Abstract
The reaction center (RC) of photosynthetic bacteria is a membrane protein complex that promotes a light-induced charge separation during the primary process of photosynthesis. In the photosynthetic electron transfer chain, the soluble electron carrier proteins transport electrons to the RC and reduce the photo-oxidized special-pair of bacteriochlorophyll. The high-potential iron-sulfur protein (HiPIP) is known to serve as an electron donor to the RC in some species, where the c-type cytochrome subunit, the peripheral subunit of the RC, directly accepts electrons from the HiPIP. Here we report the crystal structures of the RC and the HiPIP from Thermochromatium (Tch.) tepidum, at 2.2-Å and 1.5-Å resolution, respectively. Tch. tepidum can grow at the highest temperature of all known purple bacteria, and the Tch. tepidum RC shows some degree of stability to high temperature. Comparison with the RCs of mesophiles, such as Blastochloris viridis, has shown that the Tch. tepidum RC possesses more Arg residues at the membrane surface, which might contribute to the stability of this membrane protein. The RC and the HiPIP both possess hydrophobic patches on their respective surfaces, and the HiPIP is expected to interact with the cytochrome subunit by hydrophobic interactions near the heme-1, the most distal heme to the special-pair.
In photosynthetic purple bacteria, the electron transfer reactions of photosynthesis are performed by the following three components: the photosynthetic reaction center (RC), the cytochrome (Cyt) bc1 complex, and the soluble electron carrier protein. First, the RC promotes the light-induced charge separation across the plasma membrane, which results in the oxidation of the special-pair and the reduction of the quinone to the quinol. The quinol then leaves the RC and moves to the Cyt bc1 complex through the quinone pool of the plasma membrane. Second, the Cyt bc1 complex reoxidizes the quinol to the quinone, and the released electrons are transferred to the soluble electron carriers. Third, the soluble electron carriers transport the electrons to the RC through the periplasmic space. Finally, the photo-oxidized special-pair is reduced by the soluble electron carriers, and the RC comes back to the initial state. In the course of the oxidation and the reduction of the quinones, the transmembrane electrochemical gradient of the protons is formed, and its energy is used to produce ATP by ATP synthase.
Thermochromatium (Tch.; formerly Chromatium) tepidum is a purple sulfur bacterium originally isolated from the hot springs in Yellowstone National Park (1, 2) and belongs to the γ-subclass. Tch. tepidum is a thermophilic bacterium and can grow at the highest temperature of all known purple bacteria. The optimum growth temperature is 48–50°C, the maximum temperature 58°C. The RC from Tch. tepidum is stable up to 70°C in chromatophore and to 48°C in detergent-micelle (3). The RC is the first membrane protein whose three-dimensional structure has been determined at an atomic resolution (4, 5), and the crystal structures of the RCs from two mesophiles, Blastochloris (Blc.; formerly Rhodopseudomonas) viridis (6) and Rhodobacter (Rb.) sphaeroides (7–10), are now available. Compared with the RCs of these two species, the functional features unique to the Tch. tepidum RC are focused on the following two points: the thermostability of the membrane protein and the electron transfer from the soluble electron carrier proteins.
The RC of Tch. tepidum, like Blc. viridis, possesses the c-type Cyt subunit (11), which accepts electrons from the soluble electron carriers and reduces the photo-oxidized special-pair, whereas the soluble electron carriers directly reduce the special-pair in other species like Rb. sphaeroides. The Cyt subunit contains four covalently bound heme groups, which form the electron pathway to the special-pair. The soluble electron carrier proteins are classified into two groups: the c-type Cyt and the high-potential iron-sulfur protein (HiPIP) (12, 13). The HiPIP, which contains a [4Fe-4S] cluster, exists abundantly in the periplasmic space of some purple bacteria. The HiPIP shows the 2+/3+ redox transition, and its redox potential is positive and relatively high compared with that of ferredoxins with the 1+/2+ redox transition (14, 15). Which type of electron carrier is used depends on the species, and both types are involved in the electron transfer chain in some species (16, 17). For example, whereas the Blc. viridis RC uses Cyt c2 as a reductant, the Tch. tepidum RC accepts electrons from the HiPIP. In Blc. viridis, the crystal structures of the RC and Cyt c2 both have been determined independently (18, 19), and the docking site and the mechanism of the molecular recognition have been discussed (20, 21). In contrast, although the crystal structures of HiPIPs from some species have been determined (22–26), structural information on the RC whose physiological electron donor is the HiPIP is not yet available, and the mechanism of the molecular recognition has not yet been fully examined.
Here we report the crystal structures of the RC and the HiPIP from Tch. tepidum at 2.2-Å and 1.5-Å resolution, respectively. The present RC structure is the first one for the thermostable RC and is expected to show structural features with regard to thermostability of the membrane protein in comparison with other mesophiles. In addition, the structural information on the RC and the HiPIP makes it possible to discuss where and how these two proteins interact in the electron transfer reaction.
Materials and Methods
Purification and Crystallization.
The RC and HiPIP both were purified from Tch. tepidum, which was grown under luminescent conditions according to published procedures (2). The RC proteins first were solubilized from the intracytoplasmic membranes of Tch. tepidum by using the detergent lauryldimethylamine oxide (LDAO) and purified by anion-exchange chromatography as reported (27, 28). The purified RCs were charged to the anion-exchange column to exchange the detergent LDAO for n-octyl-β-d-glucopyranoside (β-OG). The HiPIPs were isolated from the soluble fraction of Tch. tepidum by using anion-exchange chromatography and gel filtration as reported (29). The RC proteins solubilized with β-OG were crystallized at 277 K by using polyethylene glycol 4000 and sodium chloride as precipitants (27, 28). The HiPIPs were crystallized at 293 K by using ammonium sulfate (29).
Data Collection and Structure Determination.
X-ray diffraction data were collected at BL-6A (Photon Factory, Tsukuba, Japan) and BL-41XU (SPring-8). At the Photon Factory, a screenless Weissenberg camera was used with a cylindrical cassette with a 286.5-mm radius (30), and the x-ray was monochromatized to 1.000 Å. The diffraction intensities were recorded on 200 × 400-mm imaging plates (Fuji) and read out on a Fuji BAS2000 scanner (31). At SPring-8, an Raxis-IV imaging-plate camera system (Rigaku, Tokyo) was used as a detector, and the x-ray was monochromatized to 0.708 Å. In data collection of the RC at SPring-8, the crystal was soaked in the solution containing 25% (wt/vol) glycerol as a cryoprotectant and frozen in a nitrogen-gas stream. The intensity data were processed with the program denzo and merged with the program scalepack (32).
The molecular replacement was carried out by using the program amore (33) in the CCP4 program suite (34). The atomic coordinates of the Blc. viridis RC (PDB code: 1PRC) (6) and the Allochromatium (Ach.; formerly Chromatium) vinosum HiPIP (1HIP) (23) were used as search models. The molecular model was built by manual fitting to the electron density map by using the programs turbo frodo (35) and o (36), followed by the structure refinement using the program x-plor (37). The electron density maps were calculated with σA weights by using the program sigmaa (38) in the CCP4 program suite so as to reduce the model bias. The structure refinement by x-plor included the simulated-annealing protocol (39) and individual B factor refinement. The topology and parameter files by Lancaster and Michel (40) were used for the prosthetic groups in x-plor, and those for the carotenoid, detergent, and lipid molecules were generated by using the program xplo2d (41), where the parameters were improved by the energy minimization in x-plor. The stereochemistry of the structure was assessed by the program procheck (42).
The rms deviation between two structures was calculated by using the program lsqman (43). The charge distribution of the molecular surface was calculated and represented by using the program grasp (44). The molecular models in the figures were drawn by using the programs molscript (45) and raster3d (46).
Results and Discussion
Crystal Structure of the Tch. tepidum RC.
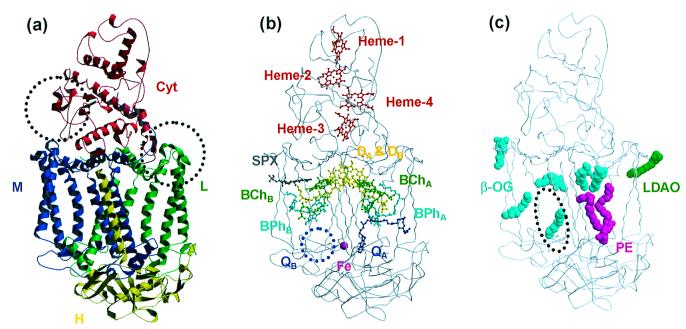
The intensity data at 2.2-Å resolution were obtained with an overall Rmerge of 9.3% and a completeness of 92.6% as shown in Table 1. The bulk solvent refinement (47) was introduced in the final stage of the refinement by using x-plor, which contributed largely to the reduction of the R values. Finally, the structure was refined at 2.2-Å resolution to a crystallographic R factor of 23.1% with a free R factor of 28.7%. The refinement statistics are shown in Table 2. The final model includes four protein subunits and six kinds of prosthetic groups (Fig. 1). Whereas the nucleotide sequence has indicated that the L, M, H, and Cyt subunits of the Tch. tepidum RC consist of 280, 324, 259, and 381 residues, respectively (48), those in the final model consist of 280, 318, 238, and 310 residues, respectively. The highly disordered regions in the electron density maps are omitted from the final structure as follows: six of the C-terminal residues in the M subunit, six of the N-terminal residues in the H subunit, and the peripheral loop region in the H subunit between Asn-H44 and Phe-H58. In the Cyt subunit, 71 of the C-terminal residues are not observed in the electron density maps. Notably, mass spectrometry also has suggested that the polypeptide chain of the Cyt subunit in the RC complex is shorter than expected from the nucleotide sequence. Over 84% of all residues are assigned in the most favored regions of the Ramachandran plot, and only eight residues appear in the generously allowed and disallowed regions. The following prosthetic groups, which form two branches (A and B branches) that are related by the pseudo 2-fold axis perpendicular to the membrane plane, are included in the final structure: a bacteriochlorophyll a dimer (DA and DB) as a special-pair, two accessory bacteriochlorophyll a monomers (BChA and BChB), two bacteriopheophytin a molecules (BPhA and BPhB), a menaquinone-8 molecule as primary quinone (QA), a nonheme-iron atom, a spirilloxanthin molecule as carotenoid, and four c-type heme groups. The secondary quinone (QB) is known to be ubiquinone, but no electron density corresponding to the ubiquinone is observed around the QB site. The spectroscopic data have suggested that spirilloxanthin has one cis-bond, which is thought to be at the position 15 based on the electron density.
Table 1.
Data collection statistics
| Data set | RC1 | RC2 | HiPIP |
| X-ray source | BL-6A(PF) | BL-41XU(SPring-8) | BL-6A(PF) |
| Wavelength (Å) | 1.000 | 0.708 | 1.000 |
| Temperature (K) | Ambient | 100 | Ambient |
| Resolution limits (Å) | 20–2.5 | 20–2.2 | 20–1.5 |
| Highest-resolution shell (Å) | 2.56–2.5 | 2.25–2.2 | 1.53–1.5 |
| Space group | P212121 | P212121 | P212121 |
| Unit cell dimensions (Å) | 134.2, 200.6, 85.0 | 133.3, 196.6, 84.2 | 47.2, 59.6, 23.6 |
| Measured reflections | 245,871 | 508,729 | 50,767 |
| Unique reflections | 63,727 | 110,923 | 10,306 |
| Rmerge (%)* | 7.7 (39.2) | 9.3 (36.2) | 6.7 (21.9) |
| Completeness (%)* | 80.3 (55.5) | 92.6 (78.6) | 86.7 (67.6) |
| l/σ(l)* | 9.4 (2.9) | 12.6 (2.6) | 13.1 (6.0) |
The value for the highest resolution shell in parenthesis.
Table 2.
Refinement statistics
| Data set | RC2 | HiPIP |
| Resolution limits (Å) | 10–2.2 | 10–1.5 |
| Highest resolution shell (Å) | 2.3–2.2 | 1.57–1.5 |
| Rwork (%)* | 23.1 (30.2) | 21.2 (24.7) |
| Rfree (%)* | 28.7 (31.2) | 23.8 (24.8) |
| No. of non-H atoms | 10,047 | 667 |
| Protein | 9,010 | 616 |
| Prosthetic group | 664 | 8 |
| Water | 188 | 43 |
| Detergent | 136 | |
| Lipid | 47 | |
| Mean B factors (Å2) | ||
| Protein | 34.9 | 7.4 |
| Prosthetic group | 27.3 | 5.2 |
| Water | 34.8 | 20.3 |
| Detergent | 53.1 | |
| Lipid | 51.7 | |
| rms deviation from idealty | ||
| Bond length (Å) | 0.011 | 0.005 |
| Bond angle (deg.) | 1.89 | 1.17 |
| Dihedral angles (deg.) | 22.9 | 26.7 |
| Improper angles (deg.) | 0.87 | 0.70 |
The value for the highest-resolution shell in parenthesis.
Figure 1.
Overall structures of the RC from Tch. tepidum. (a) The ribbon models represent four polypeptide chains: Cyt (red), L (green), M (blue), and H (yellow). The dotted circles indicate two regions where the loop conformations are different from those of the Blc. viridis RC (Left: Gln-C161–Asn-C162; Right: Gly-L56–Leu-L64). (b) The ball-and-stick models represent the configuration of the prosthetic groups (DA and DB: special-pair; BChA and BChB: bacteriochlorophyll a monomers; BPhA and BPhB: bacteriopheophytin a molecules; QA: menaquinone-8; Fe: nonheme iron; Spx: spirilloxanthin; Heme-1, -2, -3, and -4: heme c groups). The secondary quinone (QB) site (blue dotted circle) is empty because QB was not observed in the electron density maps. (c) The space-filling models represent the detergent and lipid molecules observed on the molecular surface. Six β-OG molecules are represented by cyan. One LDAO molecule and one lipid molecule (P) are represented by green and magenta, respectively. The dotted circle indicates the binding site of the cardiolipin in the Rb. sphaeroides RC structure (49).
In addition, one lipid and seven detergent molecules were observed on the molecular surface of the Tch. tepidum RC (Fig. 1c). Six of seven detergents were assigned as β-OGs. The remaining one was thought to be LDAO, which was used for the initial solubilization of the RC, because the electron density corresponding to the head group was too small to cover the large head group of β-OG, but was completely fit to the head group of LDAO. The lipid molecule could be assigned as phosphatidylethanolamine (PE), and both of the two lipidic acids were thought to have 16 carbon atoms where the presence of double bonds could not be detected in the electron density. The phosphate group of PE is bound to Arg-H31 and Lys-H35 of the RC by the electrostatic interaction, and Tyr-H39 and Gly-M256 by hydrogen bonds. There are two clefts on the molecular surface of the RC in the transmembrane region, both of which are formed between the transmembrane helix of the H subunit and the helix bundle of the L and M subunits. This PE molecule fits into one of these clefts and covers the tail groups of BChA, BPhA, and QA that protrude out of the RC complex. It recently was shown that one lipid molecule (cardiolipin) was associated with the RC complex of Rb. sphaeroides (49). This cardiolipin also was buried in the cleft formed by the transmembrane helix of the H subunit, but the binding site was located on the opposite side of this PE-binding cleft (Fig. 1c).
Comparison with the Blc. viridis and Rb. sphaeroides RCs.
Although the sequence identities of the RCs between Tch. tepidum and Blc. viridis are not so high (65.0, 59.4, 49.0, and 48.8% for the L, M, H, and Cyt subunits, respectively), the conformation of each subunit is almost conserved between these two species, showing rms deviations in the superposition of Cα atoms of 1.18 Å for the whole RC complex (0.72, 1.04, 1.16, and 1.22 Å for the L, M, H, and Cyt subunits, respectively). Conformation changes are limited to the loop regions. The region between Gly-L56 and Leu-L64 forms a large loop (Fig. 1a), which does not appear in the RCs of Blc. viridis and Rb. sphaeroides. In the primary structure, this loop region is located in the connection of the first and second transmembrane helices. Other significant conformation changes of the Tch. tepidum RC are found only in the Cyt subunit, where two long deletions exist: one located between Gln-C161 and Asn-C162, the other between Ala-C197 and Gly-C198. The former deletion causes the flipping of the loop region (Fig. 1a). The loop region of the Blc. viridis RC is close to the interface of the Cyt and M subunits, whereas that of the Tch. tepidum RC is separated from this interface. The latter deletion results only in a shortcut of the loop.
Specific interactions between the prosthetic groups and protein subunits are almost conserved, except the hydrogen bond with the ring-I keto-O atom of DB. Whereas both the RCs of Blc. viridis and Tch. tepidum have a Tyr residue in a position where it can interact with the ring-I keto-O atom of DB (Tyr-M196 in Tch. tepidum), the RC of Rb. sphaeroides has no Tyr residue in this position. The analyses of the primary structure and the Fourier transform-IR study have anticipated the presence of this hydrogen bond in the RC of Tch. tepidum (50). This hydrogen bond could provide the higher redox potential of the special-pair. In addition, this hydrogen bond could affect the delocalization of the positive charge in the oxidized special-pair.
Thermostability of the Tch. tepidum RC.
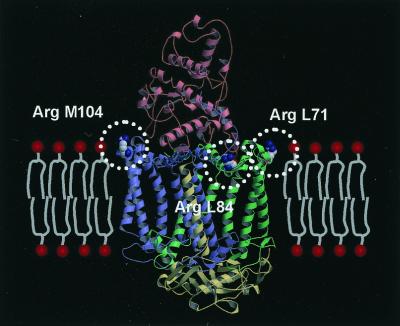
Although Tch. tepidum is a thermophilic bacterium and its RC shows some stability to higher temperature (3), no significant structural differences that might contribute to the stability of the soluble proteins were observed when the structure was compared with the RCs from mesophiles; for example, the ionic bonds and hydrogen bonds formed in the complex, the increased Pro residues in the loop region, the shortening of the loop region, and so on. The loop deletions in the Cyt subunit are not peculiar to the RC of Tch. tepidum, but common among the closely related species including mesophiles. In contrast, the sequence alignment has shown a significant difference between Tch. tepidum and mesophiles. The L and M subunits of Tch. tepidum have three more Arg residues than do the mesophiles, and it is noteworthy that all of these Arg residues are located at the interface of the transmembrane and peripheral regions, as shown in Fig. 2. Comparison with the Ach. vinosum RC (51) has revealed that these Arg residues exist only in the RC of Tch. tepidum, despite the fact that the sequence identities of the L and M subunits are 91.1 and 89.2%, respectively. The Arg and Lys residues at the membrane surface could interact with the phosphate groups of the lipids, as observed in this crystal structure, which would affect the affinity of this RC to the membrane.
Figure 2.
Three Arg residues located at the membrane surface of the RC from Tch. tepidum. In the crystal structure, all three additional Arg residues (drawn by the space-filling model) are found to be located at the membrane surface. Each subunit is colored red (Cyt), green (L), blue (M), or yellow (H). The model of the membrane is placed on the ribbon model of the RC complex, and the head groups of the phospholipids are represented by the red circles. All three Arg residues are exposed to the solvent and located in a position from which they can interact with the head groups of the phospholipids.
Crystal Structure of the Tch. tepidum HiPIP.
The intensity data at 1.5-Å resolution were obtained with an overall Rmerge of 6.7% and a completeness of 86.7% as shown in Table 1. After the refinement, the structure was refined to a crystallographic R factor of 21.2% with a free R factor of 23.8%, and all of the 83 amino acid residues and a [4Fe-4S] cluster could be assigned to the electron density maps. Over 92% of all residues were assigned in the most favored regions of the Ramachandran plot, and no residue was located in the generously allowed or disallowed regions. The final refinement statistics are shown in Table 2.
Comparison with Other HiPIPs.
The overall structure of the HiPIP from Tch. tepidum is quite similar to the previously determined crystal structures of the HiPIPs of Ach. vinosum (23, 25), showing rms deviations in the superposition of Cα atoms of less than 1.0 Å. Most of the differences in the primary structure are present in the segment between Asn-52 and Asp-57, which forms two strands of antiparallel β-sheets in the Tch. tepidum HiPIP. In the Ach. vinosum HiPIP, the β-sheet is interrupted by the hairpin loop, and this segment is one of the most flexible regions. On the other hand, the β-sheet seems more rigid in the Tch. tepidum HiPIP, because the segment is two residues shorter than that of Ach. vinosum and more hydrogen bonds are formed between the two β-strands without the hairpin loop interruption. It has been reported that the Tch. tepidum HiPIP is a little more stable than that of Ach. vinosum (52), and the deletion might reduce the flexibility of the segment and affect the thermostability of the Tch. tepidum HiPIP.
Although the crystal structures of HiPIPs of other species have shown that HiPIPs can form dimers (24, 25), the dimeric structures are significantly different among these crystal structures. In contrast, the Tch. tepidum HiPIPs exist as monomers in the crystal, and the dynamic light scattering also has suggested that the Tch. tepidum HiPIP molecules in solution do not form stable dimers (29). These findings suggest that the HiPIPs do not form a rigid dimer, where dimer-monomer exchanges can easily occur in solution.
Implication for Molecular Recognition.
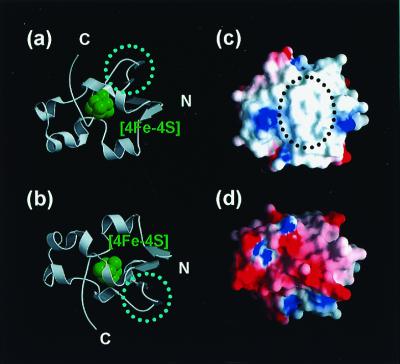
The HiPIP and the RC must possess the molecular surfaces that can complementarily interact with each other to transfer electrons. The binding site of these two proteins was screened on the basis of the charge distribution of the molecular surface. Based on the crystal structure, the Tch. tepidum HiPIP possesses a large hydrophobic patch on its surface adjacent to the [4Fe-4S] cluster, and the backside of this surface is charged and mostly acidic (Fig. 3). This hydrophobic patch is conserved among the three-dimensional structures of HiPIPs of other species. Spectroscopic study has shown that HiPIPs are capable of forming temporary dimers in solution by using the hydrophobic surface and that the electron transfer can occur between the two monomers in the dimeric structure (53, 54), suggesting that the HiPIP monomer in solution interacts temporally with the other protein via its hydrophobic patch to transfer electrons.
Figure 3.
Overall structure (a and b) and charge distribution of the molecular surface (c and d) of the HiPIP from Tch. tepidum. The molecule is viewed from the side proximal to the [4Fe-4S] cluster (a and c) and from the side distal to the [4Fe-4S] cluster (b and d). (a and b) Ribbon model with the [4Fe-4S] cluster shown in green. The dotted circles indicate the two strands of β-sheet whose conformation is different from that of the Ach. vinosum HiPIP. (c and d) Molecular surface with the charge distribution (negative charge: red; positive charge: blue). The molecular surface proximal to the [4Fe-4S] cluster forms a large hydrophobic patch, as shown in the dotted circle, whereas that of the distal side is hydrophilic and mostly negative.
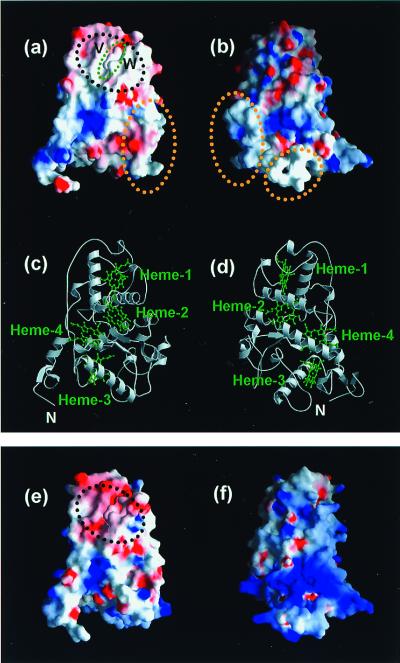
On the other hand, the Cyt subunit of the Tch. tepidum RC possesses a hydrophobic surface only around the heme-1, whereas other surfaces are mostly charged (Fig. 4). The site-directed mutagenesis of the RC of Rubrivivax gelatinosus has suggested that the electrostatic force has little influence on the interaction between the HiPIP and the Cyt subunit and that the electron transfer is inhibited in replacement of the Val and Leu residues near the heme-1 with the charged residue (21, 55). The Val residue conserved in the RC of Tch. tepidum (Val-C65) is located near the heme-1 and is indeed exposed to the solvent in the crystal structure (Fig. 4a). The Leu residue is not conserved in the Tch. tepidum RC, but the corresponding residue, Trp-C94, also contributes to the hydrophobicity of the surface around the heme-1. Considering all of these results together, the HiPIP monomer is suggested to dock to the heme-1-protruding site on the Cyt subunit for the electron transfer, by using hydrophobic interactions.
Figure 4.
Structures of the Cyt subunits of Tch. tepidum RC (a–d) and Blc. viridis RC (e and f). (a and b) Charge distribution of the Cyt subunit of Tch. tepidum around the heme-1 (a) and viewed from the backside of the heme-1 (b). The negatively charged surfaces are shown in red, and the positively charged surfaces in blue. The molecular surface around the heme-1 is mostly hydrophobic, which is shown in the black dotted circle. The protruding part of the heme-1 is shown in the green dotted circle. V and W indicate the Val-C65 and Trp-C94, respectively, which affect the molecular recognition of the RC and HiPIP. The yellow dotted circles indicate the two loop regions whose conformations are different from those of the Blc. viridis RC. (c and d) Ribbon model of the Cyt subunit of Tch. tepidum viewed from the same angle as that for a and b, respectively. Each heme is shown in green. (e and f) Charge distribution of the Cyt subunit of Blc. viridis viewed from the same angle as that for a and b, respectively. The molecular surface is generated by using the atomic coordinates of the RC from Blc. viridis (PDB code: 1PRC). In Blc. viridis, only the molecular surface around the heme-1 is acidic, which is shown in the black dotted circle, and most of the other surfaces are basic.
It also was found that the surface around the heme-1 is the best candidate for a binding site of Cyt c2 in the Blc. viridis RC (20). The heme-protruding site of Cyt c2 is positively charged (18), whereas the molecular surface around the heme-1 of the Cyt subunit of Blc. viridis is negatively charged (6) (Fig. 4). In the site-directed mutagenesis, the substitutions of acidic residues around the heme-1 inhibited the electron transfer between these two proteins (20, 21). Therefore, the electrostatic force probably dominates the interaction in the case of Cyt c2. Between these two cases, the HiPIP and Cyt c2, the binding sites of the soluble electron carrier proteins are common, but the interactions with the Cyt subunits are controlled by completely different mechanisms. As mentioned above, the conformations of the two loop regions in the Cyt subunit are largely different between the Tch. tepidum and Blc. viridis RCs. However, these conformation changes have no influence on the molecular recognition of the soluble electron carrier proteins, because these loop regions are remote from the heme-1 protruding site.
It has been thought that the heme-1 is unfavorable for accepting the electrons from the soluble electron carriers, because the redox potential of the heme-1 (Blc. viridis RC; −60 mV) is much lower to those of the soluble electron carrier proteins (Tch. tepidum HiPIP; +323 mV, Blc. viridis Cyt c2; +285 mV). The redox potential of the heme-1 of the Tch. tepidum has not been determined precisely, but is known to be far lower than that of the HiPIP. However, it was shown that the uphill electron transfer occurs in the Cyt subunit of the Blc. viridis RC (56). The electron transfer from the heme-2 to the heme-3 is stepwise. The first step, from the heme-2 to the heme-4, is thermodynamically unfavorable, but it definitely occurs. Furthermore, the uphill electron transfer is supported from the theoretical aspect (57), where proximity of the redox centers, together with thermal activation, promote the electron tunneling, even if it is endergonic. These facts suggest that the heme-1 can serve as the first electron acceptor in the electron transfer chain from the soluble electron carrier to the heme-2, and consequently, to the special-pair.
The Cyt subunits of the Tch. tepidum and Blc. viridis RCs are similar in many aspects, for example, the conformations of the polypeptide chain, the order of redox potentials of the four heme groups, and the docking site of the soluble electron carriers. In contrast, the charge distributions of their molecular surfaces are completely different and complementary to those of their respective electron carrier proteins. This might be because these RCs have changed their molecular surfaces to fit those of their electron transfer partners, while they have kept the biological architecture and function as electron conductors to the special-pair. These results give a good example of the biological molecular evolution, in terms of preservation of the folding and adaptation to the environment.
Acknowledgments
We thank Drs. N. Sakabe, A. Nakagawa, N. Watanabe, M. Suzuki, and N. Igarashi of the Photon Factory and Drs. N. Kamiya, M. Kawamoto, and Y. Kawano of SPring-8 for data collection with synchrotron radiation (proposal no. 93G065 for Photon Factory Advisory Committee, and nos. 1997B0140-NL-np, 1998A0176-CL-np, and 1999A0263-NL-np for SPring-8). We also are indebted to Dr. K. Matsuura of Tokyo Metropolitan University for useful discussion and N. Katayama, Dr. K. Inaka, F. Motojima, and K. Kitamura for crystallization and the early stages of structure determination. K.M. is a member of the Sakabe Project of TARA (Tsukuba Advanced Research Alliance), University of Tsukuba. I.F. is supported by a Grant-in-Aid for Japan Society for the Promotion of Science Fellows (no. P98450). This work was partly supported (to K.M.) by the “Research for the Future” Program (JSPS-RFTF 97L00501) from the Japan Society for the Promotion of Science, Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan, a grant from the Kansai Research Foundation for Technology Promotion, the “Ground Research for Space Utilization” Program promoted by the National Space Development Agency of Japan and the Japan Space Forum, and the REIMEI Research Resources of Japan Atomic Energy Research Institute.
Abbreviations
- RC
reaction center
- HiPIP
high-potential iron-sulfur protein
- Tch.
Thermochromatium
- Blc.
Blastochloris
- Ach.
Allochromatium
- Rb.
Rhodobacter
- Cyt
cytochrome
- β-OG
n-octyl-β-d-glucopyranoside
- LDAO
lauryldimethylamine oxide
- PE
phosphatidylethanolamine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates of the RC and HiPIP from Thermochromatium tepidum have been deposited in the Protein Data Bank, www.rcsg.org [PDB ID codes 1EYS (RC) and 1EYT (HiPIP)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240224997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240224997
References
- 1.Madigan M T. Science. 1984;225:313–315. doi: 10.1126/science.225.4659.313. [DOI] [PubMed] [Google Scholar]
- 2.Madigan M T. Int J Syst Bacteriol. 1986;36:222–227. [Google Scholar]
- 3.Nozawa T, Madigan M T. J Biochem (Tokyo) 1991;110:588–594. doi: 10.1093/oxfordjournals.jbchem.a123624. [DOI] [PubMed] [Google Scholar]
- 4.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. J Mol Biol. 1984;180:385–398. doi: 10.1016/s0022-2836(84)80011-x. [DOI] [PubMed] [Google Scholar]
- 5.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Nature (London) 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 6.Deisenhofer J, Epp O, Sinning I, Michel H. J Mol Biol. 1995;246:429–457. doi: 10.1006/jmbi.1994.0097. [DOI] [PubMed] [Google Scholar]
- 7.Yeates T O, Komiya H, Chirino A, Rees D C, Allen J P, Feher G. Proc Natl Acad Sci USA. 1988;85:7993–7997. doi: 10.1073/pnas.85.21.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ermler U, Fritzsch G, Buchanan S K, Michel H. Structure (London) 1994;2:925–936. doi: 10.1016/s0969-2126(94)00094-8. [DOI] [PubMed] [Google Scholar]
- 9.Chang C H, el-Kabbani O, Tiede D, Norris J, Schiffer M. Biochemistry. 1991;30:5352–5360. doi: 10.1021/bi00236a005. [DOI] [PubMed] [Google Scholar]
- 10.Arnoux B, Ducruix A, Reiss-Husson F, Lutz M, Norris J, Schiffer M, Chang C H. FEBS Lett. 1989;258:47–50. doi: 10.1016/0014-5793(89)81612-6. [DOI] [PubMed] [Google Scholar]
- 11.Nozawa T, Trost J T, Fukada T, Hatano M, McManus J D, Blankenship R E. Biochim Biophys Acta. 1987;894:468–476. doi: 10.1016/0005-2728(87)90126-5. [DOI] [PubMed] [Google Scholar]
- 12.Kerfeld C A, Chan C, Hirasawa M, Kleis-SanFrancisco S, Yeates T O, Knaff D B. Biochemistry. 1996;35:7812–7818. doi: 10.1021/bi952731v. [DOI] [PubMed] [Google Scholar]
- 13.Hochkoeppler A, Zannoni D, Ciurli S, Meyer T E, Cusanovich M A, Tollin G. Proc Natl Acad Sci USA. 1996;93:6998–7002. doi: 10.1073/pnas.93.14.6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Klerk H, Kamen M D. Biochim Biophys Acta. 1966;112:175–178. doi: 10.1016/s0926-6585(96)90022-9. [DOI] [PubMed] [Google Scholar]
- 15.Dus K, De Klerk H, Sletten K, Bartsch R G. Biochim Biophys Acta. 1967;140:291–311. doi: 10.1016/0005-2795(67)90470-9. [DOI] [PubMed] [Google Scholar]
- 16.Menin L, Schoepp B, Parot P, Vermeglio A. Biochemistry. 1997;36:12183–12188. doi: 10.1021/bi971163b. [DOI] [PubMed] [Google Scholar]
- 17.Menin L, Yoshida M, Jaquinod M, Nagashima K V, Matsuura K, Parot P, Vermeglio A. Biochemistry. 1999;38:15238–15244. doi: 10.1021/bi991146h. [DOI] [PubMed] [Google Scholar]
- 18.Sogabe S, Miki K. J Mol Biol. 1995;252:235–247. doi: 10.1006/jmbi.1995.0491. [DOI] [PubMed] [Google Scholar]
- 19.Sogabe S, Ezoe T, Kasai N, Saeda M, Uno A, Miki M, Miki K. FEBS Lett. 1994;345:5–8. doi: 10.1016/0014-5793(94)00389-0. [DOI] [PubMed] [Google Scholar]
- 20.Osyczka A, Nagashima K V, Sogabe S, Miki K, Yoshida M, Shimada K, Matsuura K. Biochemistry. 1998;37:11732–11744. doi: 10.1021/bi980910h. [DOI] [PubMed] [Google Scholar]
- 21.Osyczka A, Nagashima K V, Sogabe S, Miki K, Shimada K, Matsuura K. Biochemistry. 1999;38:15779–15790. doi: 10.1021/bi990907d. [DOI] [PubMed] [Google Scholar]
- 22.Breiter D R, Meyer T E, Rayment I, Holden H M. J Biol Chem. 1991;266:18660–18667. doi: 10.2210/pdb2hip/pdb. [DOI] [PubMed] [Google Scholar]
- 23.Freer S T, Alden R A, Carter C W, Jr, Kraut J. J Biol Chem. 1975;250:46–54. [PubMed] [Google Scholar]
- 24.Kerfeld C A, Salmeen A E, Yeates T O. Biochemistry. 1998;37:13911–13917. doi: 10.1021/bi9810252. [DOI] [PubMed] [Google Scholar]
- 25.Parisini E, Capozzi F, Lubini P, Lamzin V, Luchinat C, Sheldrick G M. Acta Crystallogr D. 1999;55:1773–1784. doi: 10.1107/s0907444999009129. [DOI] [PubMed] [Google Scholar]
- 26.Rayment I, Wesenberg G, Meyer T E, Cusanovich M A, Holden H M. J Mol Biol. 1992;228:672–686. doi: 10.1016/0022-2836(92)90849-f. [DOI] [PubMed] [Google Scholar]
- 27.Katayama N, Kobayashi M, Motojima F, Inaka K, Nozawa T, Miki K. FEBS Lett. 1994;348:158–160. doi: 10.1016/0014-5793(94)00534-6. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi M, Nozawa T. Bull Chem Soc Jpn. 1993;66:3834–3836. [Google Scholar]
- 29.Nogi T, Kobayashi M, Nozawa T, Miki K. Acta Crystallogr D. 2000;56:656–658. doi: 10.1107/s0907444900003127. [DOI] [PubMed] [Google Scholar]
- 30.Sakabe N, Ikemizu S, Sakabe K, Higashi T, Nakagawa A, Watanabe N, Adachi S, Sasaki K. Rev Sci Instrum. 1995;66:1276–1281. [Google Scholar]
- 31.Miyahara J, Takahashi K, Amemiya Y, Kamiya N, Satow Y. Nucl Instru Methods Phys Res Sect A. 1986;246:572–578. [Google Scholar]
- 32.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 34.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 35.Roussel A, Cambillau C. turbo frodo. Marseille, France: Biographics; 1992. [Google Scholar]
- 36.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Brünger A T. x-plor. New Haven, CT: Yale Univ.; 1992. [Google Scholar]
- 38.Read R J. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 39.Brünger A T, Krukowski A, Erickson J W. Acta Crystallogr A. 1990;46:585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- 40.Lancaster C R, Michel H. Structure (London) 1997;5:1339–1359. doi: 10.1016/s0969-2126(97)00285-2. [DOI] [PubMed] [Google Scholar]
- 41.Kleywegt G J, Jones T A. Methods Enzymol. 1997;277:208–230. doi: 10.1016/s0076-6879(97)77013-7. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 43.Kleywegt G J. Acta Crystallogr D. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 44.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 45.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 46.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 47.Jiang J S, Brunger A T. J Mol Biol. 1994;243:100–115. doi: 10.1006/jmbi.1994.1633. [DOI] [PubMed] [Google Scholar]
- 48.Fathir I, Tanaka K, Yoza K, Kojima A, Kobayashi M, Wang Z Y, Lottspeich F, Nozawa T. Photosynth Res. 1997;51:71–82. [Google Scholar]
- 49.McAuley K E, Fyfe P K, Ridge J P, Isaacs N W, Cogdell R J, Jones M R. Proc Natl Acad Sci USA. 1999;96:14706–14711. doi: 10.1073/pnas.96.26.14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivancich A, Kobayashi M, Drepper F, Fathir I, Saito T, Nozawa T, Mattioli T A. Biochemistry. 1996;35:10529–10538. doi: 10.1021/bi953047j. [DOI] [PubMed] [Google Scholar]
- 51.Nagashima K V, Hiraishi A, Shimada K, Matsuura K. J Mol Evol. 1997;45:131–136. doi: 10.1007/pl00006212. [DOI] [PubMed] [Google Scholar]
- 52.Moulis J M, Scherrer N, Gagnon J, Forest E, Petillot Y, Garcia D. Arch Biochem Biophys. 1993;305:186–192. doi: 10.1006/abbi.1993.1409. [DOI] [PubMed] [Google Scholar]
- 53.Bertini I, Gaudemer A, Luchinat C, Piccioli M. Biochemistry. 1993;32:12887–12893. doi: 10.1021/bi00210a042. [DOI] [PubMed] [Google Scholar]
- 54.Couture M M, Auger M, Rosell F, Mauk A G, Boubour E, Lennox R B, Eltis L D. Biochim Biophys Acta. 1999;1433:159–169. doi: 10.1016/s0167-4838(99)00137-5. [DOI] [PubMed] [Google Scholar]
- 55.Osyczka A, Nagashima K V, Shimada K, Matsuura K. Biochemistry. 1999;38:2861–2865. doi: 10.1021/bi982747w. [DOI] [PubMed] [Google Scholar]
- 56.Chen I P, Mathis P, Koepke J, Michel H. Biochemistry. 2000;39:3592–3602. doi: 10.1021/bi992443p. [DOI] [PubMed] [Google Scholar]
- 57.Page C C, Moser C C, Chen X, Dutton P L. Nature (London) 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]