SUMMARY
Polarization of the C. elegans zygote along the anterior-posterior axis depends on cortically enriched (PAR) and cytoplasmic (MEX-5/6) proteins, which function together to localize determinants (e.g. PIE-1) in response to a polarizing cue associated with the sperm asters. Using time-lapse microscopy and GFP fusions, we have analyzed the localization dynamics of PAR-2, PAR-6, MEX-5, MEX-6 and PIE-1 in wild-type and mutant embryos. These studies reveal that polarization involves two genetically and temporally distinct phases. During the first phase (establishment), the sperm asters at one end of the embryo exclude the PAR-3/PAR-6/PKC3 complex from the nearby cortex, allowing the ring finger protein PAR-2 to accumulate in an expanding ‘posterior’ domain. Onset of the establishment phase involves the non-muscle myosin NMY-2 and the 14-3-3 protein PAR-5. The kinase PAR-1 and the CCCH finger proteins MEX-5 and MEX-6 also function during the establishment phase in a feedback loop to regulate growth of the posterior domain. The second phase begins after pronuclear meeting, when the sperm asters begin to invade the anterior. During this phase (maintenance), PAR-2 maintains anterior-posterior polarity by excluding the PAR-3/PAR-6/PKC3 complex from the posterior. These findings provide a model for how PAR and MEX proteins convert a transient asymmetry into a stably polarized axis.
Keywords: Polarity, Embryo, Par genes, C. elegans
INTRODUCTION
Cell polarity is essential for the asymmetric divisions that generate cell diversity during development and for the functional specialization of many differentiated cell types. Recently a complex of three proteins (the PDZ domain proteins PAR-3 and PAR-6 and the atypical protein kinase C PKC-3) has emerged as a core regulator of cell polarity in cells as diverse as C. elegans zygotes, Drosophila oocytes and neuroblasts, and mammalian epithelial cells (for a review, see Doe, 2001). Although the function of these proteins in regulating cell polarity appears to have been conserved in evolution, the molecular mechanisms involved remain poorly understood.
In C. elegans, PAR-3, PAR-6 and PKC-3 are required to polarize the newly fertilized egg along its anterior-posterior (AP) axis (for a review, see Lyczak et al., 2002). This axis is established shortly after fertilization and arises in response to a cue associated with the sperm asters, which defines the posterior end of the embryo. In response to this cue, the PAR-3/PAR-6/PKC-3 complex becomes enriched on the cortex opposite the sperm asters (anterior side), while PAR-2 (a ring finger protein) and PAR-1 (a serine threonine kinase) become enriched on the cortex nearest the sperm asters (posterior side). PAR-1 in turn is required for asymmetric spindle positioning and for the asymmetric localization of several developmental regulators (Guo and Kemphues, 1995). PAR-1 exerts its influence on developmental regulators through MEX-5 and MEX-6, two redundant cytoplasmic CCCH finger proteins (Schubert et al., 2000).
The order in which these proteins function was determined by analyzing how mutations in specific pathway components affect the localization of the other components. For example, in par-2 mutants, PAR-3, PAR-6 and PKC-3 become delocalized and are found throughout the cortex (Etemad-Moghadam et al., 1995; Tabuse et al., 1998; Hung and Kemphues, 1999); similarly, in par-3, par-6 and pkc-3 mutants, PAR-2 is no longer restricted to the posterior (Boyd et al., 1996). These effects have suggested that establishment of anterior and posterior PAR domains depends on antagonistic interactions between the PAR-3/PAR-6/PKC-3 complex in the anterior and PAR-2 in the posterior. In contrast, mutations in par-1, mex-5 and mex-6 were reported not to affect PAR localization in the zygote (Etemad-Moghadam et al., 1995; Boyd et al., 1996; Tabuse et al., 1998; Hung and Kemphues, 1999; Schubert et al., 2000). These mutations, however, have a dramatic effect on the distribution of several cell fate regulators. For example, P granules and germline proteins, which in wild type become restricted to the posterior end of the zygote, remain uniformly distributed in par-1 mutants (Kemphues et al., 1988) and mex-5;mex-6 double mutants (Schubert et al., 2000). MEX-5, which in wild type localizes to the anterior, is mislocalized in par-1 mutants, but PAR-1 localization is unaffected in mex-5;mex-6 double mutants (Schubert et al., 2000). These analyses have led to the ‘sequential repression model’, whereby PAR-1 restricts MEX-5 to the anterior, and MEX-5 in turn restricts P granules and germline proteins to the posterior (Kemphues, 2000). Whether PAR-1 localizes MEX-5 by promoting its translocation to the anterior or by negatively regulating its synthesis or stability in the posterior is not known.
The actin cytoskeleton also plays a central role in the establishment of AP polarity. The cortex of the zygote undergoes extensive contractions, which drive internal cytoplasm towards, and superficial cytoplasm away from, the sperm asters, creating a fountainhead effect (Golden, 2000). Embryos treated with cytochalasin D do not develop cortical contractions or cytoplasmic flow and do not localize P granules or the germline protein PIE-1 (Hill and Strome, 1988; Hill and Strome, 1990; Reese et al., 2000). Depletion of components of the actomyosin network by RNAi (such as NMY-2, a non muscle myosin II heavy chain, and MLC-4, a myosin light chain) also blocks flow and delays P granule segregation (Guo and Kemphues, 1996; Shelton et al., 1999).
PAR proteins likely function intimately with the actin cytoskeleton to polarize the zygote, but the details of this interaction remain poorly understood. Mutations in par-2, par-3, par-5 and par-6 lead to gene-specific defects in cortical contractions and cytoplasmic flow (Kirby et al., 1990), and at least one PAR protein (PAR-1) has been shown to interact directly with a cytoskeletal component (NMY-2) (Guo and Kemphues, 1996). NMY-2 and MLC-4 depleted embryos, however, still localize PAR-2 to a small region in the posterior cortex, raising the possibility that the actin cytoskeleton is NOT required for the earliest steps of polarization (Guo and Kemphues, 1996; Shelton et al., 1999). The relationship between the sperm asters and the PARs is also poorly understood. In particular it is not known (1) whether the polarity cue associated with the sperm asters functions by recruiting PAR-2 or by excluding the PAR-3/PAR-6/PKC-3 complex, and (2) how polarity is maintained after pronuclear meeting when the sperm asters are no longer restricted to one side of the embryo.
With only two exceptions to date (P granules and PIE-1) (Hird et al., 1996; Reese et al., 2000), studies describing asymmetric localization in the zygote have relied on immunofluorescence experiments on fixed embryos. This method makes it difficult to determine localization dynamics, which must be reconstructed by comparing embryos fixed at different developmental stages. Consequently, in most cases, the temporal sequence that leads to asymmetric localization in wild type, or to lack of asymmetry in mutants, has not been determined. To address this issue, we have developed a system to express and monitor GFP fusions in live embryos (Reese et al., 2000; Strome et al., 2001). In this study, we have analyzed the localization dynamics of PAR-2, PAR-6, MEX-5, MEX-6 and PIE-1 in wild type, and in embryos lacking specific PAR or MEX activities. Our observations demonstrate that (1) polarization involves distinct establishment and maintenance phases, (2) the sperm asters function primarily by excluding the PAR-3/PAR-6/PKC-3 complex, and (3) PAR-1 negatively regulates MEX-5/6 in the posterior.
MATERIALS AND METHODS
Strains
Caenorhabditis elegans strains were derived from the wild-type Bristol strain N2 and cultured as described previously (Brenner, 1974) except that all strains were maintained at 25°C. GFP lines were created using the complex array method (Kelly et al., 1997) or by particle bombardment (Praitis et al., 2001). The GFP fusions are all amino-terminal GFP fusions under the control of the pie-1 promoter and 3′UTR, with the exception of PIE-1:GFP, which is a carboxy-terminal GFP fusion. GFP:PAR-2, PIE-1:GFP and GFP:tubulin were described previously (Wallenfang and Seydoux, 2000; Reese et al., 2000; Strome et al., 2001). Initially, four independent lines were generated for GFP:PAR-6, four for GFP:PAR-2, two for PIE-1:GFP, one for GFP:MEX-5 and one for GFP:MEX-6. Lines with the same transgene exhibited the same GFP pattern, albeit at different levels, with one exception: a single GFP-PAR-2 line showed very bright cortical signal that extended further into the anterior than in other lines. A single representative line was selected for each fusion (Table 1) and used for the experiments in this study, except for GFP:PAR-6 where two independent lines were used for the par-2(RNAi) experiments.
Table 1.
Strains used in this study
| Strain name | Description | Genotype |
|---|---|---|
| JH227 | PIE-1:GFP | axEx73[pJH3.92 PIE-1:GFP; pRF4] |
| JH1447 | GFP:MEX-6 | axEx1124[pKR2.07 GFP:MEX-6; pRF4] |
| JH1448 | GFP:MEX-5 | axEx1125[pKR2.04 GFP:MEX-5; pRF4] |
| JH1512 | GFP:PAR-6 | axIs1137[pJH7.04 GFP:PAR-6; pRF4] |
| JH1513 | PIE-1:GFP in par-3 mutant | par-3(it71) unc-32(e189)/qC1; axEx73[pJH3.92 PIE-1:GFP; pRF4] |
| JH1528 | PIE-1:GFP in par-1 mutant | par-1(ax53) dpy-11 (e224)/nT1 V; axEx73[pJH3.92 PIE-1:GFP; pRF4] |
| KK866 | GFP:PAR-2 | itIs153[pMW1.03 GFP:PAR-2; pRF4] |
| JH1473 | GFP:PAR-2 and GFP:tubulin | itIs153[pMW1.03 GFP:PAR-2; pRF4];ojIs1[pJH4.66 w/unc-119 inserted at NaeI] |
| JJ1244 | mex-5;mex-6 double mutant | mex-6(pk440); unc-30(e191) mex-5(zu199) IV/nT1 |
| KK747 | par-2 mutant | par-2(lw32) unc-45(e286ts)/sC1 III |
JH1473 was constructed by crossing WH104/+ males with KK866 hermaphrodites and selecting progeny that expressed both GFPs. WH104 was obtained by bombarding pJH4.66 unc-119 inserted at a NaeI site into unc-119 hermaphrodites (Strome et al., 2001).
The GFP:PAR-2, GFP:PAR-6 and PIE-1:GFP transgenes were tested for rescue of corresponding mutants (data not shown). All rescued, indicating that the fusions are functional.
Time-lapse microscopy
GFP localization dynamics were analyzed by time-lapse microscopy: single focal plane images, focused midway through the embryo (cross section), were collected typically at 20 second intervals over an approx. 25-minute period from pronuclear formation to the first cleavage. (Embryos that failed to divide within 35 minutes were discarded). At each time point, both Nomarski (0.01 seconds exposure) and fluorescence images (0.1 seconds exposure) were collected. Images were acquired using a Photometrics CoolSnap FX digital camera attached to a Zeiss Axioplan 2 equipped with Ludl shutters and a mercury lamp. Acquisition scripts were written using IPLab software, and acquired images were processed into QuickTime movies using 4D turnaround software (Laboratory of Optical and Computational Imaging, University of Wisconsin, Madison). Signal intensity cannot be compared between movies as the range displayed is not constant. In all movies, the maternal pronucleus is to the left and the paternal pronucleus is to the right. par-1(RNAi)mex-5(RNAi)mex-6(RNAi) embryos (5/5) had an extra maternal pronucleus – significance is not known. Movies can be viewed at ftp://www.wormbase.org/pub/wormbase/datasets/seydoux_2003.
RNA-mediated interference (RNAi)
RNAi was performed using the feeding method (Timmons and Fire, 1998), and, in the case of par-1, also by injection. Bacteria were grown overnight on NNGM plates containing 60 μg/ml ampicillin and 80 μg/ml IPTG. L4 hermaphrodites were allowed to feed for 24 hours at 25°C before video microscopy. par-2, par-3, par-5, par-6, pkc-3 (RNAi) embryos divided symmetrically; in contrast par-1(RNAi) embryos often divided asymmetrically.
Immunolocalization
Immunostainings were performed as described previously (Guo and Kemphues, 1995; Etemad-Moghadam, 1995; Hung and Kemphues, 1999; Tabuse et al., 1998).
RESULTS
Coding sequences are sufficient for localization
To analyze polarization dynamics in live embryos, we used GFP fusions to PAR-2, PAR-6, PIE-1, MEX-5 and MEX-6 (see Materials and Methods) (Wallenfang and Seydoux, 2000; Reese et al., 2000; Strome et al., 2001). The GFP:ORF fusions were driven under the control of the pie-1 promoter and 3′UTR. With the exception of MEX-6, for which there is no antibody yet, GFP distributions in transgenic lines matched those reported previously for the corresponding endogenous proteins (Movies 1–6). Thus, as is true for pie-1 (Reese et al., 2000), the par-2, par-6 and mex-5 ORFs contain all the information necessary for localization.
Localization dynamics in wild-type zygotes
Summary of zygote development
Immediately following fertilization, zygotes resume meiosis and begin synthesizing an eggshell. Zygotes are fragile during this period; therefore we typically began our time-lapse analysis after the completion of meiosis, around the time that the maternal and paternal pronuclei first appear. Before pronuclear formation, the cortex is very active and is undergoing intense ruffling throughout the length of the embryo (Fig. 1A). Following the appearance of pronuclei, ruffling stops abruptly in a small area near the sperm pronucleus. At that time, internal cytoplasm begins to flow towards the sperm pronucleus and superficial cytoplasm flows away from it (this flow is easily visualized by following the movement of individual yolk granules) (Golden, 2000). As flow proceeds, the smooth area in the posterior expands anteriorly (Fig. 1B). Eventually ruffling is confined to the anterior half of the embryo and culminates in a transient but deep invagination of the membrane (pseudocleavage furrow; Fig. 1C). After pseudocleavage, ruffling ceases entirely, the pronuclei meet in the posterior (Fig. 1D), migrate back towards the center, rotate so that the duplicated centrosomes become aligned along the AP axis (Fig. 1E), and undergo nuclear membrane breakdown. The mitotic spindle forms initially in the center of the embryo, but becomes displaced posteriorly during anaphase (Fig. 1F), resulting in a larger anterior cell (somatic blastomere AB) and a smaller posterior cell (germline blastomere P1; Fig. 1G).
Fig. 1.
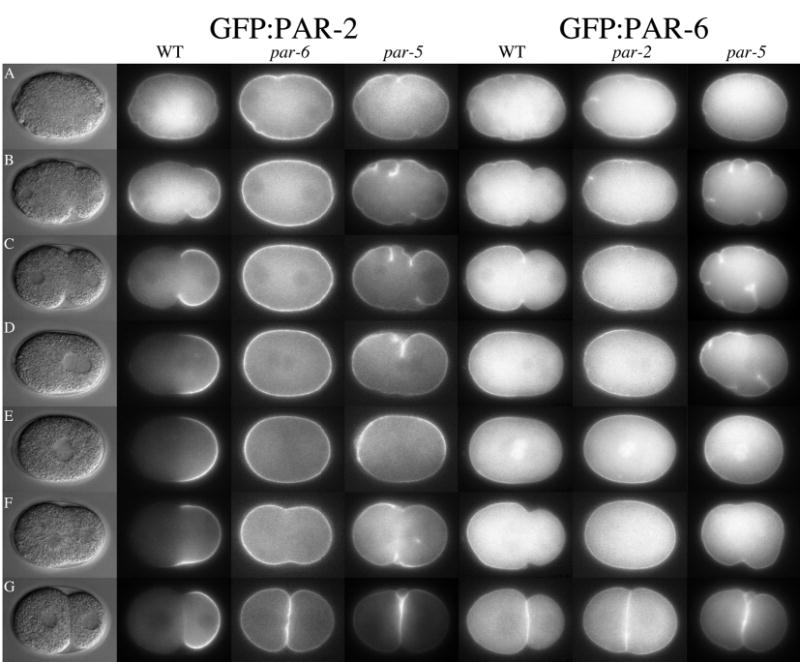
GFP:PAR-2 and GFP:PAR-6 dynamics in wild-type and mutant embryos (Movies 1–2, 10–11, 20–21). (A) Pronuclear formation. (B) Cessation of ruffling in posterior. (C) Pseudocleavage. (D) Pronuclear meeting. (E) Spindle rotation. (F) Mitosis. (G) two-cell stage. Quicktime movies of this and subsequent figures are accessible at ftp://www.wormbase.org/pub/wormbase/datasets/seydoux_2003. C. elegans zygotes are approximately 50 μm in length.
GFP:PAR-2 and GFP:PAR-6
Before pronuclear formation, GFP:PAR-2 and GFP:PAR-6 were distributed uniformly throughout the embryo. Both could be detected readily in the cytoplasm, and were also visibly enriched at the cortex (Fig. 1A, Movies 1 and 2).
The first asymmetry was seen after the appearance of pronuclei when ruffling ceased abruptly near the paternal pronucleus. At that time, GFP:PAR-2 began to increase and GFP:PAR-6 began to decrease on the cortex in the smooth region. For the next 10 minutes, as the smooth region expanded towards the anterior, the GFP:PAR-2 domain expanded along with it, whereas GFP:PAR-6 receded, becoming more prominent in the area where ruffling is maintained (Fig. 1B). By pseudocleavage (Fig. 1C), the fusions reached their final configurations on the cortex, with GFP:PAR-2 enriched in the posterior and GFP:PAR-6 enriched in the anterior. We also detected GFP:PAR-2 on centrosomes during pronuclear rotation (Movie 1) and GFP:PAR-6 in both pronuclei just before pronuclear fusion (Movie 2 and Fig. 1E). The latter localizations have not been reported previously for the endogenous proteins.
To examine the relationship between onset of polarity and formation of the sperm MTOC, we filmed embryos co-expressing GFP:PAR-2 and GFP:tubulin. In four out of the four embryos examined, we observed a good correlation, in timing and location, between appearance of the MTOC, local cessation of ruffling, and accumulation of PAR-2 on the cortex in the smooth zone. This correlation held even in embryos where the MTOC formed at a lateral position before moving to the pole. In these embryos, the GFP:PAR-2 domain initially also formed laterally, but rapidly shifted towards the pole along with the sperm pronucleus/MTOC complex (Movie 3, Fig. 2A–D).
Fig. 2.
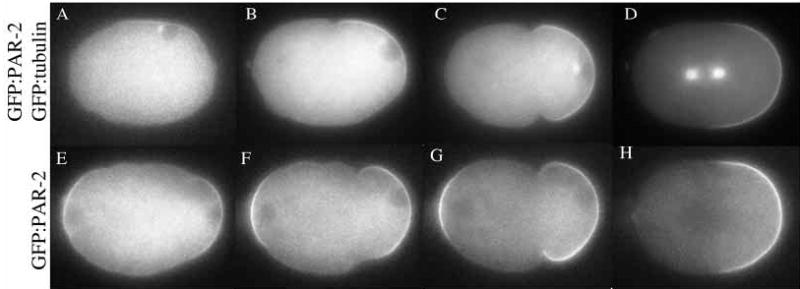
GFP:PAR-2 and GFP:tubulin dynamics in wild-type embryos. (A–D) An embryo expressing both GFP:PAR-2 and GFP:tubulin (Movie 3). (E–H) An embryo expressing GFP:PAR-2 (Movie 4).
In six out of 11 embryos, we observed GFP:PAR-2 in a second domain on the cortex next to the maternal pronucleus (Movie 4, Fig. 2E–H). This domain persisted until pseudocleavage and quickly disappeared afterwards. Transient localization in the anterior cortex was also described for endogenous PAR-2 in immunofluorescence experiments (Boyd et al., 1996). The significance of this localization is not known, but may be due to the transient influence of the meiotic spindle in this area of the cortex (Wallenfang and Seydoux, 2000).
GFP:MEX-5, GFP:MEX-6 and PIE-1:GFP
PIE-1:GFP and GFP:MEX-5 were initially uniformly distributed throughout the cytoplasm and began to increase (PIE-1) or decrease (MEX-5) in the posterior cytoplasm soon after the appearance of a smooth zone near the paternal pronucleus (Fig. 3, Movies 5 and 6). The fusion proteins also appeared to decrease (PIE-1) or increase (MEX-5) at the opposite end of the embryo. Maximal asymmetry was reached by pseudocleavage with PIE-1:GFP enriched in the posterior and GFP:MEX-5 enriched in the anterior (Fig. 3E). For both fusions, localization was not absolute, with some GFP fluorescence remaining in the opposite domain. Overall, the timing of PIE-1:GFP and GFP:MEX-5 localization was similar to that of GFP:PAR-2 and GFP:PAR-6. Small differences in timing cannot be excluded, however, since the fusions were not visualized in the same embryos.
Fig. 3.
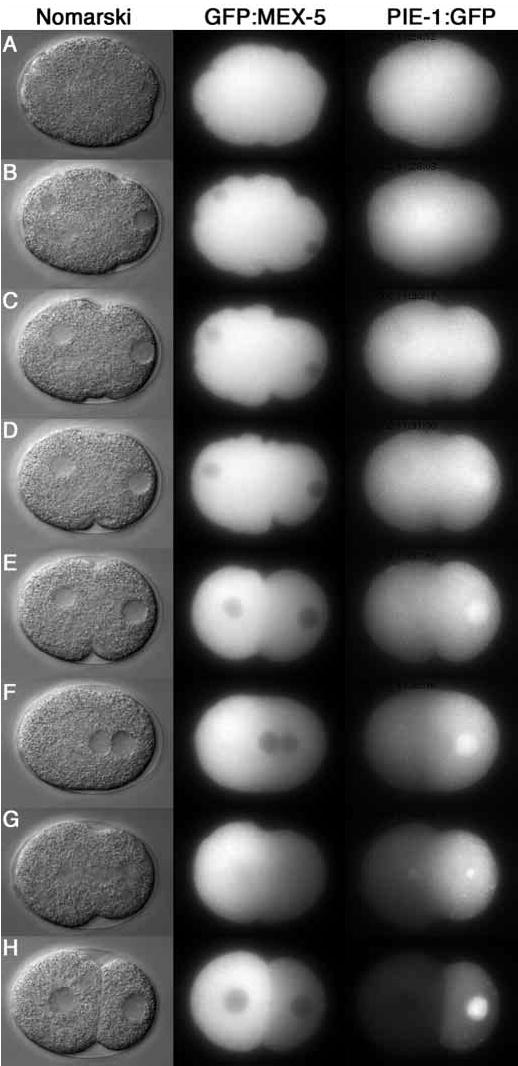
GFP:MEX-5 and PIE-1:GFP dynamics in wild-type embryos (Movies 5 and 6). (A) Pronuclear formation. (B–D) Pronuclear migration. (D) Pseudocleavage. (F) Pronuclear meeting. (G) Mitosis. (H) Two-cell stage. Note that PIE-1:GFP accumulates in the male pronucleus during pronuclear migration, on the posterior centrosome during mitosis, and in the P1 nucleus in the two-cell stage. PIE-1:GFP and GFP:MEX-5 also accumulate on the cytoplasmic P granules (best seen in the movies).
GFP:MEX-6 (Movie 7) behaved essentially like GFP:MEX-5, consistent with the fact that these proteins are 70% identical in sequence and function redundantly (Schubert et al., 2000). PIE-1:GFP, GFP:MEX-5 and GFP:MEX-6 all accumulated on granules in germline blastomeres. Localization on P granules has been reported for PIE-1 and MEX-5 (Mello et al., 1996; Schubert et al., 2000). Since antibodies specific to MEX-6 are not yet available, we do not know whether the GFP:MEX-6 pattern matches that of endogenous MEX-6.
Polarization of the zygote involves two distinct phases
To test whether the sperm asters function by recruiting PAR-2 or by excluding the PAR-3/PAR-6/PKC-3 complex from the posterior, we examined the distribution of GFP:PAR-2 and GFP:PAR-6 in embryos where par-2, par-3, par-6 or pkc-3 were inactivated by RNA-mediated interference (RNAi) (Fire et al., 1998).
In par-3(RNAi), pkc-3(RNAi) and par-6(RNAi), (n=3 time lapses for each), high levels of GFP:PAR-2 were observed throughout the cortex from pronuclear formation to the first cleavage (Fig. 1 and Movies 8–10), consistent with previous immunofluorescence observations of par-3 and par-6 mutants (Boyd et al., 1996; Watts et al., 1996). Occasionally stronger patches of GFP:PAR-2 fluorescence appeared transiently and unpredictably at one of the two poles. We conclude that, in the absence of the anterior PAR complex, PAR-2 can accumulate on the cortex but cannot become asymmetric.
In par-2(RNAi) embryos (n=21), GFP:PAR-6 started out uniformly distributed at the cortex (Fig. 1 and Movie 11). As in wild type, when ruffling stopped in the posterior, GFP:PAR-6 disappeared from that area, eventually receding to 63% egg length (average from 11 time-lapse examinations). After pronuclear meeting, however, GFP:PAR-6 crept back towards the posterior (Fig. 1E). By cytokinesis (Fig. 1F), GFP:PAR-6 was found throughout the cortex. This behavior (initial asymmetry followed by loss of asymmetry) was observed in 2 independent GFP:PAR-6 lines (total of 21 time-lapse examinations).
Because this behavior had not been noted in previous immunofluorescence studies (Hung and Kemphues, 1999) and to exclude the possibility that it was due to an artifact of RNAi or the GFP fusion, we reexamined the distribution of endogenous PAR-3, PAR-6 and PKC-3 in par-2(0) mutants by immunofluorescence using DAPI staining to stage embryos (Table 2). Those data confirmed the GFP:PAR-6 results. We conclude that PAR-3, PAR-6 and PKC-3 can accumulate at the cortex and become enriched in the anterior in the absence of PAR-2. However, they require PAR-2 to remain excluded from the posterior after pronuclear meeting. These data indicate that polarization of the cortex involves two phases: a PAR-2-independent “establishment” phase before pronuclear meeting, and a PAR-2-dependent “maintenance” phase afterwards.
Table 2.
Distribution of anterior complex components along the AP axis as determined by immunostaining
| Egg length* ± s.d. (n) at:
|
||||
|---|---|---|---|---|
| Component and genotype | Prophase | Prometaphase/metaphase | Anaphase | Telophase |
| PAR-6 in N2 | 64.2±6.3 (21) | 68.4±4.3 (12) | 69.7±3.7 (5) | 66.7±4.7 (6) |
| PAR-6 in par-2 | 65.2±8.9 (10) | 74.7±4.0 (8) | 77.6±4.7 (7) | 98.2±3.6 (4) |
| PAR-3 in N2 | 52.1±5.5 (21) | 53.8±2.8 (5) | 55.7±3.0 (6) | 64.3±2.6 (3) |
| PAR-3 in par-2 | 60.3±5.1 (5) | 73.9±1.8 (3) | 91.3±10.1 (3) | 93.7±7.7 (5) |
| PKC-3 in N2 | 55.4±2.7 (6) | 51.3±4.4 (5) | 48.2±0.3 (2) | 52.1±4.9 (2) |
| PKC-3 in par-2 | 61.1±7.7 (14) | 74.7±15.5 (5) | 75.2±12.7 (9) | 94.6±5.4 (8) |
The average distance along the long axis (anterior=0; posterior=100) at which the posterior-most cortical staining could be detected. Stage was determined by DAPI staining.
In par-3(RNAi) (n=7) and pkc-3(RNAi) (n=8) embryos, GFP:PAR-6 failed to accumulate on the cortex from the earliest stage examined (pronuclear formation; Movies 12–13). This result is consistent with previous studies, which indicated that PAR-3, PAR-6 and PKC-3 depend on each other for cortical localization (Watts et al., 1996; Hung and Kemphues, 1999; Tabuse et al., 1998). We observed, however, transient accumulation of GFP:PAR-6 in pronuclei just before mitosis, as is observed in wild type. We conclude that par-3 and pkc-3 are essential for cortical localization of PAR-6 throughout the first cell cycle, but are dispensable for PAR-6’s transient nuclear localization.
The non-muscle myosin NMY-2 and the 14-3-3 protein PAR-5 are required during the establishment phase
NMY-2
NMY-2 is required for embryonic polarity and PAR localization (Guo and Kemphues, 1996). To determine whether NMY-2 is required for the establishment or maintenance phases, we examined GFP:PAR-2 and GFP:PAR-6 in nmy-2(RNAi) embryos (Materials and Methods). nmy-2(RNAi) embryos fall into three classes, depending on the severity of loss of nmy-2 activity, from strong to weak (Guo and Kemphues, 1996).
Class I
Embryos in this class do not undergo ruffling, pseudocleavage or cytokinesis. Pronuclei form and migrate normally in these embryos, eventually resulting in the formation of a symmetric spindle (Guo and Kemphues, 1996). These phenotypes are consistent with embryos lacking actin contractility but retaining normal microtubules. In one embryo examined of this class, GFP:PAR-6 remained uniformly distributed at the cortex throughout the first cell cycle (n=1, Movie 14). GFP:PAR-2 did not become visibly enriched on the cortex (n=3), and instead was maintained throughout the cytoplasm and also on foci around centrosomes (Fig. 4; Movie 15). The lack of cortical PAR-2 in nmy-2(RNAi) embryos was dependent on PAR-6: nmy-2(RNAi);par-6(RNAi) embryos exhibited strong GFP:PAR-2 at the cortex (n=3; Fig. 4; Movie 16). We conclude that NMY-2 is required for onset of the establishment phase and that, in its absence, the PAR-3/PAR-6/PKC-3 complex occupies the entire cortex, excluding PAR-2.
Fig. 4.
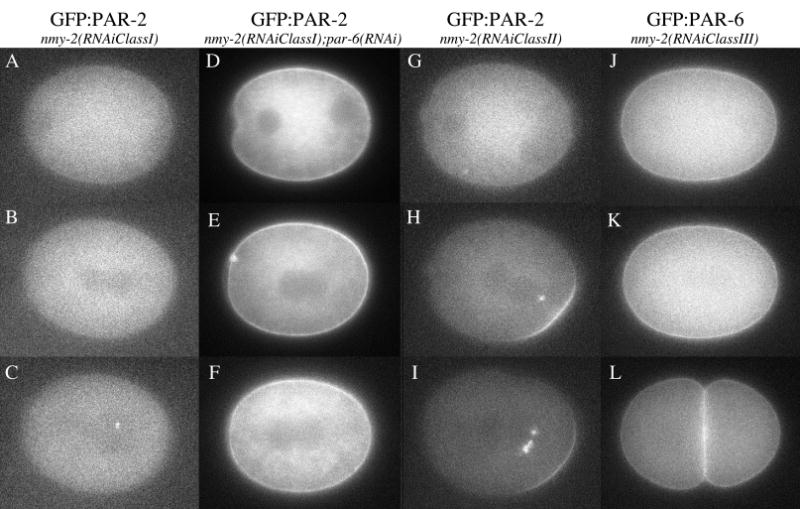
GFP:PAR-2 and GFP:PAR-6 dynamics in nmy-2(RNAi) embryos. A,D,G,J are embryos during pronuclear migration; B,E,H,K are at pronuclear meeting; C,F,I are in late mitosis; L is a 2-cell embryo. GFP:PAR-2 remains off the cortex in the most severely affected embryos (AC; Movie 15), unless par-6 is also removed (D-F, Movie 16). In more weakly affected embryos [nmy-2(RNAiClassII)], GFP:PAR-2 appears on the cortex around pronuclear meeting (H, Movie 18). Note PAR-2 accumulation in foci around pronuclei and/or spindles (C,H,I). GFP:PAR-6 remains uniformly distributed throughout the cortex even in the most weakly affected embryos (J–L, Movie 17).
Class II and Class III
These embryos developed cytokinesis furrows that were either transient (Class II) or lead to the formation of two equal size cells (Class III). Like Class I embryos, these embryos maintained GFP:PAR-6 uniformly at the cortex throughout the first cell cycle (n=2, Fig. 4, Movie 17). Unlike Class I embryos, however, Class II and Class III embryos developed one or two patches of GFP:PAR-2 at the cortex by pronuclear meeting (n=6, Fig. 3, Movies 18, 19). These patches were similar to those seen for endogenous PAR-2 in fixed nmy-2(RNAi) and mlc-4(RNAi) embryos (Guo and Kemphues, 1996; Shelton et al., 1999). Our time-lapse analysis indicates that these patches form later than in wild type (pronuclear meeting rather than pronuclear formation) and occur only in embryos with residual actin contractility (as evidenced by ingressing furrows). We conclude that the PAR-2 patches observed in nmy-2(RNAi) embryos are not the result of an early, myosin-independent step during the establishment phase, but rather correspond to a later response of PAR-2 that is still dependent on NMY-2.
In all nmy-2(RNAi) embryos (n=8), we also detected GFP:PAR-2 in foci around the pronuclei or spindle, as was reported previously for mutants that fail to polarize normally (Rappleye et al., 2002). In four out of eight time-lapse analyses, GFP:PAR-2 appeared to shuttle from the cortex to the nearest pronucleus or centrosome. The significance of this behavior is not known but suggests that PAR-2 may have an affinity for microtubules.
PAR-5
par-5 mutant embryos exhibit extensive overlap between PAR-3 and PAR-2 and between PAR-1 and PKC-3 (Morton et al., 2002), indicating that PAR-5 is essential for creating and/or maintaining distinct anterior and posterior PAR domains. To determine whether PAR-5 functions during the establishment phase or the maintenance phase, we examined GFP:PAR-2 and GFP:PAR-6 dynamics in par-5(RNAi) embryos.
We found that par-5(RNAi) embryos maintain GFP:PAR-6 (n=9) and GFP:PAR-2 (n=5) at the cortex from pronuclear formation through cleavage (Fig. 1, Movies 20–21). We observed a reproducible reduction in GFP:PAR-6 fluorescence in the posterior when ruffling ceased in that area. This reduction, however, appeared later and was not as pronounced as in wild type, with detectable levels of GFP:PAR-6 remaining in the region. After pseudocleavage, GFP:PAR-6 reappeared uniformly throughout all the cortex. During the period that GFP:PAR-6 decreased in the smooth zone, we did not observe a reproducible increase in GFP:PAR-2. Occasionally we observed patches of stronger or lower GFP:PAR-2 fluorescence in certain areas of the cortex, but these patches were transient and did not appear in a predictable pattern.
We conclude that PAR-5 during the establishment phase is required (1) for maximal response of the PAR-3/PAR-6/PKC-3 to the sperm aster cue and (2) to prevent overlap between PAR-3/PAR-6/PKC-3 and PAR-2.
PAR-1 inhibits and MEX-5 and MEX-6 promote, expansion of the posterior domain during the establishment phase
Mutations in par-1 have been reported not to affect the localization of other PAR proteins in zygotes (Boyd et al., 1996; Etemad-Moghadam et al., 1995; Hung and Kemphues, 1999). Consistent with this, we found that GFP:PAR-2 was asymmetric in par-1(RNAi) embryos, in clear contrast to what we observed in embryos lacking par-3, par-6, pkc-3 or par-5. We noted, however, that the anterior-most boundary of the PAR-2 domain was shifted towards the anterior in par-1(RNAi) zygotes (n=6; Fig. 5B; Movie 22). This displacement coincided spatially and temporally with the anteriorly displaced pseudocleavage of par-1 embryos (Kirby et al., 1990). To confirm that this effect was not an artifact of RNAi or the GFP:PAR-2 fusion, we examined par-1(b274) embryos for endogenous PAR-2 by immunofluorescent staining. We observed a similar anterior expansion of the PAR-2 domain in embryos at pronuclei meeting (6/6; data not shown).
Fig. 5.
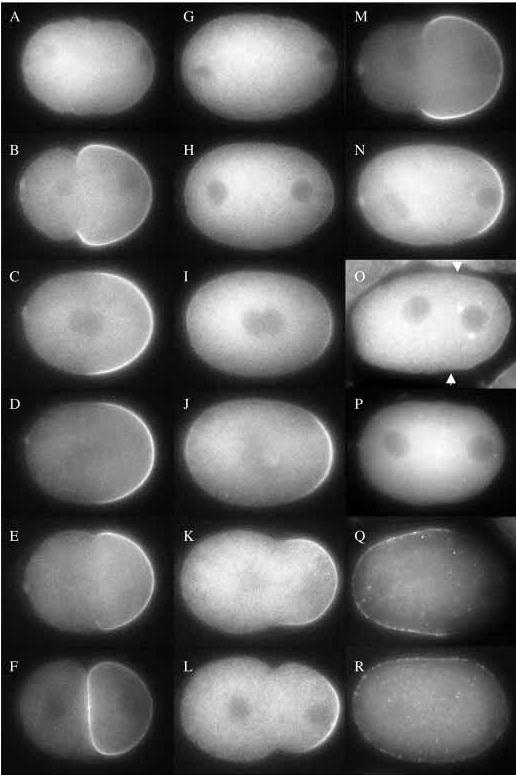
PAR-1 and MEX-5/6 function in a feedback loop to regulate expansion of the posterior domain. (A–F) par-1(RNAi) zygote expressing GFP:PAR-2 (Movie 22). (A) Pronuclear formation. (B) Pronuclear migration. (C) Pronuclear meeting. (D–E) Mitosis. (F) Two-cell stage. The GFP:PAR-2 domain is expanded at pseudocleavage compared to wild type (see Fig. 1). (G–L) mex-5(RNAi)mex-6(RNAi) zygote expressing GFP:PAR-2 (Movie 28). Stages same as in A–F. GFP:PAR-2 domain expansion is slowed down compared to wild type (see Fig. 1). (M) GFP:PAR-2 in par-1(RNAi) pie-1(RNAi) (Movie 27). Expansion of the PAR-2 domain is not suppressed (compare with B). (N) GFP:PAR-2 in par-1(RNAi) mex-5(RNAi)mex-6(RNAi) (Movie 26). Expansion of the PAR-2 domain is suppressed. Identical results were obtained in par-1(it51) mex-5(RNAi)mex-6(RNAi) embryos stained for PAR-2 (not shown). (O,P) Wild-type (O) and mex-5(zu199)mex-6(pk440) (P) zygotes stained with PAR-1 antibody. Arrows point to the ends of the PAR-1 domain in the wild-type embryo. PAR-1 is not visible on the cortex of this mex-5(zu199)mex-6(pk440) zygote. (Q,R) Wild-type (Q) and mex-5(zu199)mex-6(pk440) (R) zygotes stained with PAR-3 antibody. PAR-3 extends further posterior.
PAR-1 regulates the localization of cytoplasmic proteins, including MEX-5 and PIE-1 (Schubert et al., 2000; Tenenhaus et al., 1998). We confirmed these observations and extended them to MEX-6, by following GFP:MEX-5, GFP:MEX-6 and PIE-1:GFP dynamics in par-1(RNAi) embryos. In all cases, the fusions remained uniformly distributed (Movies 23–25). To determine whether expansion of the PAR-2 domain in par-1 zygotes was due to ectopic MEX-5/6 and/or PIE-1, we examined GFP:PAR-2 dynamics in par-1(RNAi) mex-5(RNAi) mex-6(RNAi) and par-1(RNAi) pie-1(RNAi) zygotes (Movies 26–27). We found that expansion of the PAR-2 domain was suppressed in par-1(RNAi) mex-5(RNAi) mex-6(RNAi) embryos (n=5; Fig. 5N), but not in par-1(RNAi) pie-1(RNAi) embryos (n=3; Fig. 5M). Suppression of PAR-2 expansion was also observed in par-1(it51) mex-5(RNAi) mex-6(RNAi) embryos stained for endogenous PAR-2 (5/5; data not shown). These results indicate that expansion of the PAR-2 domain in par-1 zygotes is due to ectopic MEX-5 and MEX-6.
To explore further the role of MEX-5/6, we examined PAR-2 dynamics in embryos depleted of these two proteins by RNAi. mex-5;mex-6 double mutants were reported not to affect PAR asymmetry (Schubert et al., 2000). Consistent with those results, we found that GFP:PAR-2 was asymmetric in most mex-5(RNAi) mex-6(RNAi) zygotes (5/7). However, GFP:PAR-2 dynamics were clearly aberrant in those embryos. In wild type, the GFP:PAR-2 domain expands quickly and reaches its maximal domain (close to 50% egg length) by pseudocleavage (Fig. 1). In contrast, in five out of seven mex-5(RNAi)mex-6(RNAi) embryos, the GFP:PAR-2 domain remained small (33% egg length) throughout pronuclear migration and began to expand only after pronuclear meeting, eventually reaching its normal distribution just before the first cleavage (Fig. 5G–L, Movie 28). This pattern was essentially opposite that seen in par-1(RNAi) embryos, where the PAR-2 domain extends further than normal (Fig. 5A–F). In the other 2 embryos examined (2/7), PAR-2:GFP was never seen at the cortex (Movie 29). Unlike most mex-5(RNAi);mex-6(RNAi) embryos, which divide asymmetrically, these two embryos divided symmetrically. These defects were specific to mex-5 and mex-6, as pie-1(RNAi) embryos exhibited normal GFP:PAR-2 dynamics and divided asymmetrically (n=3, Movie 30).
To verify that the mex-5/6 results were not an artifact of RNAi or GFP fusions, we re-examined the distribution of endogenous PAR-1 and PAR-2 in mex-5(zu199)mex-6(pk440) double mutants by immunofluorescence, using DAPI staining to stage embryos. This analysis confirmed that establishment of the PAR-1/PAR-2 domain is delayed in many mex-5(–)mex-6(–) zygotes, and occasionally does not occur at all [Fig. 5P; 18/27 zygotes had PAR-1 domains smaller than wild type; similar results were also obtained for PAR-2 (not shown)]. Examination of two-cell mex-5(zu199)mex-6(pk440) double mutants confirmed that these embryos occasionally undergo a symmetric first cleavage (19/133).
We conclude that MEX-5 and MEX-6 are required to promote rapid and consistent expansion of the posterior domain during the establishment phase. MEX-5/6 could affect growth of the posterior domain directly by promoting PAR-1/PAR-2 localization there, or indirectly, by excluding the anterior PARs from that region. To distinguish between these possibilities, we analyzed GFP:PAR-6 dynamics in par-1(RNAi) and mex-5(RNAi);mex-6(RNAi) embryos. We found that five out of six par-1 embryos had a smaller GFP:PAR-6 domain at pseudocleavage (Movie 31), and one out of mex-5(RNAi);mex-6(RNAi) embryos maintained uniform PAR-6:GFP throughout the first cell cycle (Movie 32). Similarly, 11 out of 19 mex-5(zu199)mex-6(pk440) had expanded PAR-3 domains (Fig. 5R). These phenotypes are unlikely to be a secondary consequence of the smaller PAR-2 domain observed in these embryos, since par-2 is not essential to exclude anterior PARs from the posterior during the establishment phase (Fig. 1). We conclude that MEX-5/6 regulate expansion of the posterior domain by helping to exclude anterior PARs from that region.
In the course of analyzing GFP:PAR-6 dynamics in par-1(RNAi) embryos, we noticed that PAR-6 asymmetry became significantly less pronounced after pronuclear meeting (Movie 31). Whereas par-2(RNAi) embryos lose all GFP:PAR-6 asymmetry after pronuclear meeting, par-1(RNAi) embryos appeared to regain asymmetric GFP:PAR-6 at the two-cell stage. This is consistent with previous analyses, which demonstrated that par-1 mutants maintain anteriorly enriched PAR-3 and PAR-6 even after undergoing a symmetric cleavage (Boyd et al., 1996; Etemad-Moghadam et al., 1995). We conclude that PAR-1 contributes to PAR asymmetry during the maintenance phase, although unlike PAR-2, it may not be essential after cleavage.
PAR-1 inhibits MEX-5/6 activity and/or levels
The opposite phenotypes of par-1 and mex-5/6 mutants during the establishment phase, and the fact that mex-5/6 are epistatic to par-1 (Fig. 5) indicate that PAR-1 negatively regulates MEX-5/6. Two models could account for this negative regulation. PAR-1 could inhibit MEX-5/6 by causing them to relocate to the anterior, or PAR-1 could inhibit MEX-5/6 by negatively regulating their activity or level in the posterior. The latter predicts that mutants with uniform PAR-1 would have low MEX-5/6 activity and/or levels throughout the zygote. We have obtained evidence in support of this prediction, using PIE-1:GFP to monitor MEX-5/6 activity.
MEX-5 and MEX-6 are required for PIE-1 asymmetry (Schubert et al., 2000). As expected, PIE-1:GFP remained uniform in mex-5(RNAi)mex-6(RNAi) embryos (n=4; Movie 33). [In contrast, GFP:MEX-5 localized normally in pie-1(RNAi) embryos (n=3; Movie 34)]. During these analyses, we noticed that the cytoplasmic:nuclear ratio of PIE-1 is also regulated by MEX-5/6. In wild-type embryos, during pronuclear migration, PIE-1:GFP accumulated progressively with time in the sperm pronucleus (posterior), but not in the maternal nucleus (anterior, Fig. 3D). After cleavage, PIE-1:GFP accumulated in the P1 nucleus (posterior) but remained excluded from the AB nucleus (anterior, Fig. 3H). This difference is dependent on MEX-5/6: PIE-1:GFP was found in both pronuclei in mex-5(RNAi)mex-6(RNAi) zygotes (n=4, scored at pronuclear meeting stage), and in neither pronucleus in par-1(ax54) zygotes (n=6), where MEX-5 and MEX-6 remain uniformly distributed (Fig. 6). We confirmed that PIE-1’s low nuclear:cytoplasmic ratio in par-1 mutants was dependent on MEX-5/6, by removing MEX-5/6 by RNAi in par-1(ax54) embryos. As expected, we detected PIE-1:GFP in both pronuclei in par-1(ax54)mex-5(RNAi)mex-6(RNAi) zygotes (n=8; Fig. 6).
Fig. 6.
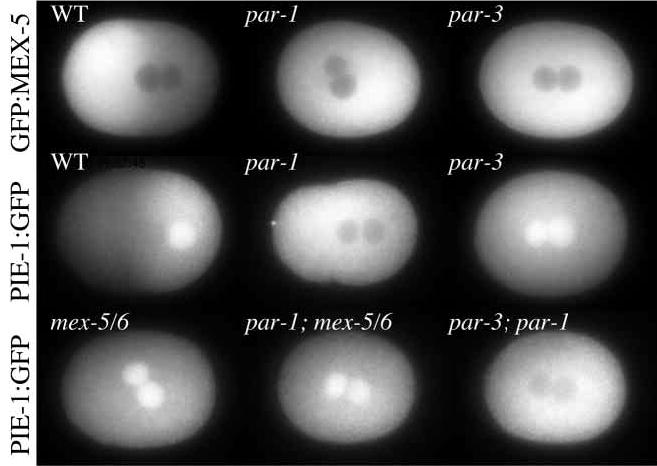
Regulation of PIE-1’s nuclear:cytoplasmic ratio by MEX-5/6 is under PAR-1 control. GFP:MEX-5 is primarily cytoplasmic in all genotypes. In contrast, the nuclear:cytoplasmic ratio of PIE-1:GFP varies depending on the genotype as described in text.
In par-3 embryos, PAR-1 is uniformly distributed throughout the cortex (Guo and Kemphues, 1995) and MEX-5 (and presumably MEX-6) remains uniformly distributed throughout the cytoplasm (Fig. 6) (see also Schubert et al., 2000). Strikingly, we found that PIE-1:GFP accumulates in both pronuclei in par-3(it71) zygotes (n=4), as is seen in mex-5(RNAi)mex-6(RNAi) embryos (Fig. 6). This observation indicated that MEX-5/6 is not active, or does not accumulate to a critical threshold level, in par-3 zygotes. A likely explanation for this result is that uniformly distributed cortical PAR-1 reduces MEX-5/6 activity or level equally throughout the cytoplasm. If so, removal of PAR-1 in par-3 mutants should restore MEX-5/6 activity, or accumulation, and keep PIE-1 out of the pronuclei. In agreement with this prediction, par-3(it71);par-1(RNAi) double mutants exhibited the same low PIE-1 nuclear:cytoplasmic ratio that is observed in par-1 mutants (n=10; Fig. 6). We conclude that PAR-1 can inhibit MEX-5/6 activity even under conditions where it does not create MEX-5/6 asymmetry. These observations suggest that PAR-1 acts on MEX-5/6 by negatively regulating their activity, level, or both.
DISCUSSION
Polarization of the zygotic cortex involves two distinct phases
We have used GFP fusions to monitor polarization dynamics in live embryos. Based on these analyses, we propose that polarization of the C. elegans zygote occurs in two phases: an ‘establishment’ phase before pronuclear meeting and a ‘maintenance’ phase after pronuclear meeting. During the establishment phase, signaling between the sperm asters and the actin cytoskeleton displace the PAR-3/PAR-6/PKC-3 complex from the posterior cortex, allowing PAR-2 to accumulate there. In the maintenance phase, PAR-2 prevents the PAR-3/PAR-6/PKC-3 complex from re-entering the posterior domain (Fig. 7).
Fig. 7.
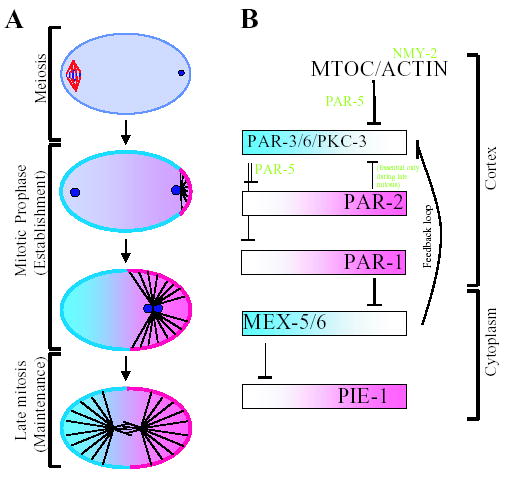
Model for polarization of the C. elegans zygote. (A) Graphic depiction of the establishment and maintenance phases during polarization of the zygote. Proteins that localize to the anterior are in blue (PAR-3, PAR-6, PKC-3 on the cortex, MEX-5 and MEX-6 in the cytoplasm). Proteins that localize to the posterior are in pink (PAR-2 and PAR-1 on the cortex, PIE-1 in the cytoplasm). During meiosis, all are uniformly distributed throughout the zygote (purple color). Circles: pronuclei, Black lines: microtubules. (B) Sequential repression model [modified from Kemphues (Kemphues, 2000)]. Lines with bars depict antagonistic interactions, whereas lines with arrows depict positive interactions. In this model, we show that PAR-1 is restricted to the posterior by the anterior PARs. This hypothesis is supported by the observation that, in late one-cell embryos, PAR-1 is present throughout the cortex in par-3 mutants and par-3;par-2 double mutants (Etemad-Moghadam et al., 1995; Boyd et al., 1996). Further support for this model awaits analysis of PAR-1 dynamics in live embryos.
Establishment
Previous studies have implicated the sperm-derived MTOC as the most likely source for the spatial cue that initially polarizes the zygote (Sadler and Shakes, 2000; O’Connell et al., 2000; Wallenfang and Seydoux, 2000). Our time-lapse analysis supports this view. Formation of the MTOC correlates temporally and spatially with the earliest evidences of polarity: (1) cessation of ruffling, (2) enrichment of GFP:PAR-2, and (3) loss of GFP:PAR-6 in the posterior cortex. Our data also demonstrates that the primary effect of the polarizing cue is to clear the PAR-3/PAR-6/PKC-3 complex from the posterior cortex. This effect does not require PAR-2. In contrast, restriction of PAR-2 to the posterior requires PAR-6, PAR-3 and PKC-3, suggesting that PAR-2 does not sense the polarity cue directly but instead responds to local displacement of the anterior complex.
The establishment phase requires the class II non-muscle myosin, NMY-2: nmy-2(RNAi) prevents PAR-6 (and presumably associated PAR-3 and PKC-3) from sensing the polarity cue, causing it to remain uniformly distributed throughout the cortex. In NMY-2-depleted embryos, PAR-2 is prevented from accumulating at the cortex by PAR-6 (and/or its partners). This ‘default’ state of PAR-6 on/PAR-2 off is also observed in mutants lacking sperm asters (O’Connell et al., 2000; Wallenfang and Seydoux, 2000) and in mutants where the MTOC detaches from the cortex prematurely (Rappleye et al., 2002). These observations suggest that the initial symmetry-breaking event involves signaling between the MTOC and the actin cytoskeleton. Consistent with this view, one of the earliest signs of polarization is cessation of ruffling in the cortex nearest the MTOC. Cessation of ruffling correlates with MTOC formation, but does not appear to require PAR activity (cessation of ruffling was observed in all par mutants examined in this study). These observations suggest that modification of the actin cytoskeleton may be an obligatory step before the onset of PAR asymmetry. We propose that signaling from the MTOC modifies the actin cytoskeleton locally, which causes the PAR-3/PAR-6/PKC-3 complex to become destabilized, allowing PAR-2 to accumulate in its place.
The establishment phase also requires the 14-3-3 protein PAR-5. In its absence, PAR-6 responds only weakly, if at all, to the polarity cue and PAR-2 is no longer excluded from the cortex by the PAR-3/PAR-6/PKC-3 complex. Cessation of ruffling in the posterior, however, still occurs in par-5(RNAi) embryos, suggesting that PAR-5 is not required for the initial MTOC/actin cytoskeleton interaction. Although this interpretation is complicated by the fact that residual PAR-5 activity may persist in par-5(RNAi) embryos (Morton et al., 2002), we propose that PAR-5 functions primarily by regulating the ability of the PAR-3/PAR-6/PKC-3 complex to (1) exclude PAR-2 and (2) respond to changes in the cytoskeleton. The presence of a potential 14-3-3 binding motif in PAR-3 (Morton et al., 2002) is consistent with the possibility that PAR-5 regulates the PAR-3/PAR-6/PKC-3 complex by binding to it directly.
A feedback loop during the establishment phase
Surprisingly, we found that the predominantly cytoplasmic MEX-5 and MEX-6 also play a role during the establishment phase. In the absence of MEX-5 and MEX-6, the posterior domain occasionally does not form (15–30% of embryos), and frequently (50% or more of embryos) is slow to reach its final configuration. These observations indicate that, although MEX-5 and MEX-6 are not absolutely required for PAR localization in the zygote (Schubert et al., 2000), they do play a role in ensuring a robust response by the PAR-3/PAR-6/PKC-3 complex to the MTOC/actin cytoskeleton signal.
We have found that this aspect of MEX-5/6 function is negatively regulated by PAR-1. In par-1 mutants, MEX-5 and MEX-6 cause the posterior domain to extend further towards the anterior during the establishment phase. Since PAR-1 itself becomes enriched in the posterior domain, one attractive possibility is that PAR-1 and MEX-5/6 participate in a feedback loop that limits expansion of the posterior domain. We propose the following model. At the beginning of the establishment phase, MEX-5 and MEX-6 levels are high throughout the zygote and help clear the PAR-3/PAR-6/PKC-3 complex from the region nearest the sperm asters. This clearing allows PAR-2 and PAR-1 to accumulate on the cortex, which in turn reduces MEX-5/6 activity and/or levels in the surrounding cytoplasm. Eventually, MEX-5/6 levels become too low to fuel further expansion of the posterior domain. We do not yet know whether the partial penetrance of the mex-5(–);mex-6(–) phenotype is due to redundancy with other factors, or is indicative of a minor role for the feedback loop in regulating PAR asymmetry.
Maintenance
The finding that the sperm-derived MTOCs play a role in initiating polarity raised the question of how polarity is maintained after pronuclear meeting, when the pronuclei/centrosome complex rotates and microtubules invade the anterior end of the embryo (Fig. 7). Our observations provide an answer: PAR-2. We have found that in the absence of PAR-2, PAR-3 and PAR-6 and PKC-3 can become asymmetric before pronuclear meeting, but return into the posterior domain afterwards. This finding demonstrates two points: (1) the PAR-6/PAR-3/PKC-3 complex no longer responds to the MTOC-dependent cue after pronuclear meeting, and (2) PAR-2 is required after pronuclear meeting, but not earlier, to exclude the PAR-6/PAR-3/PKC-3 complex from the posterior. We propose that pronuclear meeting (and/or the end of prophase) triggers a change in the cytoskeleton, or in the PAR-6/PAR-3/PKC-3 complex, which turns off the MTOC-dependent polarity signal, or the ability to respond to it. From that point on, PAR-2 becomes essential to keep PAR-6/PAR-3/PKC-3 out of the posterior cortex. It is intriguing that PAR-6 briefly localizes to nuclei at pronuclear meeting, raising the possibility that it becomes modified at that time.
The existence of distinct establishment and maintenance phases is also supported by the observation that cdc-42 is required after prophase, but not earlier, for PAR-3, PAR-6 and PKC-3 asymmetry (Gotta and Ahringer, 2001). Our analysis of GFP:PAR-6 dynamics in par-1(RNAi) embryos suggests that PAR-1 also contributes to maintenance of PAR asymmetry after pronuclear meeting. How PAR-2, CDC-42 and PAR-1 function together to maintain the balance between anterior and posterior PAR domains remains to be determined.
Asymmetric protein localization in C. elegans zygotes
During the establishment phase, MEX-5, MEX-6 and PIE-1 asymmetries appear in the cytoplasm with approximately the same temporal dynamics as PAR asymmetries on the cortex. In agreement with previous studies, we have found that anterior localization of MEX-5 and MEX-6 is dependent on PAR-1, and posterior localization of PIE-1 is dependent on PAR-1 and on MEX-5/6. Since PAR-1 is not essential for asymmetric localization of other PARs during the establishment phase, these findings are consistent with PAR-1 being the PAR protein most directly required for localization of cytoplasmic factors.
PAR-1 could localize MEX-5 and MEX-6 by promoting their translocation to the anterior or by negatively regulating their activity and/or stability in the posterior. We have obtained evidence in support of the latter by analyzing the effect of delocalized PAR-1 on MEX-5/6 activity. In these experiments, we used the nuclear:cytoplasmic ratio of PIE-1:GFP as a readout for MEX-5/6 activity. We found that MEX-5/6 activity is low in par-3(–) zygotes where PAR-1 is delocalized. This low activity is dependent on PAR-1: zygotes lacking both par-3 and par-1 have high MEX-5/6 activity (Fig. 6). We conclude that PAR-1 can inhibit MEX-5/6 activity even under conditions where MEX-5/6 do not become asymmetric. We do not know yet whether this inhibition depends on lowering MEX-5/6 levels, inhibiting their activity, or a combination of both, as attempts to quantify protein levels in vivo have been unsuccessful thus far.
These results suggest that restriction of PAR-1 to the posterior may be necessary to generate cytoplasmic asymmetries, but conflict with previous evidence. In par-2 mutant embryos, PAR-1 is not detectably cortical or asymmetric (Boyd et al., 1996), yet P granules (Boyd et al., 1996) and PIE-1 (Tenenhaus et al., 1998) become asymmetric in the zygote. One possibility is that asymmetric activation of PAR-1 still occurs in these mutants in the absence of localization. Future experiments monitoring PAR-1 dynamics in live embryos will be needed to clarify the roles of PAR-2 and PAR-1 in regulating cytoplasmic asymmetries.
In addition to negatively regulating MEX-5/6 in the posterior, PAR-1 also causes MEX-5 and MEX-6 to accumulate in the anterior. We do not yet know the mechanism that mediates this enrichment. A possibility, which is consistent with our data, is that local action of PAR-1 destabilizes MEX-5/6 in the posterior, causing them to accumulate only in the anterior (owing to on-going translation of these proteins from maternal RNA). Accumulation of MEX-5 and MEX-6 in the anterior would in turn allow PIE-1 and other germline factors to accumulate only in the posterior. This outcome is reminiscent of the effect of the MTOC/actin cytoskeleton signal, which destabilizes or displaces the PAR-6/PAR-3/PKC-3 complex from the posterior cortex, causing it to accumulate only in the anterior, which in turn allows PAR-2 and PAR-1 to accumulate in the posterior. An attractive possibility is that local changes in stability among competing proteins is the dominant mechanism mediating asymmetric localization in C. elegans zygotes. The finding that the localizations of PAR-2, PAR-6, MEX-5, MEX-6 and PIE-1 do not require sequences in untranslated regions (this study) (Reese et al., 2000) already suggests that mechanisms acting at the protein, rather than the RNA, level prevail in the zygote. A challenge for the future will be to determine whether these mechanisms regulate protein stability, movement, or both.
We thank Christian Malone for the GFP:tubulin line, Kim Reese for the GFP:MEX-5 and GFP:MEX-6 fusions, Matt Wallenfang for the GFP:PAR-2 fusion and Lesilee Rose for critical reading of the manuscript. This project was funded by NIH grants R01GM64537 to G. S. and R01HD27689 to K. K. and by a David and Lucile Packard Fellowship for Science and Engineering to G. S.
References
- Boyd L, Guo S, Levitan D, Stinchcomb DT, Kemphues KJ. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122:3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Cell polarity: the PARty expands. Nat Cell Biol. 2001;3:E7–E9. doi: 10.1038/35050684. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Golden A. Cytoplasmic flow and the establishment of polarity in C. elegans 1-cell embryos. Current Opinion in Genetics and Development. 2000;10:414–420. doi: 10.1016/s0959-437x(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Goldstein B. Embryonic polarity: a role for microtubules. Curr Biol. 2000;10:R820–822. doi: 10.1016/s0960-9822(00)00790-9. [DOI] [PubMed] [Google Scholar]
- Gotta M, Ahringer J. Axis determination in C. elegans: initiating and transducing polarity. Curr Opin Genet Dev. 2001;11:367–373. doi: 10.1016/s0959-437x(00)00206-9. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature. 1996;382:455–458. doi: 10.1038/382455a0. [DOI] [PubMed] [Google Scholar]
- Hill DP, Strome S. An analysis of the role of microfilaments in the establishment and maintenance of asymmetry in Caenorhabditis elegans zygotes. Dev Biol. 1988;125:75–84. doi: 10.1016/0012-1606(88)90060-7. [DOI] [PubMed] [Google Scholar]
- Hill DP, Strome S. Brief cytochalasin-induced disruption of microfilaments druing a critical interval in one-cell C. elegans embryos alters the partitioning of developmental instructions to the two-cell embryo. Development. 1990;108:159–172. doi: 10.1242/dev.108.1.159. [DOI] [PubMed] [Google Scholar]
- Hird SN, Paulsen JE, Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122:1303–1312. doi: 10.1242/dev.122.4.1303. [DOI] [PubMed] [Google Scholar]
- Hung TJ, Kemphues KJ. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development. 1999;126:127–135. doi: 10.1242/dev.126.1.127. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K. PARsing embryonic polarity. Cell. 2000;101:345–348. doi: 10.1016/s0092-8674(00)80844-2. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kirby C, Kusch M, Kemphues K. Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev Biol. 1990;142:203–215. doi: 10.1016/0012-1606(90)90164-e. [DOI] [PubMed] [Google Scholar]
- Lyczak R, Gomes JE, Bowerman B. Heads or tails: cell polarity and axis formation in the early Caenorhabditis elegans embryo. Dev Cell. 2002;3:157–166. doi: 10.1016/s1534-5807(02)00226-5. [DOI] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Morton DG, Shakes DC, Nugent S, Dichoso D, Wang W, Golden A, Kemphues KJ. The Caenorhabditis elegans par-5 gene encodes a 14-3-3 protein required for cellular asymmetry in the early embryo. Dev Biol. 2002;241:47–58. doi: 10.1006/dbio.2001.0489. [DOI] [PubMed] [Google Scholar]
- O’Connell K, Maxwell K, White J. The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote. Dev Biol. 2000;221:55–70. doi: 10.1006/dbio.2000.9714. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye CA, Tagawa A, Lyczak R, Bowerman B, Aroian RV. The anaphase-promoting complex and separin are required for embryonic anterior-posterior axis formation. Dev Cell. 2002;2:195–206. doi: 10.1016/s1534-5807(02)00114-4. [DOI] [PubMed] [Google Scholar]
- Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Sadler PL, Shakes DC. Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development. 2000;127:355–366. doi: 10.1242/dev.127.2.355. [DOI] [PubMed] [Google Scholar]
- Schubert CM, Lin R, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- Shelton CA, Carter JC, Ellis GC, Bowerman B. The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior-posterior polarity, and body morphology during Caenorhabditis elegans embryogenesis. J Cell Biol. 1999;146:439–451. doi: 10.1083/jcb.146.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Powers J, Dunn M, Reese K, Malone CJ, White J, Seydoux G, Saxton W. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 1998;125:3607–3614. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Schubert C, Seydoux G. Genetic requirements for PIE-1 localization and inhibition of gene expression in the embryonic germ lineage of Caenorhabditis elegans. Dev Biol. 1998;200:212–224. doi: 10.1006/dbio.1998.8940. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Wallenfang MR, Seydoux G. Polarization of the anterior-posterior axis of C. elegans is a microtubule-directed process. Nature. 2000;408:89–92. doi: 10.1038/35040562. [DOI] [PubMed] [Google Scholar]
- Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC, Priess JR, Kemphues KJ. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development. 1996;122:3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]


