human transcripts, a fact that is indicated by the green dotted link linking exons 2 and 4 (CI138_HUMAN). The elephant transcript ENSLAFT00000011080 does contain this exon; however, because of the gap in the elephant sequence, only the exon length can be inferred from the corresponding human transcript, so the exon sequence is composed entirely of ‘N’s in the transcript and ‘X’s in the corresponding translation. Interestingly, in human a shorter alternative transcript is also annotated with a missing third exon (Q5VZT6_HUMAN); however, the form with the third exon appears to be conserved across mouse, rat and dog, suggesting that it is likely to be conserved in elephant too.
This strategy has been developed and piloted on the initial 3× coverage WGS assembly of the cow (Bos taurus) genome. The current cow assembly is 6× coverage and its gene set has been generated using the standard Ensembl pipeline. A new higher quality 7.1× coverage cow assembly, that incorporates sequence data from BAC libraries, has just been released. When the gene build on this assembly is complete it will be possible to carry out a more detailed assessment of the quality and value of gene builds on low-coverage genomes by comparison to standard gene builds on high-quality assemblies. In the mean time some statistics for exon coverage for both the gene builds on the 3× and 2× assemblies are shown in Table 1. For all of these gene builds the human genome has been used as the reference, making side by side comparison possible. Since all these genomes are mammals, it is anticipated that the vast majority of human coding exons should be found in each. The number of exons annotated therefore gives an idea of the quality of gene builds possible given the genome assembly quality. The first two columns of Table 1 show the percentage of base pairs of human exons that can be aligned to the sequence of the target genome by the WGA BLASTz step and the percentage completely missed. These values are a few percentage points less than the theoretical expected percentage coverage for 2× and 3× WGS sequencing of 88% and 95%, respectively, but broadly in line with expectations (20). The last two columns show the equivalent figures for the final gene set after it has been filtered as a result of the gene build process and are significantly lower. In the current gene build process, after raw alignments are chained into gene-scaffolds and the best-in-genome match found, predicted gene structures are removed if >50% of the original exons are missing. These criteria are adopted as in our view where such partial genes are built they contain an unacceptable likelihood of error and are better discarded. Most likely, many of these partial genes result from assembly artifacts; however, this will be better understood after the cow gene set comparisons are carried out. In the mean time it is worth noting the significant difference between the final exon coverage for the initial 3× cow genome and the other four 2× genomes. It appears that dropping from 3× to 2× sequence coverage roughly doubles the fraction of exons missing from resulting gene sets when the filtering we believe necessary to achieve acceptable gene set quality is applied.
Table 1.
Completeness of gene builds on low sequence coverage genomes
| Genome | Raw unfiltered | Filtered chained (final gene set) | ||
|---|---|---|---|---|
| Base pair covered (%) | Exons missed (%) | Base pair covered (%) | Exons missed (%) | |
| Cow (3×) | 91.4 | 6.8 | 80.0 | 15.5 |
| Elephant | 80.0 | 17.6 | 64.8 | 30.3 |
| Rabbit | 82.0 | 16.7 | 65.7 | 30.8 |
| Armadillo | 77.0 | 21.3 | 59.4 | 36.6 |
| Tenrec | 83.2 | 15.1 | 69.1 | 27.0 |
This table shows fraction of base pairs of human Ensembl gene set exons covered by raw alignments to WGS scaffolds (first column) and in the filtered, chained gene-scaffolds presented as the final gene set (third column). Fraction of exons completely missed in each case is also shown (second and fourth column, respectively). All genomes are 2× WGS assemblies, except for cow which is 3× (see text).
The code developed for the projection build has also been applied in the gene build for the new chimpanzee (Pan troglodytes) genome assembly, which has greatly improved the gene set. For chimpanzee the gene-scaffold generation step is skipped as the assembly is high coverage.
Improved pipeline for genomes that are evolutionarily distant from main sources of transcript data
The completeness of each gene set depends on the quantity of transcript data either specific for the genome in question, or evolutionarily close enough to be reliably aligned. Although there are a number of large chordate genomes with little organism-specific transcript data, most are mammals and so evolutionarily close to the huge amounts of transcript data from human and mouse. The current major exceptions are chicken (Gallus gallus) and opossum (Monodelphis domestica). Two gene sets have been produced on the chicken genome assembly and been made available though the Ensembl website. For both gene builds the standard Ensembl gene build pipeline was used; however, for the most recent gene build (December 2005) the pipeline was significantly customized to improve gene set quality. Investigations showed that mapping distantly related cDNAs and ESTs onto the chicken genome using the standard pipeline was creating very extended gene structures, incorrectly linking transcripts of some adjacent genes. Gene building on vertebrate sized genomes is very expensive, so the pipeline contains optimizations to reduce the CPU cost. One such strategy is the creation of ‘mini’ sequences (11) which remove regions thought to be intronic from the final alignment step. In the case of chicken, because of the low similarity of transcripts from other organisms being mapped to the genome sequence, this step was removing some exons and causing gene artefacts in some regions. Removing the mini sequence step and optimizing other parameters resulted in a greatly improved gene set. This process was in part developed in parallel for the opossum genome. Although opossum is nearer to other mammals than chicken, adopting this approach led to similar quality improvements. The downside is a ∼5-fold increase in the CPU cost of the gene build; however, this has been at least partly offset by optimizations to the genewise program (9) itself which is used for this final alignment step.
Converging to the reference gene set for the high-quality reference genomes of human and mouse
Analysis showing the progressive and significant improvement in the quality of human and mouse gene sets generated by the Ensembl system were presented in past year's report (17). More recently a blind test assessment of gene set quality took place under the ENCODE project (21). The EGASP gene prediction competition (22) was held to compare a variety of automatic gene prediction methods with a curated and experimentally validated human gene set generated by the Sanger Havana group (3) as part of the Gencode consortium (23). The results confirmed the high accuracy of Ensembl gene predictions, with Ensembl ranked as the best or close to the best over a variety of different evaluation criteria. However, the best predicted transcript for each gene still differed to the annotated Gencode reference in 30% of cases. When all annotated alternative transcripts were considered, transcript accuracy was only 40–50%. Even allowing for errors in the Gencode set, this is a significant gap. For further details of the evaluation the reader is referred to the special issue of genome biology devoted to evaluation of the EGASP experiment, especially (22). While the EGASP evaluation will lead to some further improvements to the Ensembl gene build system for human and mouse, we recognize that there are likely to be limits on how much more the accuracy of automatic methods on these very high-quality genomes can be improved. Human and mouse already have extensive species-specific experimental data, and there are diminishing returns from each increase in the complexity of the gene build logic to handle remaining hard classes of gene, such as dense clusters of duplicated genes.
For the human genome, Ensembl and Havana have for some time been collaborating as part of the CCDS consortium which includes the RefSeq group at NCBI (2) and the UCSC genome group (1). CCDS is a stable set of protein-coding gene structures for which all consortium members agree on to the base pair. The initial CCDS set contained 13 142 loci which corresponded to ∼60% of the 22 000 protein-coding loci annotated in Ensembl being accepted (for further details and statistics concerning CCDS sets, see http://www.ncbi.nlm.nih.gov/CCDS/). The CCDS process is being extended to the mouse genome now that the assembly (NCBI build 36) is largely composed of finished sequence. While CCDS only addresses the CDS region of gene structures, the Gencode experience illustrates the value of making greater use of the full Havana curated annotation including UTRs, noncoding transcripts and pseudogenes. As a result the Havana and Ensembl gene build groups are now working towards a single integrated gene set that combines automatic and curated annotation, the first version of which was released in Ensembl 38 (April 2006). In this first iteration 12 000 Havana curated full length protein-coding transcripts were incorporated into Ensembl gene entries directly. Because of the problem of reconciling differences between slightly different annotations that probably represent the same transcript, this initial process led to some transcript duplication. We are working to progressively refine the merging process to address this. The process is providing improvements in both directions. In many cases Havana transcripts are more complete as its possible to set lower alignments thresholds since manual review filters out false positives. However, because it is very slow, manual annotation always risks being out of date, so Ensembl transcripts can also be longer when they have used more recent transcript evidence. Ensembl is also identifying regions where the quality of annotation from the automatic gene build pipeline is low so that they can be prioritized for manual curation by the Havana group. There is still a long way to go in this process as Havana annotation is only available for ∼50% of the human genome and ∼20% of the mouse genome; however, it is hoped that with this system of prioritization it should be possible to more rapidly improve the overall quality of human and mouse gene sets.
IDHistoryView and stable identifier discovery
A key utility of all gene sets, regardless of how they are generated, is stability of identifiers between releases. Ensembl manages to map the vast majority of stable identifiers between gene builds; however, changing assemblies, evidence, algorithms and logic inevitably lead to previously predicted genes being absent from a subsequent release or gene splits and merges events. The Ensembl core schema now fully supports an archive of obsolete transcript entries and a new view IDHistoryView has been introduced to allow the history of an identifier to be viewed. Using this interface it should be possible to discover the fate of any Ensembl gene stable identifier back to Ensembl 1.2 (2001) and see how the sequence of the gene structure has changed (the version of transcript and exon stable identifiers is increased if the sequence of that feature changes). The page contains links to Ensembl archive versions to allow users to view older gene structures as they were previously presented.
Variation resources
In the previous report on the Ensembl project (17), major improvements to handle large scale variation data were described. One component of this was a software infrastructure to efficiently store resequencing data. This year we have exploited this system to take advantage of the extensive collection of mouse DNA sequence reads, including those recently released by Celera, data from dbSNP and resequencing data generated by Perlegen Sciences for the US National Institutes of Environmental Health Sciences (NIEHS). These were processed at the Sanger institute using the well-established SNP calling algorithm ssahaSNP (24) to compute >50 million SNPs from common laboratory Mus musculus strains, which were then merged with dbSNP release 126. The resulting data were incorporated into the Ensembl variation schema and a new transcript-centric display TranscriptSNPView was developed to show this variation in a strain-specific way (25). Figure 3 shows an example of this display. TranscriptSNPView is also available in dog (Canis familiaris) Ensembl to provide access to SNPs determined from 16 different strains as part of the dog sequencing project (26). As well as providing an organized view of these data through a web interface, the underlying data storage structure and variation software API makes it easy for bioinformaticians to write custom programs to analysis these complex data. For example, the API makes it possible to view variation from the point of view of any strain with all necessary coordinate transformation being carried out transparently. With resequencing data being generated at an ever increasing rate we anticipate incorporating strain-specific data for a variety of species in the future. Specifically rat (Rattus norvegicus) and chicken (G.gallus) variation data will be available through TranscriptSNPView before the end of 2006.
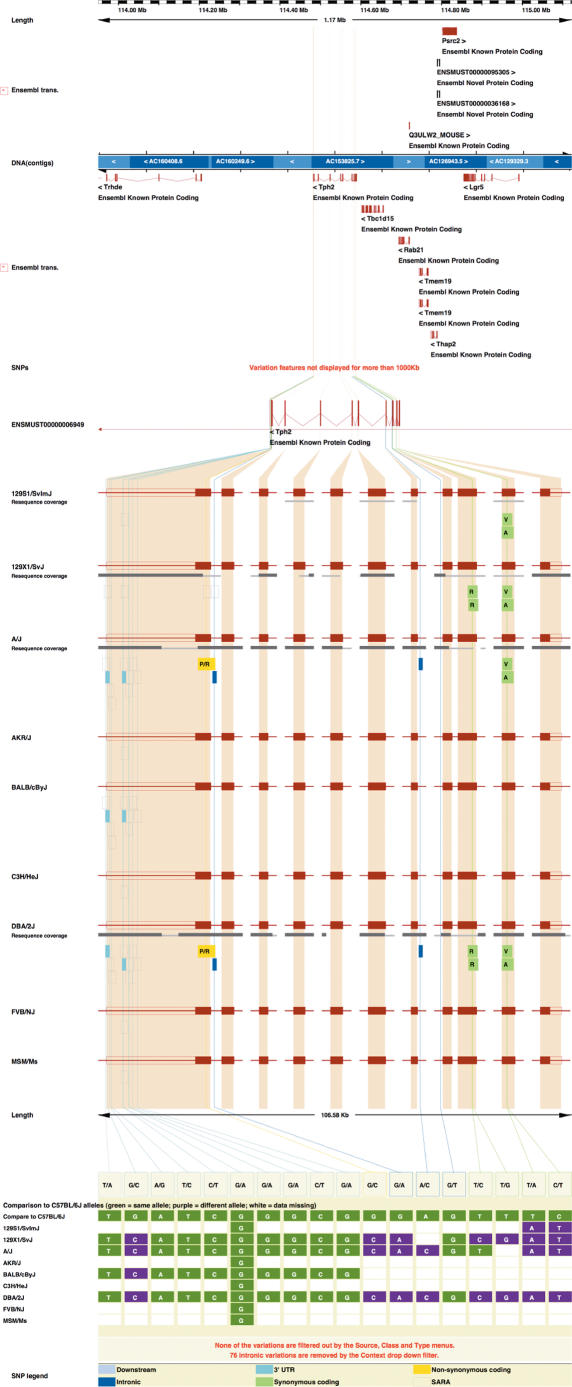
Figure 3.
Figure shows the sequence variation across mouse strains for the transcript ENSMUST00000006949 in the new view TranscriptSNPView. Strain-specific SNPs were calculated by aligning mouse reads from different strains against the reference genome as described previously (25). This gene-centric view collapses the size of introns to focus on variation within exons, which are shown separately for each strain with the consequences of any SNPs on their coding sequence. The extent of resequencing coverage is also shown for each strain.
Comparative genomics
The major improvement in comparative genomics has been the June 2006 release switch of the ortholog/paralog prediction pipeline to one based on protein tree calculations from one based on best reciprocal similarity relationships. This is a major change and has required a completely new pipeline, schema, API and display. In the new pipeline maximum-likelihood phylogenetic unrooted gene trees are built using the algorithm PHYML (27) from multiple protein sequence alignments generated using MUSCLE (28) for each gene family containing sequences from all species. Gene families are generated by calculating best reciprocal relationships between translations of all genes followed by single linkage clustering. Finally each gene tree is reconciled with the species tree using the RAL algorithm (29) to call duplication events on internal nodes and to root the tree. The advantage of a gene tree based pipeline is that it is able to identify complex one-to-many and many-to-many relationships between genes resulting from ancient duplication events, unlike best reciprocal methods. The new structure of orthology/paralogy relationships permeate the entire Ensembl site; however, the biggest visual change is the new GeneTreeView display as shown in Figure 4. Predicted gene trees of course suffer issues of the prediction algorithm's parameters not being ideal for all cases and of bad sequence alignments introducing errors, just as does automatic gene annotation: in a proportion of cases the predicted tree will be worse than could be obtained by manual curation. As a result a similar relationship between Ensembl automation and curation is developing for gene trees as for gene sets. Ensembl has started to collaborate with the TreeFam project (30), a curated resource of gene trees, and is currently investigating ways to integrate available curated data into the automatic pipeline.
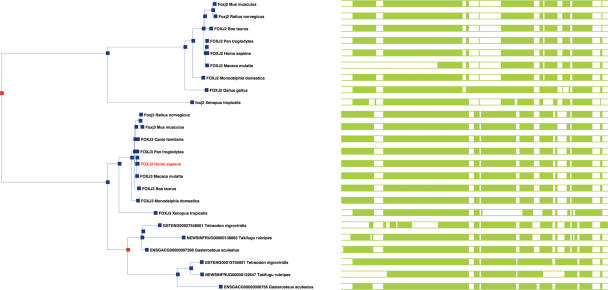
Figure 4.
Figure shows the gene tree panel from the GeneTreeView web page for the human FOXJ3 gene, generated by the Ensembl gene orthology/paralogy prediction pipeline. Most of the ortholog relationships are one to one; however, there is a one-to-many relationship to the fish lineage, where the gene appears to have duplicated. The relationship to the paralogous gene FOXJ2 can also be seen, where the orthologs in the fish lineage appear to have been lost. The full web page (not shown) includes links to view the tree structure in the Java applet ATV and view the protein sequence alignment upon which it is based in Java applet Jalview. The green bars represent the alignments of the protein translations upon which the tree is based, where shaded blocks represent aligned regions. Poor and fragmented alignments can be the cause of erroneous placements of genes in the tree, so the visualization of the alignment is useful when interpreting the tree.
One of the hidden consequences of switching to this more robust predictive model is the ability to use the more reliable implied functional relationships between genes to propagate functional labeling between them. Previously Ensembl genes were described as ‘Known’ if there was some known functional description attached to supporting organism-specific transcription sequence (such as from UniProt). If the only annotated supporting evidence was from another organism, the gene would be labeled as ‘Novel’. From the February 2006 release, a third category of genes was introduced called ‘Known by projection’. For these genes, a functional description has been projected from GO terms via the orthology mapping. Although these are predictions, we are confident that the annotation is sufficiently accurate to be very useful.
Web usability and web service integration
Following the major redesign of the website reported past year (17), this has been a year of consolidation for the website, adding completely new views, such as TranscriptSNPView, GeneTreeView and IDHistoryView and working on backend infrastructure that will allow new functionality, such as users logins to be added over the coming year.
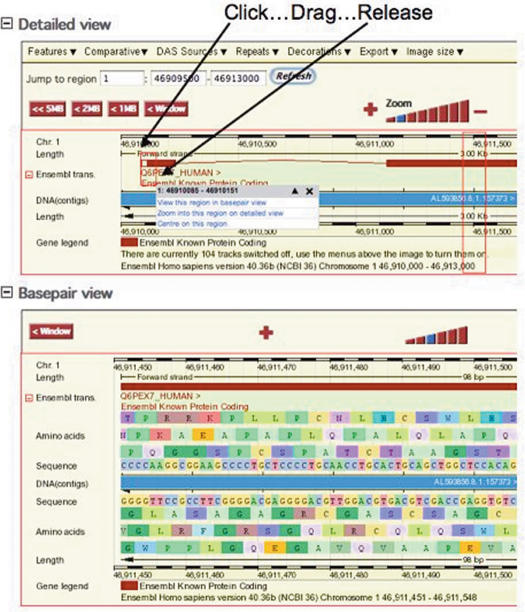
A major ongoing issue for the website is improving interactivity. One improvement to at least partly address this is a drag and zoom functionality that has been added to ContigView using JavaScript as shown in Figure 5. This greatly improves the interactivity of the display; however full scrolling functionality, as popularized by sites such as maps.google.com, will require the adoption of new web technologies for which extensive redevelopment is required. As a step towards this some of the information displayed in the popup menus on the ContigView pages are now fetched asynchronously using the AJAX (Asynchronous JavaScript and XML) protocol. Previously, descriptions were not included at all in these popups since it would make the pages too large, so this is added functionality made possible by the new protocols.
Figure 5.
Figure shows a portion of the human genome in ContigView showing the transcript Q6PEX7_HUMAN. ContigView has a ‘Basepair view’ panel allowing DNA sequence and six frame translation to be examined, which is shown centered on the second exon of this transcript. The introduction of AJAX functionality to ContigView greatly simplifies navigation to precise locations in ‘Basepair view’. Click and drag in any ContigView panel and a red box is drawn. Upon mouse release a popup appears. In this example a region around the start of translation has been selected in ‘Detailed view’. The mouse gesture that the user needs to perform of ‘click…drag…release’ is shown by the annotation on the figure. Clicking on the first option in the popup would reposition base pair view around this feature. This functionality greatly improves the interactivity of the web interface and will be progressively incorporated into other Ensembl views.
FUTURE DIRECTIONS
There are a number of areas which Ensembl is anticipating over the coming year. The increase in the number of 2× genomes will require continuous development of both gene building and comparative genomics pipelines. In comparative genomics we are also concentrating on providing other clade-specific multiple alignments, starting with the telosts. We will be collaborating with the TreeFam group (30) to provide better gene level comparative genomics across the genomes we handle. We envisage a steady growth of whole genome assays of DNA-binding proteins using techniques such as Chromatin immunoprecipitation on DNA microarrays (ChIP/chip) and other functional studies of genome sequence over the next year. We see ArrayExpress (31) as a natural archive of the experimental results such as ChIP/chip, but Ensembl as the display and integration engine; our goal is to move beyond just the display of the ChIP/chip results towards providing a ‘Regulatory Build’ integrating appropriate information. Finally we foresee growth in the variation data both in human and in other species, in particular resequencing data. We hope to integrate more resequencing information currently available in the trace archive (http://trace.ensembl.org/, http://www.ncbi.nlm.nih.gov/Traces/) with many of the species in Ensembl and present it in a friendly manner.
Acknowledgments
The Ensembl project is principally funded by the Wellcome Trust with additional funding from EMBL, NIH–NIAID and BBSRC. We are grateful to Gudmundur Arni Thorisson, to users of our website and to the developers on our mailing lists for much useful feedback and discussion. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashurst J.L., Chen C.K., Gilbert J.G., Jekosch K., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., et al. The Vertebrate Genome Annotation (Vega) database. Nucleic Acids Res. 2005;33:D459–D465. doi: 10.1093/nar/gki135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarz E.M., Antoshechkin I., Bastiani C., Bieri T., Blasiar D., Canaran P., Chan J., Chen N., Chen W.J., Davis P., et al. WormBase: better software, richer content. Nucleic Acids Res. 2006;34:D475–D478. doi: 10.1093/nar/gkj061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasprzyk A., Keefe D., Smedley D., London D., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birney E., Andrews D.T., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., et al. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birney E., Clamp M., Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuff J.A., Coates G.M., Cutts T.J., Rae M. The Ensembl computing architecture. Genome Res. 2004;14:971–975. doi: 10.1101/gr.1866304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curwen V., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M., Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyras E., Caccamo M., Curwen V., Clamp M. ESTGenes: alternative splicing from ESTs in Ensembl. Genome Res. 2004;14:976–987. doi: 10.1101/gr.1862204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potter S.C., Clarke L., Curwen V., Keenan S., Mongin E., Searle S.M., Stabenau A., Storey R., Clamp M. The Ensembl analysis pipeline. Genome Res. 2004;14:934–941. doi: 10.1101/gr.1859804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Searle S.M., Gilbert J., Iyer V., Clamp M. The otter annotation system. Genome Res. 2004;14:963–970. doi: 10.1101/gr.1864804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stabenau A., McVicker G., Melsopp C., Proctor G., Clamp M., Birney E. The Ensembl core software libraries. Genome Res. 2004;14:929–933. doi: 10.1101/gr.1857204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stalker J., Gibbins B., Meidl P., Smith J., Spooner W., Hotz H.R., Cox A.V. The Ensembl website: mechanics of a genome browser. Genome Res. 2004;14:951–955. doi: 10.1101/gr.1863004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D. Evolution's cauldron: duplication, deletion and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lander E.S., Waterman M.S. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 21.The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 22.Guigó R., Flicek P., Abril J.F., Reymond A., Lagarde J., Denoeud F., Antonarakis S., Ashburner M., Bajic V.B., Birney B., et al. EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol. 2006;7:S2. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl. 1):S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ning Z., Cox A.J., Mullikin J.C. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham F., Rios D., Griffiths M., Smith J., Ning Z., Cox T., Flicek P., Marin-Garcin P., Herrero J., Rogers J., et al. TranscriptSNPView: a genome-wide catalog of mouse coding variation. Nature Genet. 2006;38:853. doi: 10.1038/ng0806-853a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 27.Guindon S., Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 28.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dufayard J.F., Duret L., Penel S., Gouy M., Rechenmann F., Perriere G. Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- 30.Li H., Coghlan A., Ruan J., Coin L.J., Heriche J.K., Osmotherly L., Li R., Liu T., Zhang Z., Bolund L., et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brazma A., Kapushesky M., Parkinson H., Sarkans U., Shojatalab M. Data storage and analysis in ArrayExpress. Meth. Enzymol. 2006;411:370–386. doi: 10.1016/S0076-6879(06)11020-4. [DOI] [PubMed] [Google Scholar]