Abstract
WSB-1 is a SOCS-box-containing WD-40 protein of unknown function that is induced by hedgehog signaling in embryonic structures during chicken development. Here we show that WSB-1 acts as an E3 ubiquitin ligase for the thyroid-hormone activating type 2 iodothyronine deiodinase (D2). The WD-40 propeller of WSB-1 recognizes a novel 18-amino acid loop in D2 that confers metabolic instability, while the SOCS-box domain mediates its interaction with an ubiquitinating catalytic core complex, modeled as Elongin BC-Cul5-Rbx1 (ECSWSB-1). In the developing tibial growth plate, hedgehog-stimulated D2 ubiquitination via ECSWSB-1 induces parathyroid hormone related peptide (PTHrP), thereby regulating chondrocyte differentiation. Thus, ECSWSB-1 mediates a novel mechanism by which "systemic" thyroid hormone can effect local control of the hedgehog-PTHrP negative feedback loop and thus skeletogenesis.
Keywords: ubiquitin ligase, thyroid hormone, Sonic/Indian Hedgehog, deiodinase
In all vertebrates, thyroid hormone can be activated or inactivated by iodothyronine deiodinases. D2 generates T3 from the prohormone T4 in the perinuclear space, increasing the supply of T3 to the cell nucleus. Targeted disruption of the Dio2 gene in mice impairs cochlear development, pituitary thyroid-stimulating hormone (TSH) feedback, and adaptive thermogenesis, while myocardial overexpression of D2 causes chronic cardiac-specific thyrotoxicosis 1.
Regulation of D2 activity is achieved primarily by ubiquitination. For endoplasmic reticulum (ER)-resident proteins such as D2 1, this regulatory mechanism is referred to as ER-associated degradation (ERAD) 2,3. Ubiquitination inactivates D2 and targets the protein for degradation in the proteasomes, a process that is accelerated during deiodination of T4 4,5. As a consequence, the half-life of D2 can vary from 12 to 300 minutes depending on the rate of T4 deiodination. Remarkably, inactive ubiquitinated D2 can be reactivated by the pVHL-interacting deubiquitinating enzyme-1 (VDU1) 6. This dynamic, reversible mechanism integrates developmental, environmental, and homeostatic signals to control thyroid hormone action.
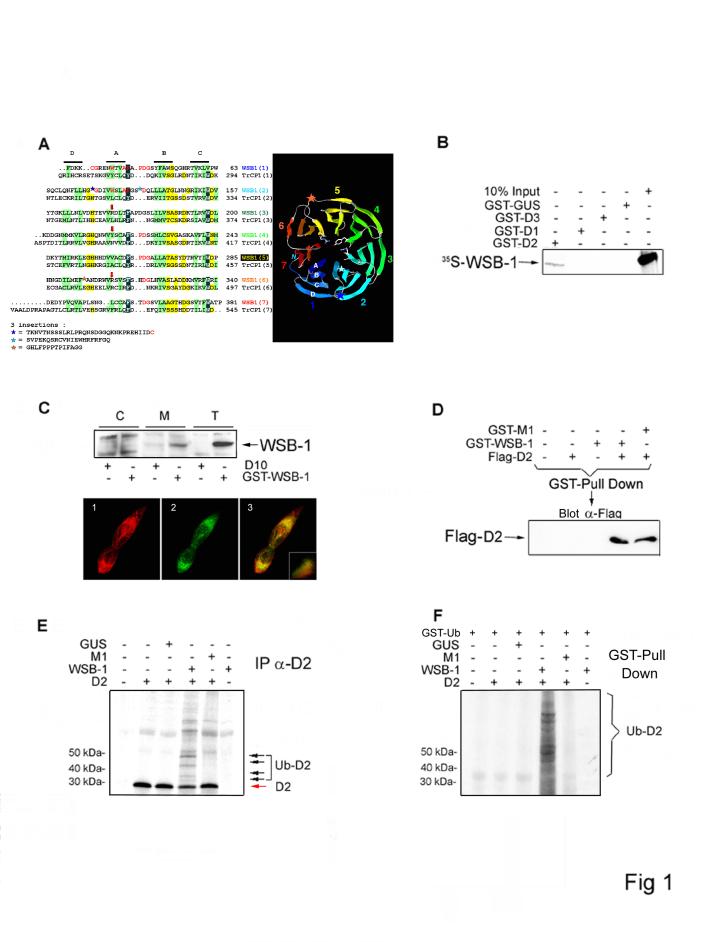
To characterize the D2 ubiquitinating complex, we performed a yeast two-hybrid screen of a human brain library with D2 6. WSB-1 7 (BC021110.1; also known as SWiP-1 8) was identified as a D2-interacting protein (Fig. 1A). The physical interaction between WSB-1 and D2 was confirmed in vitro, as bacterially expressed GST-D2 interacted with mouse 35S-WSB-1 in GST pull-down assays, while the nonubiquitinated deiodinases D1 and D3 did not (Fig. 1B). Functional studies in HEK-293 cells, which endogenously express WSB-1 but not D2, revealed that WSB-1 was predominantly present in the microsomal pellet and co-localized with transiently expressed D2 in the perinuclear region (Fig. 1C). D2-WSB-1 interaction was confirmed by coprecipitation in sonicates of cells transiently expressing both proteins (Fig. 1D).
Figure 1.
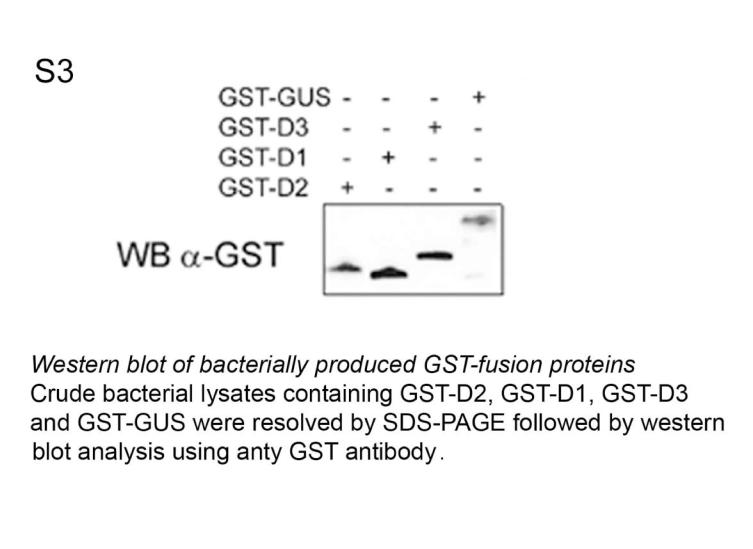
WSB-1 is a D2-interacting E3 ubiquitin ligase. (A) The alignment of the WD-40 repeats of WSB-1 with those of TrcP1 was refined using HCA 9. Positions of the four strands are indicated, with hydrophobic amino acids colored green. Large insertions are symbolized with stars, with the corresponding amino acid sequences indicated below. Arrows indicate the three amino acids located in position A+2 within strand A. Included in the far right is a ribbon representation of the WSB-1 propeller model conceived on the basis of the alignment. (B) Crude bacterial lysates containing GST-D2, GST-D1, GST-D3 or GST-GUS (Arabidopsis Thaliana β-glucouronidase) were incubated with 35S-labelled in vitro translated WSB-1. After precipitation by glutathione-Sepharose, co-precipitated proteins were resolved by SDS-PAGE. Ten percent of the 35S-WSB-1 input was loaded in the far right lane. GST-fusion proteins were measured by western blot and normalized for each reaction (Fig. S3). (C) Cytosolic (C) and microsomal (M) fractions of HEK-293 cells transiently expressing GST-WSB-1 were resolved by SDS-PAGE and analyzed by western blot using GST antibody. The positive control (T) was total cell lysate and negative controls were obtained from cells transfected with D10 plasmid DNA. Below is an immunofluorescence confocal microscopy of HEK-293 cells transiently co-expressing FLAG-D2 and GFP-WSB-1, using anti-FLAG (1; red) and anti-GFP (2; green) antibodies, respectively. The superimposition image of the same field is shown in 3. The inset is the distribution spectrum of the image pixels. (D) GST pull down of HEK-293 cells transiently expressing FLAG-D2, GST-WSB-1 and/or GST-M1. Pellets were resolved by SDS-PAGE and analyzed by western blot with anti-FLAG antibody. (E) 35S-labeled D2 was used in the in vitro ubiquination assay in a reticulocyte lysate system containing 10 μM ubiquitin aldehyde, energy solution and 30 μM ubiquitin. Bacterially expressed WSB-1 or M1 proteins were added as indicated after normalization by Coomassie-blue stained SDS-PAGE. Ubiquitinated D2 was immunoprecipitated using D2 antiserum (IP α-D2), and the pellets were resolved by SDS-PAGE. The black arrows indicate the ubiquitin-35 S-D2 conjugates (Ub-35S-D2). (F) Similar experiment to (E) except that 30 μM GST-ubiquitin (GST-Ub) was used and samples were processed for GST pull down.
The structure-function relationship of the D2-WSB-1 interaction was then examined by using hydrophobic cluster analysis (HCA) to analyze the WSB-1 sequence 9. A five-amino acid sequence in WSB-1 links the SOCS-box motif to a propeller-like structure formed by seven WD-40 repeats (Fig. 1A). Two large insertions in the second repeat were previously assigned as pieces of an eighth repeat 7, but actually do not possess features characteristic of the more conserved WD-40 inner β-strands A,B,C (Fig. 1A). These insertions might constitute additional strands in the propeller blades, as observed in other WD-40 propeller proteins 10. Screening of the Protein Data Bank (PDB) for WSB-1 related structures identified β-TrCP1, one of the interchangeable E3 ubiquitin ligases of the Skp1-Rbx1-Cdc53 (SCFβ-TrCP1) ubiquitinating complex essential for the NF-κB, Wnt/Wingless, and Hedgehog signaling pathways 11. In β-TrCP1, a seven-bladed WD-40 propeller is linked to an F-box motif. The structural relationship with WSB-1 is preserved because SOCS-box proteins are both structurally and functionally related to F-box proteins 12,13. In both β-TrCP1 and Cdc4, a related WD-40 propeller/F-box E3 ubiquitin ligase, the F-box motif allows for interaction with the Skp1 component of the SCF constant catalytic core. These observations indicate that WSB-1 contains the structural hallmarks of a D2-specific E3 ubiquitin ligase, with the WD-40 propeller mediating substrate recognition and the SOCS box the interaction with other components of the catalytic core complex.
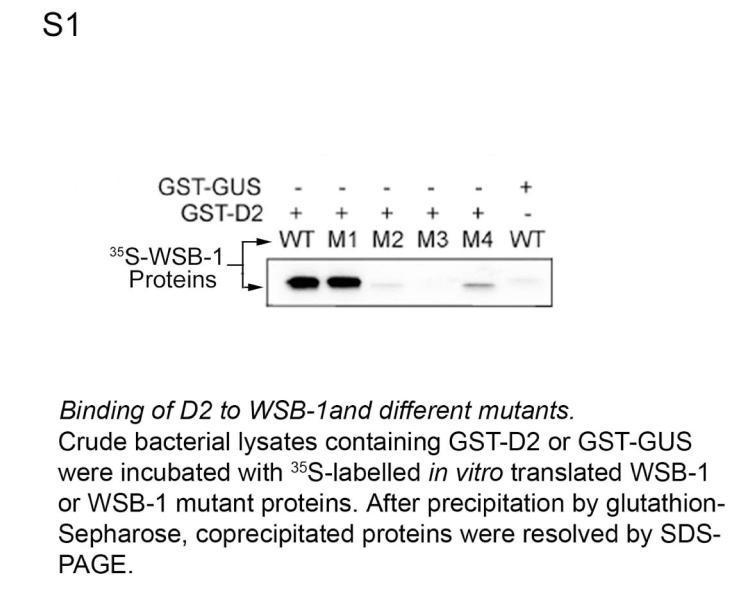
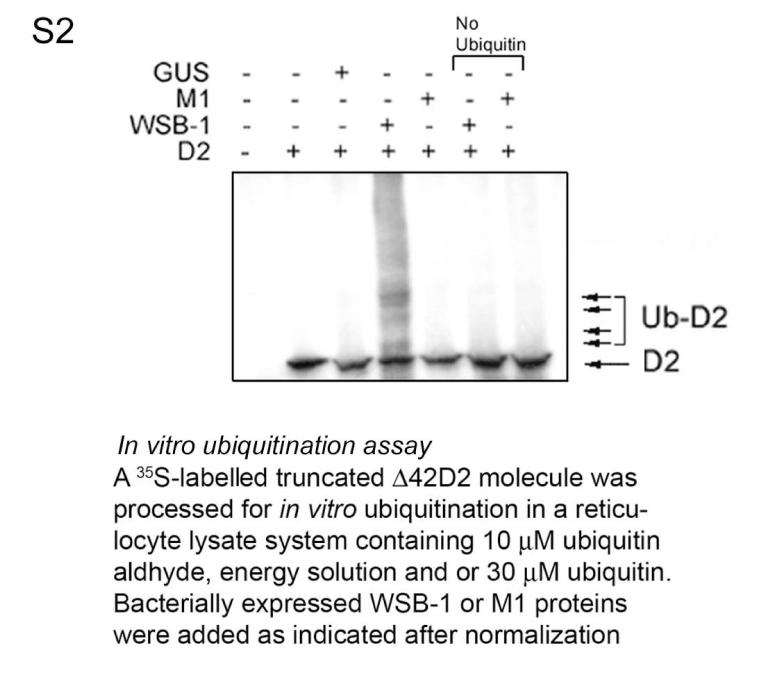
To test this hypothesis we first sought to demonstrate that D2 ubiquitination in vitro requires WSB-1. In these studies we also used M1, a mutant WSB-1 with a critical disruption in the BC box that prevents binding to the catalytic core complex 14 but does not affect binding to D2 (Figs. 1D, S1). Bacterially expressed WSB-1 was incubated with in vitro translated catalytically active 35S-labeled D2 in a cell lysate prepared to support ubiquitination 15. Immunoprecipitation with D2 antiserum revealed higher-molecular weight 35S-D2 bands in a typical ladder pattern only when ubiquitin and WSB-1, but not M1, were present in the reaction mixture (Figs. 1E, S2). D2 ubiquitination was confirmed using GST-ubiquitin (Fig. 1F).
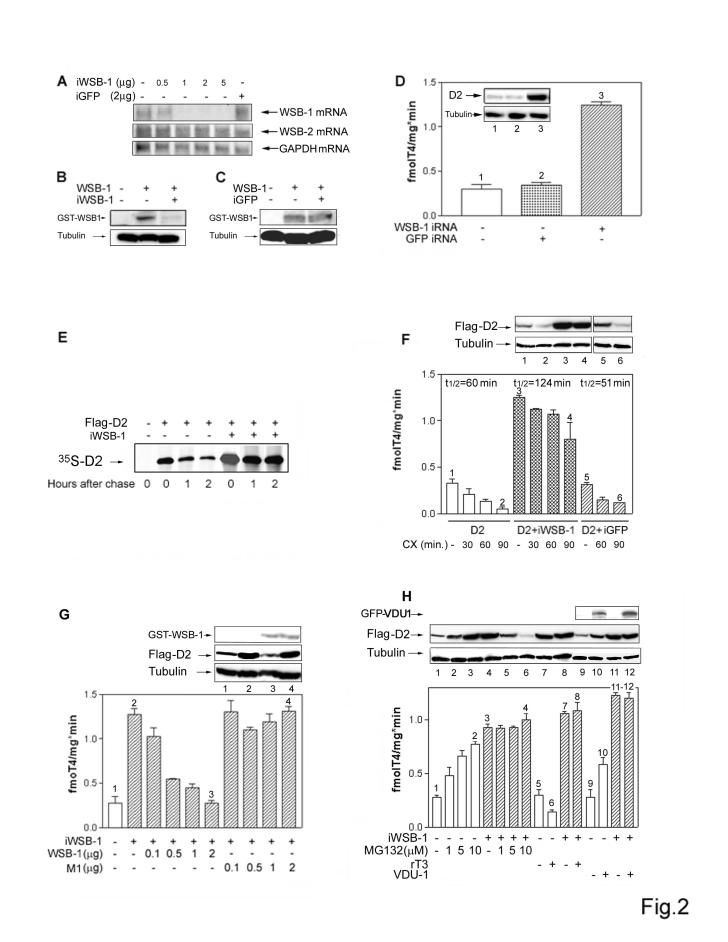
The hypothesis that WSB-1 is a D2-specific E3 ubiquitin ligase is supported by functional studies in which RNAi-mediated knockdown of WSB-1 (Fig. 2A-C) caused a 5-fold increase in steady-state D2 level (Fig. 2D). This increase in D2 resulted from a 2-3-fold prolongation of its half-life (Fig. 2E-F) and was reversed in a dose-dependent fashion by co-transfection with WSB-1, but not M1, encoding plasmid DNA (Fig. 2G). To explore whether alternate E3 ligases for D2 exist, the effect of WSB-1 knockdown was also assessed after treatment with the proteasome inhibitor MG132 or reverse T3 (rT3), a D2 substrate that, like T4, accelerates the rate of D2 ubiquitination. During WSB-1 knock down MG132 treatment did not result in additional increases in D2 (Fig. 2H) nor exposure to rT3 decreased D2 (Fig. 2H). Furthermore, the expected increase in D2 seen during VDU-1 overexpression was abrogated during WSB-1 knockdown (Fig. 2H). These results demonstrate that WSB-1 is the primary E3 ubiquitin ligase for D2.
Figure 2.
WSB-1 is the primary E3 ubiquitin ligase for D2. (A) WSB-1 knockdown was achieved by transfecting HEK-293 cells with the indicated amounts of pSilencer 2.1-U6 neoplasmid (Ambion, Austin TX) encoding a 19-base pair double-stranded oligonucleotide targeting the WSB-1 coding sequence (iWSB-1). Northern blot analysis was used to measure endogenous levels of WSB-1, WSB-2 (a structurally related WD-40 SOCS-box-containing protein 7 that does not affect D2) and GAPDH mRNA. Also shown are HEK-293 cells transfected with a pSilencer 2.1-U6 neoplasmid encoding a 19-base pair double-stranded oligonucleotide targeting the GFP coding sequence (iGFP). (B) Western blot was used to document reduced WSB-1 levels in cells co-transfected with iWSB-1 and GST-WSB-1 (0.2 ug). (C) iGFP was used to document the specificity of the WSB-1 knock down by western blot. (D) D2 activity was measured in HEK-293 cells transiently expressing D2 and/or iWSB-1 or iGFP-1. The inset contains a western blot of sonicates of cells transiently expressing FLAG-D2 vector studied under identical conditions. Here and below, numbers match the gel lanes with the bars. (E) HEK-293 cells transiently expressing FLAG-D2 or FLAG-D2 and iWSB-1 were pulse labeled with 35S-methionine-cysteine and chased with unlabeled medium for the indicated times. Total cell extracts were immunoprecipitated with anti-FLAG antibody. Pellets were resolved by SDS-PAGE. The sample in the far left lane is from cells transfected with D10 vector. (F) D2 activity was measured in HEK-293 cells transiently expressing D2, D2 and iWSB-1, or D2 and iGFP treated with vehicle or cycloheximide (CX; 100 μM) for the indicated times. The approximate activity half-life under each condition is indicated above the columns. Here and below, western blotting was used to quantify D2 or other proteins as indicated. (G) D2 activity was measured in HEK-293 cells co-transfected with the indicated amounts of iWSB-1, WSB-1 or M1 expressing plasmid DNA. (H) D2 activity in HEK-293 cells transiently expressing D2, co-expressing D2 and iWSB-1 or D2 and VDU-1, treated with vehicle or the indicated concentrations of MG132 or rT3 for one hour. For all experiments in B-F, results are the mean ± standard deviation (SD) of 4-6 samples.
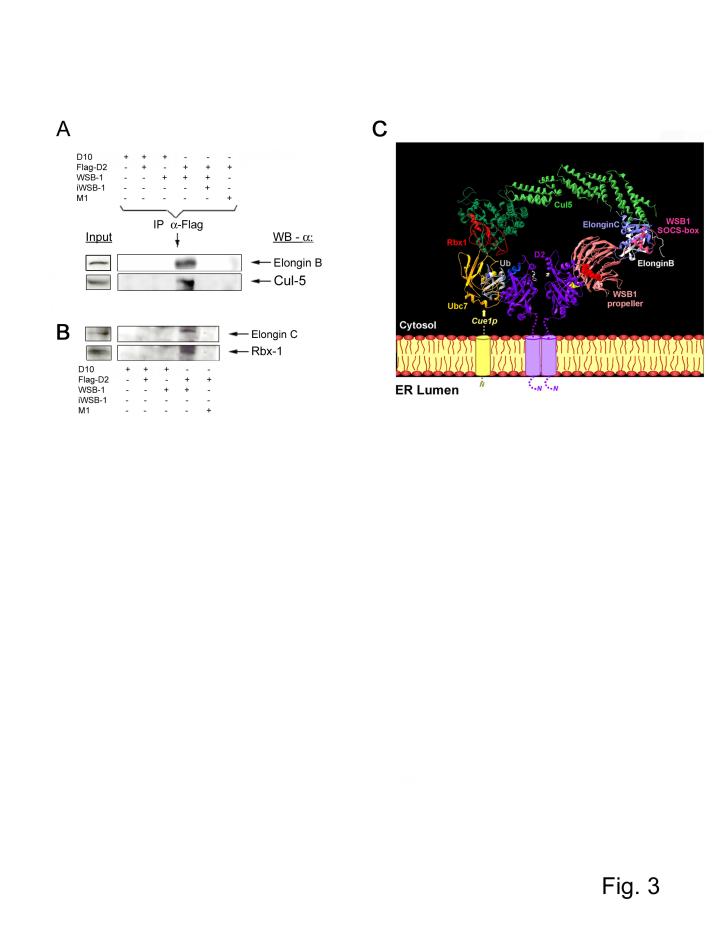
The BC box motif found in SOCS-box proteins allows binding with the Elongin-BC multifunctional protein complex, which in turn is capable of assembling with the E2-activating Cul5/Rbx1 module to reconstitute potential ubiquitin ligases in vitro 14,16. We confirmed that endogenous Elongin B, Elongin C, Cul5 and Rbx1 are present in the D2 catalytic core complex, as these proteins co-immunoprecipitated with D2 only when FLAG-D2 was co-expressed with functional WSB-1 protein, and not M1 (Fig. 3A,B). The resulting model for the Elongin BC, Cullin, SOCS-box protein (ECSWSB-1) complex was derived by assembling WSB-1 with these adaptor subunits based on defined structures: Cul1-Rbx1-Skp1-Fbox(Skp2) 17, Ubc1/ubiquitin 18, Elongin BC-VHL complex 19 and the TrcP1-beta-catenin-Skp1 (Fig. 3C) 20. Ubc7 was integrated in the ECSWSB-1 (Fig. 3C) based on the structure for RING-type E3 c-Cbl / UbcH7 E2 21, with superimposition of the RING domain of c-Cbl with Rbx1, and substitution of HuUbc7 (pdb 1FBV, 21) by Ubc7 (pdb 2UCZ, 22). Binding of Rbx1 to either Ubc7, which is associated to the ER surface through interaction with its membrane-bound receptor Cue123, or Ubc6 24, an integral ER membrane protein that also supports D2 ubiquitination 25,26, could explain the recruitment of the ECSWSB-1 complex to the ER. Finally, ubiquitin was docked on Ubc7 according to the structure of Ubc1/ubiquitin (pdb 1FXT, 18).
Figure 3.
Composition of the ECSWSB-1 catalytic core complex. (A-B) Lysates of HEK-293 cells transiently expressing the indicated plasmid combination were pulled down with anti-Flag antibody and pellets were processed for western blotting as indicated. (C) In order to assemble the ECSWSB-1 ubiquitinating complex, Elongin C (pdb 1LM8; 14) was docked on the N-terminal part of Skp1 within the structure of the Cul1-Rbx1-Skp1-Fbox(Skp2) complex (pdb 1LDK, 13) and a model of Cul5, which shares 30 % sequence identity with Cul1, was used. The Cul5-Rbx1-Elongin C-Elongin B-VHL complex was reconstructed using the VHLα domain as template for the SOCS-box motif of WSB-1, because such motifs bind Elongin C 12,13. Despite a similar fold for the F-box of Skp2 and the α-domain of VHL, the binding mode with Skp1/ElonginC is different 20. Only the first three helices of the α-domain (three-helix cluster) constitute the common core between F-box, VHL α-domain, and SOCS-box 27.The TrcP1-β-catenin-Skp1 complex (pdb 1P22; 20) was used as template after superimposition of Skp1 on the Skp1/ElonginC structure of the complex described above. The model of the WSB-1 propeller was superimposed on the structure of the TrcP1 propeller and adjusted manually to join the C-terminus of the propeller to the N-terminal end of the SOCS-box through a predicted 5 amino acid loop in WSB1. In contrast to TrcP1, WSB-1 does not have a long linker, so that the propeller is near to the SOCS-box motif. The N-terminal part of WSB-1, which cannot be modeled at this stage, should correspond to a helical extension, located on the Elongin C side of the propeller. Data on the structure of Cue1p are not available and its interaction with Ubc7 is indicated with a dotted line and an arrow. D2 is represented as a dimer 28 with the T4 near the active center (white). The 18-amino acid loop of the right D2 subunit, which can not be modeled at this stage and for which only the start and end positions are indicated in yellow, is oriented towards WSB1, in which the 3 critical amino acids (R174, R315, Y218) are shown as red spheres 25.
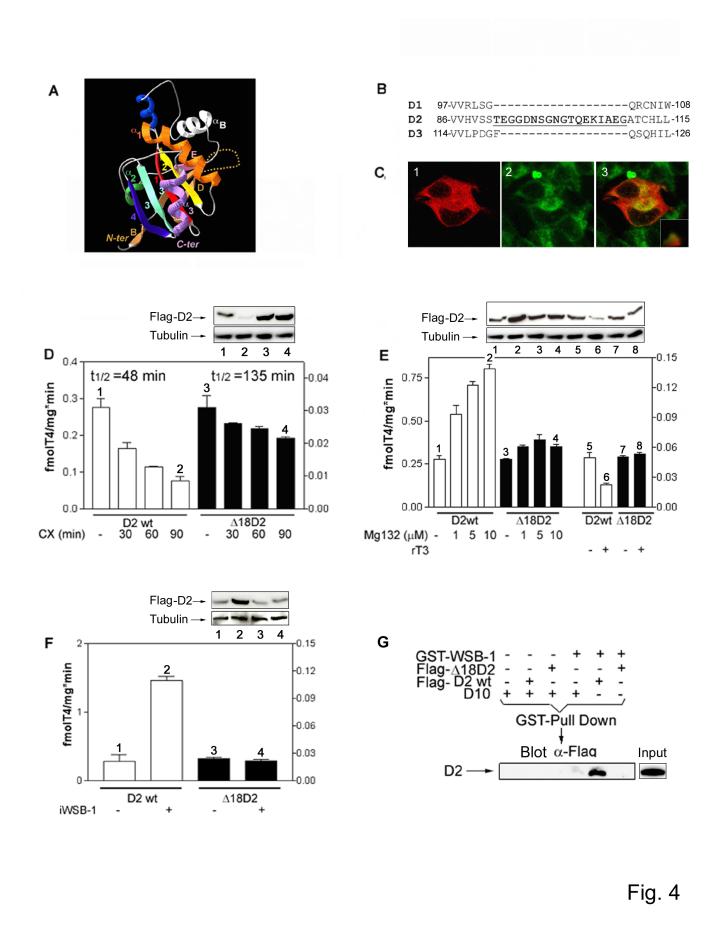
To orient D2 relative to the ECSWSB-1 complex, we sought to identify potential WSB-1 recognition sequences in D2. We compared three-dimensional models of D2 and the nonubiquitinated D1 and D3 enzymes, finding that D2 has a unique 18-amino acid sequence with the typical characteristics of a large loop (Fig. 4A-B) 27. A deletion mutant lacking this loop (Δ 18D2) was still present in the ER (Fig. 4C) and retained D2 activity with a normal substrate affinity (Km-T4 ∼5 nM), but displayed a lower turnover rate (Fig. 4D-F). This Δ 18D2 deletion mutant was found to have a 2.8-fold longer half-life (Fig. 4D) while being insensitive to MG132 (Fig. 4E) and rT3-induced loss of protein and activity (Fig. 4E). Δ 18D2 was not affected by WSB-1 knockdown (Fig. 4F), a finding explained by the inability of Δ 18D2 to bind with WSB-1 (Fig. 4G). These data indicate that the 18-amino acid loop is critical for D2 recognition by WSB-1. D2 was integrated in the model by orienting the 18-amino acid loop proximal to the WSB-1 propeller D-A/B-C side (Fig. 3C). The top face (D-A/B-C side) of the propeller has a clear line of sight to the E2 ubiquitin conjugase, suggesting that D2 is held between these two enzymes. In this model, a physical gap between Ubc7 and D2 exists, prompting us to integrate D2 in its dimeric form 28. This is suported by the known Ubc7 binding interaction with the IDUA-αB-βE-α2-β3 domain in D2 (Fig. 4A) 25. Thus, positioning a second D2 monomer in the putative dimer formation not only fills the existent gap but places the αB-βE D2 motif in condition of mediating the D2-Ubc7 interaction (Fig. 3C).
Figure 4.
D2 contains an 18-amino acid destruction loop. (A) Three-dimensional model of D2 shown in a similar orientation as in Fig. 3C (right subunit). D2 has a single N-terminal transmembrane segment and several clusters, typical of α-helices or β-strands, corresponding to core secondary structures of the deiodinase globular domain. A striking feature is the presence of the thioredoxin (TRX) fold, defined by the βαβ and ββαmotifs. Within the canonical TRX fold, the relationship between the βαβ and ββα motifs is locally interrupted by interfering elements, one of which is highly conserved and shares striking similarities with the lysosomal enzyme α-L-iduronidase (IDUA). The active center, which contains the amino acid selenocysteine, is a pocket defined by the β1-α1-β2 motifs of the TRX-fold and the IDUA-like insertion. The dotted line illustrates the destruction loop in D2. (B) Amino acid sequence of human D1, D2, and D3 detailing the 18-amino acid destruction sequence (underlined) in D2. (C) Immunofluorescence confocal analysis of HEK-293 cells transiently expressing FLAG-Δ18D2 and co-stained with anti-FLAG antibody (1; red) and ER-tracker (2; green). The superimposition image of the same field is shown in 3. The inset is the distribution spectrum of the image pixels. (D) D2 activity in HEK-293 cells transiently expressing D2 or Δ18D2 (units on the right Y axis), treated with vehicle or 100 μM CX for the indicated times. Here and below, western blotting was used to quantify D2 or other proteins as indicated. Numbers match the gel lanes with the bars. (E) D2 activity in HEK-293 cells transiently expressing D2 or Δ18D2, treated with vehicle, MG132 or 30 nM rT3 for 1 h. (F) D2 activity in HEK-293 cells co-transfected with D2, Δ18D2 and/or iWSB-1 expression plasmids. For all experiments in D-F, results are the mean ± SD of 4-6 samples. (G) HEK-293 cells were co-transfected with FLAG-D2 or FLAG-Δ18D2 and GST-WSB-1 expression plasmids. Whole cell extracts were used in a GST pull down, and pellets were resolved by SDS-PAGE and western analysis probed with FLAG antibody. In the far right lane a sample containing total cell lysate is shown.
In other WD-40 propeller E3 ubiquitin ligases, substrate recognition is dependent on "super sites" for protein-protein interaction, the most common being the second positions of each blade (the A+2 positions of the inner strand A), which are frequently occupied by Arg residues that interact with phosphate groups in the substrate 10,20. Arginine occupies two A+2 sites in WSB-1, i.e., Arg174 (third WD-40 repeat) and Arg315 (seventh WD-40 repeat). Mutation of these sites (Arg174Ala (M3) and Arg315Ala (M4)) abrogated interaction with D2 (Fig. S1). A third A+2 position in WSB-1 occupied by Tyr (Tyr218) was also found to be critical for D2 binding via mutation to Ala (Tyr218Ala) (Fig. S1). These data confirm that the WSB-1 WD-40 propeller super sites are critical for D2 recognition.
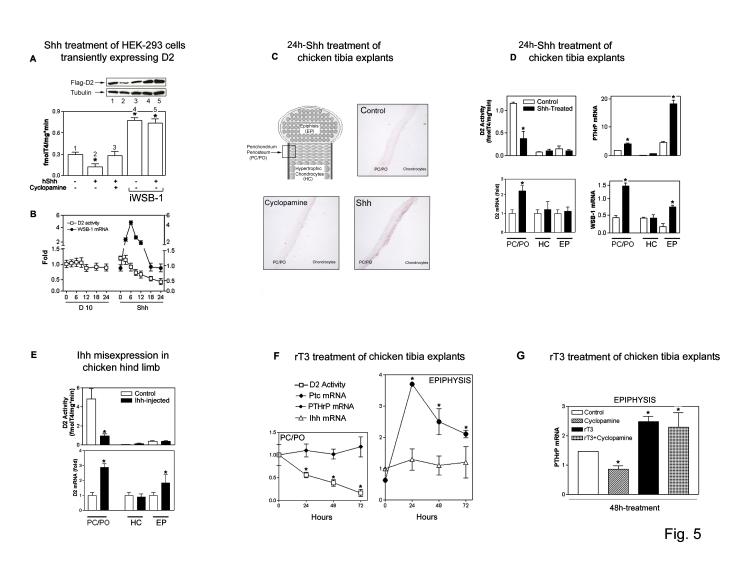
WSB-1 expression is induced by Shh in developing limb buds and other embryonic structures 8. To test the hypothesis that hedgehog regulation of the ECS WSB-1/D2 mechanism for control of thyroid hormone activation is physiologically relevant in this setting, we used an HEK-293 cell-based transient D2 expression system. Exposure to Shh for 24 h induced a loss (∼50%) of D2 (Fig. 5A) that was preceded by a transient ∼5-fold increase in WSB-1 mRNA (Fig. 5B), while D2 mRNA levels remained unchanged (not shown). This effect was blocked by the hedgehog-specific antagonist cyclopamine (Fig. 5A), and importantly, Shh failed to down-regulate D2 during WSB-1 knock down (Fig. 5A).
Figure 5.
Hedgehog signaling decreases D2 by inducing WSB-1. (A) D2 activity was measured in HEK-293 cells transiently expressing D2 co-cultured for 24 h with an equivalent number of HEK-293 cells transiently expressing Shh or D10 vector. Vehicle or 5μM cyclopamine were used as indicated. iWSB-1 was employed as indicated. D2 was quantified by western blotting and numbers match gel lanes to bars. *p<0.05 vs. vehicle-treated cells (ANOVA). (B) Time-course of D2 activity and WSB-1 mRNA levels by real-timer PCR in cells prepared as in (A). Results are expressed relative to time zero and the loss of D2 activity negatively correlates with time (p<0.0001). In A-B, results are the mean ± SD of 4-6 samples. (C) WSB-1 in situ hybridization of chicken tibiotarsi from day 12 chicken embryos cultured in serum-free medium containing 740 pM T4 and treated for 24 h with vehicle, Shh (4 μg/ml) or cyclopamine (5μM). The bone area shown analyzed is labeled as PC/PO in the schematic of the growth plate. At least 4 tibias were examined for each experimental condition with similar results. Hybridization with the sense probe was negative (not shown). (D) D2 activity and different mRNA levels in tissues of tibiotarsi isolated and processed as in (C): perichondrium/periosteum (PC/PO), hypertrophic chondrocytes (HC), and bone epiphysis (EP). Results are the mean ± SD of 4 embryos. *p<0.05 vs. untreated tibiotarsi by Student's t-test. (E) D2 activity and mRNA levels in day 8 chicken embryo hind limb misexpressing Ihh. The uninfected tibia was used as control. Results are the mean ± SD of 3 embryos. *p<0.05 vs. uninfected tibial tissue by Student's t-test. (F) D2 activity and different mRNA levels in tissues of tibiotarsi isolated and cultured as in (C), and treated with 20 nM rT3 for the indicated times. Results are the mean ± SD of 4 embryos. *p<0.05 by Student's t-test vs. untreated tibiotarsi. (G) PTHrP mRNA levels in tibiotarsi isolated and cultured as in (C); treatments were 20 nM rT3 and/or 5μM Cyclopamine. Results are the mean ± SD of 2 embryos. *p<0.05 vs. untreated tibiotarsi by one way ANOVA.
In the developing tibial growth plate, Indian Hedgehog (Ihh) is produced by chondrocytes leaving the proliferative pool, inducing the secretion of PTHrP from perichondrial cells and chondrocytes near the ends of the skeletal elements, which acts to maintain chondrocyte proliferation such that hypertrophic differentiation is limited to the cells furthest from where PTHrP is produced 29. Ihh has similar biological properties and stimulates the same target genes as Shh 30 such that activation of the hedgehog pathway by either Ihh or Shh would be expected to induce WSB-1-mediated D2 ubiquitination. The applicability of this system for our studies was confirmed by the finding of both D2 and WSB-1 expression in the perichondrial/periosteal (PC/PO) sheaths surrounding the developing chondrocytes, with much lower levels of D2 in the chondrocytes themselves (Fig. 5C-G).
Exposure of 12-day old chicken tibial explants30 to Shh led to a striking induction of WSB-1 gene expression detected via in situ hybridization, particularly in cells contained in the inner layer of the PC/PO (Fig. 5C). In addition, the tibiae were manually dissected into three regions after exposure to Shh: the non-articular perichondrium and periostium (PC/PO); the prehypertrophic chondrocytes, proliferating chondrocytes and periarticular perichondrium (which cannot be cleanly removed from the articular cartilage) collectively referred to here as the epiphysis (EP); and the hypertrophic chondrocytes (HC). Treatment with Shh led to the expected high level induction of PTHrP specifically in the EP fraction containing the periarticular perichondrial cells (Fig 5D).
Hedgehog signaling is necessary and sufficient for high level PTHrP expression in the periarticular perichondrium 30,31. This induction is believed to be indirect, mediated by changes in expression of an, as of yet, unidentified secondary signal downstream of Ihh in the perichondrium 30. Strikingly, we observed a greater than 50 % decrease in D2 activity in the PC/PO fraction following Shh treatment, whereas D2 activity was much lower in the HC and EP cells with no significant modulation by Shh. The decrease in D2 activity in the PC/PO cells correlated with a concomitant 3-fold increase in WSB-1 mRNA levels (Fig 5D). WSB-1 mRNA was also increased approximately 3-fold in the EP tissue which, as noted above, does not exhibit significant D2 activity. This result is consistent with the fact that the proliferating chondrocytes are also targets of Ihh 31, but unlike the perichondrium do not participate in the PTHrP feedback loop 32.
The physiological consequence of a decrease in D2 activity in the growth plate would be the generation of local hypothyroidism, as seen in other D2-expressing tissues 1. To test this, we took advantage of the fact that Dio2 expression is negatively regulated by thyroid hormone 1 but not by Shh signaling (not shown). D2 mRNA was increased two fold in the Shh-treated PC/PO cells (Fig. 5D), as would be expected if the cells were relatively hypothyroid. In HC and EP cells, where basal D2 activity is low, D2 mRNA levels were not affected by Shh treatment (Fig. 5D). These findings are consistent with previous observations that hypertrophic chondrocyte differentiation is impaired and PTHrP mRNA levels are increased in the growth plate of young rats with systemic hypothyroidism 33, a strikingly similar phenotype to that seen with Ihh misexpression 30. In addition, thyroid hormone inhibits chondrocyte clonal expansion and proliferation while simultaneously promoting HC differentiation 34.
To verify that the hedgehog induced decrease in D2 activity reflects a physiological mechanism, we measured D2 activity following Ihh misexpression in 8 day-old chick embryos in ovo. Four days after infecting limbs with a replication-competent retroviral vector carrying Ihh, more than 80 % of D2 activity had been lost in the PC/PO sheath (Fig 5E). As with the tibia explants, Ihh misepression resulted in significant increase of D2 mRNA (Fig. 5E). Taken together, these results are consistent with a model where Ihh acts on the perichondrium to induce WSB-1-expression, thereby decreasing D2 activity and thus thyroid hormone signaling, finally causing an up-regulation of PTHrP expression in the periarticular perichondrium. To verify these epistatic relationships, we utilized rT3, an inactive metabolite of thyroid hormone that accelerates ECSWSB-1-mediated D2 ubiquitination (see Fig. 2H) but does not bind the thyroid hormone receptor. After 24-72 h of incubation with rT3, we observed a 50-80 % decrease in D2 activity in PC/PO sheaths (Fig. 5F). Remarkably, a 2.5- to 5.5-fold induction of PTHrP mRNA levels was observed in EP cells (Fig. 5F), and the magnitude of this induction was similar to that seen in Shh-treated tibia explants (Fig. 5D). Neither Ihh nor its downstream target Ptch1 was induced by rT3 treatment (Fig. 5F). The ability to induce PTHrP expression independently of hedgehog signaling in this manner strongly suggests that the decrease in D2 activity downstream of Ihh is normally responsible for PTHrP induction. Treatment of chicken tibia explants with cyclopamine led to a decrease in PTHrP expression due to its ability to block endogenous Ihh activity (Fig. 5G). Strikingly, PTHrP mRNA induction by rT3 was not sensitive to cyclopamine (Fig. 5G), confirming that D2 acts downstream of Ihh in this pathway. Whether the induction of PTHrP is a direct response to hypothyroidism or results from T3-induced changes in other signaling elements remains to be clarified.
Methods
Reagents. Unless otherwise specified all reagents were obtained from Sigma (St. Louis, MO) or Calbiochem (La Jolla, CA). Mouse monoclonal Flag antibody (M2) and mouse monoclonal α-Tubulin (DM1A) antibody were from Sigma. Mouse monoclonal antibody to Elongin C was from Transduction Laboratories (Greenland, NH). Goat polyclonal antibodies to Cul 5 (H-300), to Rbx-2 (N-15) and to Elongin B (A-19) were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibody to Rbx1 was from Neomakers (Fremont, CA).
Constructs. The C-terminal FLAG-D2 and C-terminal FLAG-CysD2 (Sec133C, Sec266C), GST-D1 and GST-D3 plasmids have been described 5. A 1266-nt PCR product, corresponding to the mouse WSB-1 cDNA coding region, was subcloned into pDEST27 plasmid (Invitrogen, Inc., Gaithersburg, MD); to generate the WSB-1 mutant constructs M1(L387P and C391F), M2(Y218A), M3(R174A) and M4(R315A), we used recombinant PCR with two sets of oligos, and the final PCR products were re-inserted into pDEST15 plasmids (Invitrogen). The truncated Δ42D2 construct, lacking the 42 N-terminal amino acids in the coding region of human CysD2 (1-42), was obtained by PCR, and the resulting fragment was subcloned into the pCI vector (Promega, Madison, WI). The Δ18D2 was constructed by engineering a 18 aa (92-109) deleted D2 fragment by Vent PCR, inserting it between the XcmI sites of the wild type human D2, followed by overlap-extension PCR to mutate the selenocysteine 133 and 266 to cysteine. The human Shh cDNA was subcloned into the D10 plasmid.
Transfections and D2 activity assay. HEK-293 cells were transfected by the CaPO4 method or by using LipofectAMINE Plus reagents (Invitrogen). Transiently expressed D2 has previously been shown to retain the ubiquitination properties of endogenously expressed D2 4,5. Unless specified, cells were harvested 48 h later. For activity half-life studies, the above protocol was modified by treatment either with vehicle (dimethylsulfoxide) or with 100 μM CX for 0-90 min before harvesting. For substrate-induced degradation studies, the above protocol was modified by treatment either with vehicle (40 mM NaOH) or with 30 nM rT3 for 1 h before harvesting. Wherever D2 activity was measured, cells were transfected with 0.1 μg FLAG-D2 plasmid; for the western blot analysis cells were transfected with 1 μg FLAG-CysD2 plasmid. In all experiments hGH (TKGH) was used as a control for the transfection efficiency, as described previously 5. D2 activity was measured as described previously 5.
35S pulse-chase and immunoprecipitation studies. 35S incorporation was performed as previously described 6,28. Immunoprecipitation (IP) with FLAG antibody was performed as previously described 5. An in vitro coupled transcription/translation system (Promega) and L-u-methionine/cysteine Easytag (Perkin Elmer) were used to generate radioactive recombinant proteins. Ten μl of the indicated in vitro translated WSB-1, M1, M2, M3 or M4 was added to the GST-D2, GST-D1, GST-D3 or GST-GUS (Arabidopsis thaliana β-glucoronidase) previously bound to GST resin, and the GST pull-down was performed as previously described 25. Pellets were resolved in a 10% SDS PAGE gel, dried, and exposed to autoradiography for 35S-labeled band detection. For the in vivo co-immunoprecipitation experiments, HEK-293 cells were transiently transfected with GSP-WSB-1 (5 μg), GST-M1 (5 μg), FLAG-CysD2 (5 μg) or D10 (to normalize for DNA transfection amounts). After 48 h cells were harvested and the pellets were resuspended in lysis buffer containing 1 X PBS, 5 mM EDTA, 0.5% Triton X-100 (Sigma), type I protease inhibitors cocktail (Calbiochem) and 1 mM PMSF for IP. Cell lysates with each transfection combination were incubated with Glutathione Sepharose™ 4B (Amersham, Uppsala Sweden), and IP pellets were washed five times with lysis buffer, resuspended in sample loading buffer, boiled for 5 min, resolved in a 10% SDS-PAGE, and used for western blot analysis for FLAG-D2 detection with FLAG M2 monoclonal antibody (Sigma) at dilution 1:4000 5, using the BM® chemiluminescence western blotting kit (Roche) according to the manufacturer's instructions.
In vitro D2 ubiquitination assay. For optimal translation efficiency and to avoid nonspecific ubiquitination via the unfolded protein response, a catalytically active D2 (Δ42D2) truncated in the short N-terminal transmembrane domain was in vitro translated in a reticulocyte lysate system in the presence of L-35S-methionine/cysteine. After 90-min incubation at 30°C, addition of 1 mM unlabeled methionine/cysteine mix terminated the reaction. Ubiquitination reactions were carried out in a total volume of 30 μl containing 5 μl of GST-fusion protein from bacterial cultures in the assay buffer containing 10 μM Ubiquitin Aldehyde (Boston Biochem, Boston MA), 1X Energy Regeneration Solution (Boston Biochem), 30 μM Ubiquitin (Boston Biochem), or in some experiments 30 μM GST-Ubiquitin and the in vitro translated 35S-Δ42D2. The reaction mixture was incubated at 37°C for 1 h. Ub-D2 was then immunoprecipitated with D2 antiserum (IP α-D2), and the pellets were resolved by SDS-PAGE. When GST-ubiquitin was used, the samples were processed by GST pull down (IP α-GST). Samples were boiled for 5 min and analyzed by SDS-PAGE. Western blot analysis was performed to normalize the expression levels of the GST-fusion proteins used in the assays.
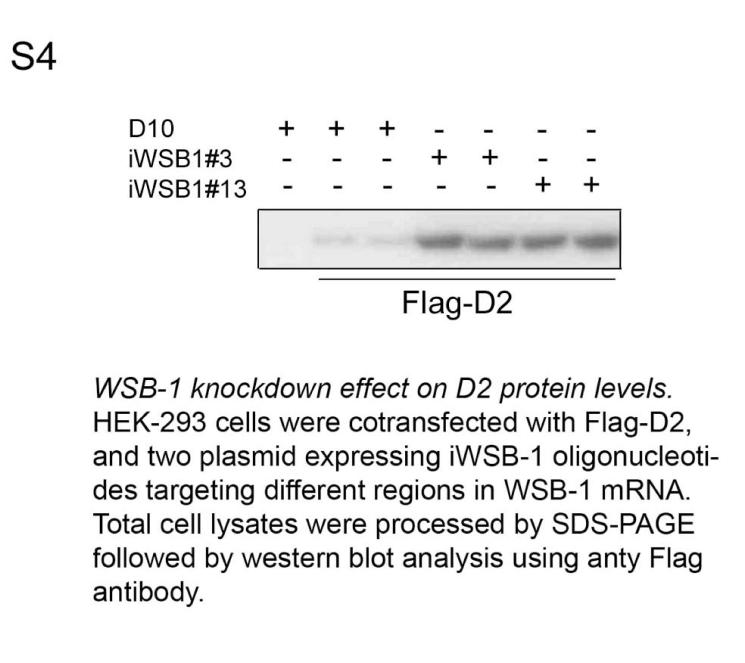
WSB-1 RNAi. To knockdown WSB-1 gene expression by RNAi a complementary 60-nt oligonucleotide with 5' single-stranded overhangs (#3, 5'-aatggcatcgcttcagatttg-3') was designed and cloned into the pSilencer™ neo plasmid (Ambion, Austin TX). In a typical experiment of WSB-1 knock down by RNAi, HEK-293 cells grown to 60-70 % confluence on a 60 mm plate were transfected with 2 μg iWSB-1 using LipofectAMINE Plus reagents (Invitrogen) and studied 48 h later. Both WSB-1 mRNA and protein levels were dramatically reduced under these conditions, which is typical of a classical RNAi (Fig. 2A-C). Unintentional effects on the expression of other genes was minimal as evidenced by stable WSB-2 mRNA levels (a structurally related WD-40 SOCS-box-containing protein 7 that does not affect D2) during iWSB-1. Titration of the iWSB-1 effect indicated that transfection with relatively low amounts of plasmid DNA (1-2 μg) was sufficient to achieve WSB-1 knock down (Fig. 2A). Using a second oligonucleotide sequence targeted to a different site in the WSB-1 mRNA, #13, 5'-tggcatcgcttcagatttg-3', resulted in a similar WSB-1 knock down efficiency, as evidenced by a sharp increase in D2 (Fig. S4). As an additional control, cells were transfected with 2 μg of pSilencer™ neo plasmid encoding an oligonucleotide sequence targeted to the GFP mRNA (iGFP), provided by the manufacturer, with no effects on D2 expression (Fig. 2D).
Infection of chicken tibia with IHH. RCAS-IHH 35 was injected in embryonic day 8 chicken right hind limbs. The infected and uninfected limbs were harvested on day 12 and the tibiotarsi were surgically removed. Each tibiotarsus was dissected into four parts as described below. The thin membrane around the condensation was removed first and this part is referred to as PC/PO. As inferred by the appearance and the site of blood vessel invasion, the tibia was divided into two parts. The region above the site of blood vessel invasion up to the suture is referred to as Epiphysis (Ep). Epiphysis as defined here consists of the prehypertrophic chondrocytes, proliferating chondrocytes and periarticular perichondrium (which cannot be cleanly removed from the articular cartilage). The region below is referred to as hypertrophic chondrocytes (HC). This tissue contains some ossified tissue as well. The dissections were conservative in order to avoid cross-contamination. The harvested tissues from each tibia were divided into two parts and put directly into liquid nitrogen. Later they were processed for real-time RT-PCR and measurement of D2 activity.
Organ culture of day 12 chick tibiotarsus. Tibiotarsi isolated from day 12 chicken embryos were cultured essentially as described 6. DMEM was supplemented with 0.1% BSA and 740 pM T4. In some experiments this medium was supplemented with 4 μg/ml Shh (R&D Systems Inc., Minneapolis MN), 5μM Cyclopamine or 20 nM rT3, as indicated. The tibiotarsi were harvested after 24-72 h, dissected, and processed as described above.
Other analytical methods. Immunofluorescence confocal microscopy was performed as described previously. In situ hybridization studies were performed as described previously 30 using specific chicken WSB-1 probes kindly provided by Dr. Daniel Vasiliauskas, New York University. Subcellular fractionation and Quantitative Real-Time Polymerase Chain Reaction were performed as described previously 17.
Acknowledgements
We thank Drs. P. Reed Larsen and Domenico Salvatore for helpful insights and comments on the manuscript. This work was supported by DK058538, DK56246 and TW006467 NIH grants.
References
- 1.Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR. Biochemistry, cellular and molecular biology and physiological roles of the iodothyronine selenodeiodinases. Endocrine Reviews. 2002;23:38–89. doi: 10.1210/edrv.23.1.0455. [DOI] [PubMed] [Google Scholar]
- 2.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–78. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 3.Hampton RY. ER-associated degradation in protein quality control and cellular regulation. Curr Opin Cell Biol. 2002;14:476–82. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- 4.Steinsapir J, Harney J, Larsen PR. Type 2 iodothyronine deiodinase in rat pituitary tumor cells is inactivated in proteasomes. J Clin Invest. 1998;102:1895–1899. doi: 10.1172/JCI4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gereben B, Goncalves C, Harney JW, Larsen PR, Bianco AC. Selective proteolysis of human type 2 deiodinase: a novel ubiquitin- proteasomal mediated mechanism for regulation of hormone activation. Mol Endocrinol. 2000;14:1697–1708. doi: 10.1210/mend.14.11.0558. [DOI] [PubMed] [Google Scholar]
- 6.Curcio-Morelli C, et al. Deubiquitination of type 2 iodothyronine deiodinase by pVHL-interacting deubiquitinating enzymes regulates thyroid hormone activation. J Clin Invest. 2003;112:189–196. doi: 10.1172/JCI18348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilton DJ, et al. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc Natl Acad Sci U S A. 1998;95:114–9. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasiliauskas D, Hancock S, Stern CD. SWiP-1: novel SOCS box containing WD-protein regulated by signalling centres and by Shh during development. Mech Dev. 1999;82:79–94. doi: 10.1016/s0925-4773(99)00014-3. [DOI] [PubMed] [Google Scholar]
- 9.Callebaut I, et al. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol Life Sci. 1997;53:621–45. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phospho-dependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–56. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 11.Maniatis T. A ubiquitin ligase complex essential for the NF-kappa B, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 12.Stebbins CE, Kaelin WG, Jr., Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–61. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 13.Schulman BA, et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–6. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 14.Kamura T, et al. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 1998;12:3872–81. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomon V, Lecker SH, Goldberg AL. The N-end rule pathway catalyzes a major fraction of the protein degradation in skeletal muscle. J Biol Chem. 1998;273:25216–22. doi: 10.1074/jbc.273.39.25216. [DOI] [PubMed] [Google Scholar]
- 16.Kamura T, et al. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J Biol Chem. 2001;276:29748–53. doi: 10.1074/jbc.M103093200. [DOI] [PubMed] [Google Scholar]
- 17.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton KS, Ellison MJ, Shaw GS. Identification of the ubiquitin interfacial residues in a ubiquitin-E2 covalent complex. J Biomol NMR. 2000;18:319–27. doi: 10.1023/a:1026773008237. [DOI] [PubMed] [Google Scholar]
- 19.Min JH, et al. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–9. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 20.Wu G, et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–56. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 21.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–9. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 22.Cook WJ, Martin PD, Edwards BF, Yamazaki RK, Chau V. Crystal structure of a class I ubiquitin conjugating enzyme (Ubc7) from Saccharomyces cerevisiae at 2.9 angstroms resolution. Biochemistry. 1997;36:1621–7. doi: 10.1021/bi962639e. [DOI] [PubMed] [Google Scholar]
- 23.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–9. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 24.Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–9. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 25.Kim BW, et al. ER-associated degradation of the human type 2 iodothyronine deiodinase (D2) is mediated via an association between mammalian UBC7 and the carboxyl region of D2. Mol Endocrinol. 2003;17:2603–2612. doi: 10.1210/me.2003-0082. [DOI] [PubMed] [Google Scholar]
- 26.Botero D, et al. Ubc6p and Ubc7p are required for normal and substrate-induced endoplasmic reticulum-associated degradation of the human selenoprotein type 2 iodothyronine monodeiodinase. Mol Endocrinol. 2002;16:1999–2007. doi: 10.1210/me.2002-0135. [DOI] [PubMed] [Google Scholar]
- 27.Callebaut I, et al. The iodothyronine selenodeiodinases are thioredoxin-fold family proteins containing a glycoside hydrolase-clan GH-A-like structure. J Biol Chem. 2003;278:36887–36896. doi: 10.1074/jbc.M305725200. [DOI] [PubMed] [Google Scholar]
- 28.Curcio-Morelli C, et al. In vivo dimerization of types 1, 2, and 3 iodothyronine selenodeiodinases. Endocrinology. 2003;144:3438–3443. doi: 10.1210/en.2002-220960. [DOI] [PubMed] [Google Scholar]
- 29.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–6. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 30.Vortkamp A, et al. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–22. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 31.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–86. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- 33.Stevens DA, et al. Thyroid hormones regulate hypertrophic chondrocyte differentiation and expression of parathyroid hormone-related peptide and its receptor during endochondral bone formation. J Bone Miner Res. 2000;15:2431–42. doi: 10.1359/jbmr.2000.15.12.2431. [DOI] [PubMed] [Google Scholar]
- 34.Robson H, Siebler T, Stevens DA, Shalet SM, Williams GR. Thyroid hormone acts directly on growth plate chondrocytes to promote hypertrophic differentiation and inhibit clonal expansion and cell proliferation. Endocrinology. 2000;141:3887–97. doi: 10.1210/endo.141.10.7733. [DOI] [PubMed] [Google Scholar]
- 35.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]