Abstract
Objective
To determine whether APOE genotype influences brain response and whether nonverbal stimuli generate findings comparable with those of previous studies that used verbal stimuli. The relationship between APOE genotype and blood oxygenation level dependent (BOLD) brain response was examined during a picture-encoding task in nondemented older adults.
Methods
Twenty nondemented participants with normal episodic memory function were divided into two groups based on the presence (n = 10) or absence (n = 10) of the APOE ε4 allele. Picture learning was completed during functional MRI in a blocked design alternating between experimental (novel pictures) and control (repeated picture) conditions.
Results
Nondemented older adults with an APOE ε4 allele showed greater magnitude and extent of BOLD brain response during learning of new pictures relative to their matched ε3 counterparts. Different patterns and directions of association between hippocampal activity and learning and memory performance were also demonstrated.
Conclusions
The results suggest that brain response differences are not due to poorer general memory abilities, differential atrophy, or brain response during control conditions, but instead appear to be directly influenced by APOE genotype. Results are consistent with a compensatory hypothesis wherein older adults at genetic risk for Alzheimer disease by virtue of the APOE ε4 allele appear to require additional cognitive effort to achieve comparable performance levels on tests of episodic memory encoding.
Studies of nondemented older adults who later develop Alzheimer disease (AD) show a subtle decline in episodic memory prior to emergence of the obvious cognitive and behavioral changes required for a clinical diagnosis of the disease.1-7 Often this decline in episodic memory is evident some years prior to the development of dementia and has been shown to predict the subsequent development of AD.1-4,8,9 In addition, the ε4 allele of the gene coding for APOE is linked to an increased risk of developing late-onset AD.10,11 There have been a variety of brain changes associated with the APOE ε4 allele in AD, including structural12,13 and functional brain changes,14 as well as subtle neuropsychologic deficits.8,9,15 Further, studies suggest that the pathologic burden of AD may begin decades—not years—prior to the diagnosis of AD and may be influenced by one's APOE genotype.16
Given the increasing emergence of treatments for dementia, sensitive and reliable markers of incipient dementia are needed to enhance our ability to detect AD in its earliest stages when potential neuroprotective agents might be most effective.17 fMRI offers considerable promise as a noninvasive technique for detection of early brain changes associated with an incipient dementia. fMRI study of episodic memory in at-risk older adults would have direct relevance to the early pathologic changes in medial temporal and prefrontal cortices in AD18 and provide a useful assay of activity in susceptible brain regions during the preclinical period of AD.
Initial work19 demonstrated an increase in the intensity and extent of brain activation in nondemented middle-aged and older adults with the APOE ε4 allele during verbal learning, suggesting a compensatory mechanism wherein APOE ε4 carriers utilize additional cognitive resources to bring memory-related performance to a normal level. However, differential atrophy or response to baseline conditions between genotype groups were not assessed, and APOE ε4 genotype and poorer memory function were confounded to some degree, thereby clouding the issue of whether the differential brain response was due to APOE genotype or simply due to poorer memory function. To determine the influence of the APOE ε4 allele in ostensibly healthy older adults with comparable episodic memory function, and to determine whether pictoral stimuli generate similar findings, we examined the relationship between APOE genotype and blood oxygen level dependent (BOLD) brain response during a picture-encoding task in nondemented older adults.
Methods
Participants
We studied 20 right-handed older adults from a larger pool of 80 volunteers participating in a longitudinal study of aging, all of whom were ostensibly healthy and living independently at the time of scanning. Participants were recruited through newspaper advertisements and community lectures (i.e., no clinic-based or medical referral sources) and selected without regard to ethnicity or race. Written informed consent was obtained from all participants. All participants were considered normal based on extensive medical, neurologic, laboratory, and neuropsychologic evaluations. Participants with a history of alcoholism, drug abuse, learning disability, neurologic, or psychiatric illness (including depression) are routinely excluded from the pool of normal control participants. No participant reported a significant level of depressive symptoms on the Geriatric Depression Scale (i.e., ≥10).20 Thus, participants did not demonstrate deficits on cognitive screening or formal memory testing, nor did they demonstrate significant affective disturbance, functional impairments, or difficulties with activities of daily living (table 1).
Table 1.
Demographic, global cognitive, and learning and memory characteristics of the APOE ε4 and ε3 groups
|
APOE |
|||
|---|---|---|---|
| Variables | ε4, n = 10 | ε3, n = 10 | p Value |
| Demographics | |||
| Age, y | 76.2 (4.8) | 75.7 (5.8) | 0.84 |
| Education, y | 14.9 (2.5) | 15.3 (2.1) | 0.70 |
| Women/men | 6/4 | 5/5 | 0.65 |
| Global cognition | |||
| DRS total (144 points possible) | 140.6 (2.2) | 141.3 (1.9) | 0.45 |
| Learning and memory | |||
| DRS memory subscale | 24.4 (0.7) | 24.4 (0.7) | 0.89 |
| WMS-R immediate recall | 25.9 (7.2) | 24.6 (3.9) | 0.62 |
| WMS-R delayed recall | 22.8 (7.9) | 19.6 (5.3) | 0.31 |
| CVLT List 1–5 total recall | 53.8 (6.1) | 44.4 (11.6) | 0.04 |
| CVLT long delay free recall | 11.5 (2.6) | 9.4 (4.0) | 0.18 |
| CVLT recognition memory, % | 94.7 (3.7) | 89.6 (8.2) | 0.09 |
| Post-MRI recognition memory accuracy for the pictures, % | 79.2 (11.8) | 72.3 (13.1) | 0.28 |
All 80 participants were genotyped for APOE allele type using a PCR-based method11 (APOE ε4% = 25%). From the larger group 20 participants were then selected for scanning based on their demographic characteristics and APOE genotypes and divided into two groups on the basis of the presence (n = 10) or absence (n = 10) of the APOE ε4 allele. All non-ε4 participants were homozygous for the ε3 allele (ε3/ε3); two of the ε4 participants were homozygotes (ε4/ε4) and the remaining eight participants were ε3/ε4 heterozygotes. As shown in table 1, the two groups did not differ on the basis of mean age, education, sex distribution, or on global cognitive functioning (all p values > 0.45). Furthermore, on memory testing, the two groups did not differ on variables from either the Wechsler Memory Scale–Revised21 or the California Verbal Learning Test (CVLT),22 with the exception of the summary learning measure from the CVLT (p = 0.04) wherein the ε4 group outperformed the ε3 group. Nonetheless, both groups' mean scores were well within normal limits on this measure as well as on the other measures (see table 1).
Stimuli and procedure
A picture-encoding paradigm23 was completed during scanning in a blocked design alternating between experimental and control conditions. In the experimental (“ENCODE”) condition, participants viewed and were asked to learn and remember 48 color photographs of indoor and outdoor scenes presented one at a time. In the control condition (“REPEAT”), one color photograph of autumn leaves was used as a repeated picture. Each block consisted of six trials, comprising picture presentation (2,500 msec) and an intertrial interval (500 msec). A 3-second warning screen reminded participants which type of block was to follow. Thus, each block lasted 21 seconds. Additionally, four blocks of fixation baseline trials were presented (“FIXATE”), one in the beginning, two in the middle, and one at the end of the trial blocks. The pictoral stimuli were presented to the participants using an LCD projector, back-projected onto a screen at the participant's feet. In the scanner, participants pressed a button on each trial to indicate they were attending to the pictures. Motor responses were made using a fiber-optic button box and were recorded by the MicroExperimental Lab2 software package (Psychological Software Tools, Pittsburgh, PA). Approximately 10 minutes after imaging, all participants completed a two-choice recognition testing for the 48 photos.
Anatomic and functional whole-brain imaging
All scans were whole-brain acquisitions conducted with a 1.5 T GE imager. High-resolution T1-weighted anatomic images were collected with either an SPGR sequence (124 slices acquired in the sagittal plane; 1.2 mm slice thickness; 256 × 256 matrix; field of view [FOV] = 250 mm; resulting in a 1 mm2 in-plane resolution) or a SPIRAL sequence (128 slices with 8 echoes; resolution was 1.25 mm in plane, and 1.33 to 1.42 mm through plane; 192 × 192 matrix using 16 spiral interleaves; FOV = 240 mm). Functional data from 156 whole brain images of BOLD signal intensity were acquired axially using a gradient-recalled echoplanar imaging (EPI) sequence. Seven-millimeter-thick slices sampled the entire brain (20 to 22 slices acquired in the axial plane; repetition time [TR] = 3,000 msec; echo time = 40 msec; flip angle = 90 degrees; 64 × 64 matrix; FOV = 240 mm; 3.75 mm2 in-plane resolution).
Individual participant data analysis path
All analyses were conducted with Analysis of Functional NeuroImages (AFNI) software,24 and all participant datasets were analyzed in the following scripted manner. A three-dimensional brick was created from the structural scan slices, and the three-dimensional bricks were warped into a standardized coordinate space (Talairach-Tournoux).25 A three-dimensional brick of image data was also created for each TR down the time course of the EPI scanning sequence. Effects of small movements were minimized with the AFNI three-dimensional volume registration program and, very infrequently when excessive motion occurred, by inserting the mean value of adjacent repetitions in voxels with residual motion. A threshold value was used to exclude low intensity values generally located outside the brain.
The comparison of interest was the difference in activation levels while viewing new pictures vs a repeated picture (i.e., ENCODE – REPEAT). A predicted and shifted (up to 6 seconds) trapezoidal reference function was cross-correlated with the motion-corrected MR time course data in each voxel within the three-dimensional functional brick. Datasets were then resampled into 4 mm3 voxels and written into Talairach-Tournoux space.25 Finally, to reduce noise and account for correlations between adjacent voxels and variations in anatomy, functional datasets were blurred with a 7 mm FWHM kernel.
Group data analysis path
T-maps were created for each APOE genotype group to determine whether the mean intensity value difference between the new and repeated picture conditions in each voxel was different from zero. A between-group t-map was also generated to determine whether pattern of average intensity value differences between the picture conditions differed between the two APOE genotype groups. A cluster thresholding technique was used to determine which areas of activation on the t-maps were significant by thresholding at a p value of 0.025. The cluster size was predetermined as a region of at least 13 contiguous significant voxels (i.e., 832 mm3 volume), which protected for a whole-brain p value of 0.05. This correction is appropriate to protect the hypothesis that when no activation is present anywhere within the brain a chance volume of activation will occur less than 2.5% of the time.
Search regions of interest (ROIs)
Encoding-related brain response was also examined within each group and compared between the two groups in each voxel of four bilateral search regions: hippocampus, parahippocampal gyrus, inferior prefrontal cortex (Brodmann's area [BA] 44/45), and fusiform gyrus. Search regions were selected from previous studies of novel picture encoding.23,26-29 Coordinates for each search region, as well as its extent, were determined by Talairach Daemon software.30 Search regions were used to mask each participant's three-dimensional dataset of fit coefficients. To examine group differences, we compared mean fit coefficient between groups in all voxels within each search region via independent-samples t-tests. Clusters of brain response were considered significantly different from zero within groups or significantly different between the two groups if they contained at least seven contiguous voxels, each with a p < 0.025 per voxel (i.e., 448 mm3 volume). This threshold and cluster volume pairing was found to protect a search-region-wise p = 0.059 in a Monte Carlo simulation (AlphaSim, AFNI). Between-subject variability at each voxel within the search regions also was compared between groups via analysis of variance (ANOVA). We identified areas of significant BOLD response within each group by comparing fit coefficients of voxels within each search region to zero (single-sample t-test). For the statistical maps of brain response presented in the figures, the magnitude of between- or within-group effect was expressed as an effect size statistic (eta2) and given a valence based on its direction (range = −1.0 to + 1.0).26
Post hoc analyses
Several post hoc analyses were conducted to aid interpretation of the observed group differences.26,31 First, the contrast of ENCODE vs FIXATE was compared between groups in a control region (primary visual cortex, BA 17) in order to explore the specificity of observed differences to encoding-related brain regions and to address the potential concern that methodologic or broad physiologic factors influenced the between-group findings.32 Second, the contrast of REPEAT vs FIXATE was compared between groups to explore whether there were significant baseline differences between genotype groups that could limit interpretation of our primary contrast (ENCODE vs REPEAT). Third, correlations between hippocampal brain response and recognition memory task performance were computed, as were correlations between hippocampal brain response and learning on the CVLT.
Finally, whole brain segmentation of same-session structural MRI scans was undertaken to address the potential concern that any observed differences between APOE genotype groups in brain response may have arisen from broad differences in cerebral atrophy.33 Following bias correction,34 each scan was processed with one or more of the following automated skull-stripping methods: FreeSurfer's Hybrid Watershed Algorithm,35 Brain Extraction Tool,36 and 3dIntracranial in AFNI.24,37 All scans were then processed through ANFI's24 3dIntracranial program to remove remaining skull37 and manually edited to remove residual non-brain material. Tissue segmentation was performed using TriComp, a locally developed set of programs designed to fit a three-compartment Gaussian mixture model to the signal intensity histogram of the skull stripped T1-weighted images. Means, SDs, and weights for gray matter, white matter, and CSF compartments were estimated using the downhill-simplex method to minimize the residual sum of squares.38 Total intracranial volume was also derived, and thus proportions of each compartment were calculated.
Results
Behavioral data: Recognition memory for the novel pictures
There were no significant differences in performance between those with an APOE ε4 allele and those homozygous for the APOE ε3 allele on forced-choice recognition testing of the novel pictures (see table 1). Both groups averaged better than 70% accuracy for recognition of the pictures following their scanning session (APOE ε4 group = 79.2%, SD = 11.8; APOE ε3 group = 72.3, SD = 13.1, p = 0.28).
Functional MR imaging data: Brain response to the encoding task
Within-genotype group whole brain analysis: APOE ε4 group
In multiple brain regions, the APOE ε4 group showed significantly greater BOLD brain response while learning new pictures compared to viewing a repeated picture. The majority of these sites were located in large, bilateral regions of the occipital/fusiform gyri, although the areas of greater BOLD response were spread throughout multiple brain regions (e.g., precuneus, frontal, temporal, and cingulate gyri) in all four lobes of the cerebrum (see table 2 and figure 1A for details).
Table 2.
Areas of significantly greater brain response during encoding of novel vs repeated pictures in nondemented older adults
| Brain region | Subregion | Volume, mm3 | Coordinates* of maximum intensity voxel | Eta2 for encode vs repeat |
|---|---|---|---|---|
| APOE ε4 group | ||||
| R fusiform gyrus | BA 19 | 119,552 | 38R, 65P, 8I | 0.93 |
| L precuneus | BA 7 | 10,368 | 2L, 69P, 40S | 0.83 |
| L superior frontal gyrus | L middle frontal gyrus | 5,248 | 30L, 11A, 48S | 0.94 |
| L middle frontal gyrus | BA 46 | 3,008 | 46L, 35A, 16S | 0.70 |
| R medial frontal gyrus | BA 8/R cingulate gyrus | 2,944 | 2R, 27A, 40S | 0.71 |
| R red nucleus | R thalamus | 2,816 | 2R, 25P, 4I | 0.74 |
| L paracentral lobule/BA 6 | BA 4 | 2,816 | 2L, 33P, 60S | 0.67 |
| R superior temporal gyrus | BA 21 | 1,216 | 54R, 7A, 12I | 0.56 |
| R cingulate gyrus | BA 24 | 960 | 10R, 17P, 36S | 0.56 |
| APOE ε3 group | ||||
| L middle occipital gyrus | BA 18 | 14,016 | 26L, 81P, 0I | 0.81 |
| R middle occipital gyrus | BA 19 | 6,656 | 30R, 65P, 4S | 0.76 |
| L caudate tail | L cingulate gyrus | 3,712 | 22L, 41P, 20S | 0.74 |
| L medial frontal gyrus | BA 10/L superior frontal gyrus | 3,200 | 2L, 67A, 4I | 0.79 |
| R inferior frontal gyrus | BA 46 | 1,408 | 38R, 35A, 4S | 0.70 |
| R cingulate gyrus | R posterior cingulate | 1,088 | 14R, 41P, 24S | 0.68 |
| L caudate | L parahippocampal gyrus/L hippocampus | 1,024 | 34L, 37P, 0I | 0.71 |
| R caudate | R caudate tail | 896 | 22R, 33P, 16S | 0.64 |
From Talairach and Tournoux.25
BA = Brodmann area; P = posterior; I = inferior; S = superior; A = anterior.
Figure 1.
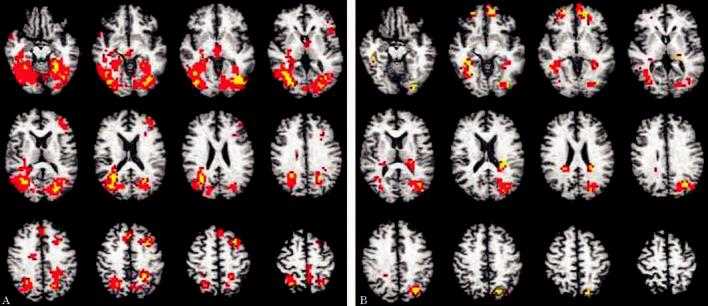
Magnitude and direction of brain response to the task overlaid onto axial slices of a representative anatomic image in Talairach space (slices span from 12 inferior to 54 superior in 6 mm increments). Activations shown include voxels significant at p < 0.025 that are contained within a cluster of 13 or more voxels. Color scale represents effect sizes for the within-subjects difference between conditions (ENCODE vs REPEAT) as measured by eta2 (red voxels: 0.50 < η2 ≤ 0.75; yellow voxels: 0.76 < η2 & 1.0 [η2 indexes the effect size for the magnitude of the difference between the observed response and 0]). See also table 2 for areas of significant activation. (A) APOE ε4 participants. (B) APOE ε3 participants.
Within-genotype group whole brain analysis: APOE ε3 group
Unlike the findings in those with an APOE ε4 allele, participants in the APOE ε3 group showed relatively fewer regions of greater BOLD brain response during the new vs repeated picture conditions. The majority of these regions also were located in bilateral occipital/fusiform cortices, left medial frontal and right inferior frontal gyri. In addition, the APOE ε3 group uniquely demonstrated greater encoding-related brain response in bilateral caudate nuclei, and left parahippocampal cortex and left hippocampus (see table 2 and figure 1B for details).
Between-genotype groups whole brain analysis
Individuals with an APOE ε4 allele showed greater differential BOLD brain response while encoding new pictures vs viewing a repeated picture in a number of brain regions (see table 3 and figure 2). Specifically, clusters (i.e., 13 contiguous significant voxels) of significantly greater BOLD response were observed in six regions, including bilateral fusiform gyri, right superior parietal cortex, left pyramis/uvula, left middle frontal gyrus, and the medial frontal gyrus (see table 3 and figure 2).
Table 3.
Whole brain analysis regions of significantly greater brain response in APOE ε4 vs APOE ε3 groups during novel picture encoding
| Brain region | Sub region | Volume, mm3 | Coordinates* of maximum intensity voxel | Eta2 for ε4 vs ε3 |
|---|---|---|---|---|
| L fusiform gyrus | BA 37 | 7,552 | 46L, 53P, 20I | 0.59 |
| R fusiform gyrus | BA 19 | 5,440 | 34R, 69P, 16I | 0.52 |
| R superior parietal lobule | BA 7 | 2,496 | 6R, 65P, 56S | 0.42 |
| L pyramis/uvula | Cerebellum | 1,216 | 6L, 69P, 24I | 0.38 |
| L middle frontal gyrus | BA 6 | 1,088 | 26L, 11A, 56S | 0.33 |
| Medial frontal gyrus | BA 8 | 960 | 2R, 27A, 40S | 0.37 |
From Talairach and Tournoux.25
BA = Brodmann area; P = posterior; I = inferior; S = superior; A = anterior.
Figure 2.
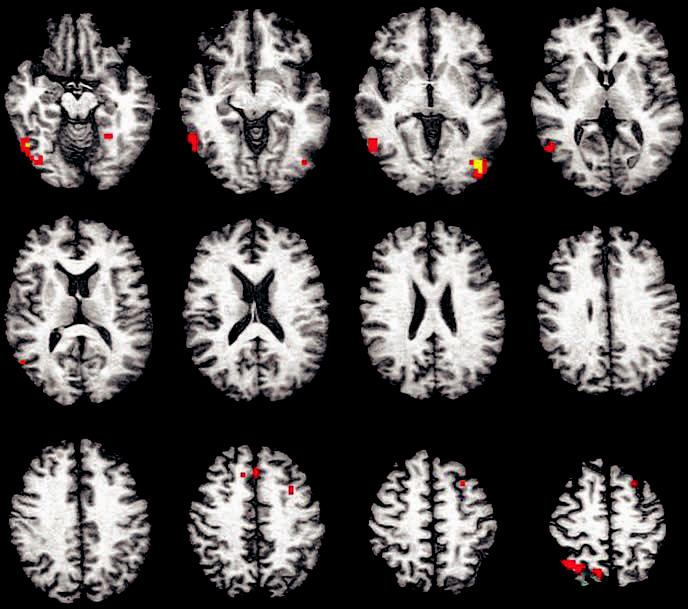
Clusters of significant difference between APOE ε4 and ε3 participants for encoding-related brain response overlaid on a representative anatomic image in Talairach (1988) space (slices span from 12 inferior to 54 superior in 6 mm increments). Activations shown include voxels significant at p < 0.025 that are contained within a cluster of 13 or more voxels. Color scale represents effect sizes for the ε4 – ε3 difference in fit coefficient as measured by eta2 (signed to reflect the direction of the contrast) (red voxels: 0.50 < η2 ≤ 0.75; yellow voxels: 0.76 < η2 < 1.0).
Search ROI analyses
Within the search regions examined, the between-subjects tests of encoding-related BOLD brain response were greater in the APOE ε4 group relative to the APOE ε3 group in left inferior frontal, bilateral fusiform, and right hippocampal and parahippocampal cortices. However, in clusters within the left hippocampus and parahippocampal cortex, the APOE ε3 group showed a larger positive response (i.e., greater signal intensity during novel pictures than during repeated pictures), whereas the APOE ε4 group showed either a small positive response or no difference in signal intensity between the two conditions (see table E-1 on the Neurology Web site at www.neurology.org).
Post hoc analyses
Control analyses
Analysis of the response of voxels within BA 17 revealed a strong positive bilateral response to the encoding condition compared with fixation for both APOE ε4 and ε3 groups, with no significant clusters of group difference observed within this search region. We also found no evidence for differentially lower BOLD response among the ε4 group in any of the six regions identified in table 3 to the repeat condition compared with fixation (data available upon request).
Correlations
Patterns of correlation between individual differences in encoding-related brain response and subsequent recognition memory performance were examined for the pictures as well as for word-list learning as indexed by the CVLT. Given the small sample sizes, each of the reported r-values below do not attain significance (all p values > 0.05). Rather, they are reported solely for descriptive purposes. In the left hippocampus, the APOE ε4 group demonstrated a positive correlation between the mean fit coefficient and recognition memory (r = 0.57), whereas the ε3 group did not (r = 0.07). However, an opposite pattern of correlation was observed for the right hippocampus (ε4: r = 0.01; ε3: r = 0.33). Contrasting findings were also demonstrated in the correlations between hippocampal fit coefficients and performance on the summary learning variable from the CVLT (List A Trials 1 to 5 Total Recall). Specifically, in the left hippocampus, the APOE ε4 group demonstrated little correlation between the mean fit coefficient and CVLT learning (r = 0.02), whereas the ε3 group evinced a relatively larger association (r = 0.28). However, an opposite pattern and direction of association was observed for the right hippocampus (ε4: r = −0.46; ε3: r = 0.53). A Fisher's z transformation of the difference between these two correlation coefficients was significant (zdiff = 2.03, p < 0.05). Given concerns over ROIs derived from standard space for structures as variable as the hippocampus (i.e., volumes derived from native space may be more appropriate), we also found comparable patterns of association when the most intense voxel located in hippocampal native space was used instead of the mean fit coefficient in standard space (e.g., right hippocampus: ε4: r = −0.72; ε3: r = 0.13).
Segmentation
Total intracranial volume, gray and white matter volumes, and CSF volume were analyzed in a series of four 2 × 2 ANOVAs (APOE genotype [ε4 vs ε3] by MRI scan type [SPGR vs SPIRAL sequence]). Results revealed main effects for scan type on two of the four brain volumes measured (total intracranial volume: F (1,18) = 4.75, p = 0.05; gray matter: F < 1, p = 0.37; white matter: F (1,18) = 1.68, p = 0.21; CSF: F (1,18) = 21.58, p < 0.001), but no main effects for APOE genotype on any of the four measures (total intracranial volume: F (1,18) = 1.06, p = 0.32; gray matter: F < 1, p = 0.97; white matter: F < 1, p = 0.65; CSF: F (1,18) = 3.05, p = 0.10), and no significant interactions between APOE genotype and scan type (all Fs < 1, ps = 0.41 to 0.97). Overall, the two APOE genotype groups did not differ on any of the whole-brain segmentation measures, indicating comparable volumes of gray and white matter, CSF volumes, as well as proportion scores (see table E-2 on the Neurology Web site at www.neurology.org).
Discussion
In multiple brain regions, nondemented older adults with an APOE ε4 allele and normal learning and memory capabilities showed greater magnitude and extent of BOLD brain response during picture learning relative to their ε3 counterparts (e.g., bilateral fusiform and medial frontal gyri, left inferior and middle frontal, right superior parietal, and right hippocampal and parahippocampal cortices). In addition, the APOE ε4 group showed lower brain response in the left hippocampus, relative to the APOE ε3 group. Both groups were performing equally well across many learning and memory measures. Both groups demonstrated comparable volumes of gray and white matter and CSF, all of which were also quite consistent with gray, white, and CSF segmentation volumes of older adults reported by others,39 thereby demonstrating the robustness of our segmentation procedures and comparability of our groups to other nondemented older adult samples. Also, we found no evidence for differentially lower BOLD response among the ε4 group in any of the six regions identified in the ENCODE vs REPEAT contrast (see table 3). Our ε4 and ε3 groups also showed a strong positive bilateral response to the encoding condition in primary visual cortex (BA 17)—an area whose contribution to the encoding task should not differ between groups. Thus, it appears that BOLD response differences in nondemented older adults are not accounted for by differential memory abilities, differential atrophy, baseline differences in the control (i.e., Repeat) condition, or broad physiologic differences per se, but instead appear to be more directly influenced by one's APOE genotype.
Results are consistent with a compensatory hypothesis wherein APOE ε4 persons appear to require additional cognitive effort to achieve the same level of performance. Indeed, several functional neuroimaging studies have shown that BOLD brain response associated with the performance of memory tasks is more diffuse in patients with early AD than in normal older individuals, suggesting the need to recruit areas outside of the usual structures that mediate memory.40-42 It may be that, after an initial decline in memory proficiency following damage to MTL structures, patients in the preclinical stage of AD are able to effectively recruit compensatory brain resources (e.g., frontal and temporal cortical regions important for executive functions and semantic memory) to halt or slow further memory decline for a period of time. A similar compensatory response in certain brain-derived neurotrophic factors43,44 or cholinergic activity45 may also attenuate memory changes for a time. Given that adequate cholinergic activity and neurotrophic mechanisms are partly responsible for the maintenance of neuronal function and structural integrity, these findings suggest that, under conditions of progressive neurodegeneration, the MTL stimulates the overexpression of certain cholinergic and neurotrophic factors as a possible mechanism of compensation. As the disease progresses, however, each of these additional resources becomes compromised and the patient exhibits a period of rapid decline in episodic memory abilities.15 Attendant declines in brain activity42 and cholinergic activity45 in these critical brain regions for episodic memory function would then be expected in clinically evident AD.
Several functional MRI studies are also consistent with the compensation hypothesis, although none specifically investigated the role of APOE genotype. Rombouts et al.46 found increases in fusiform and frontal—but not MTL—activation during face encoding following rivastigmine administration. Machulda et al.47 showed that MCI and AD patients had less MTL activation during encoding than normal subjects but similar activation on a sensory task. Grady et al.41 also demonstrated that AD patients used additional neural resources in prefrontal and posterior cortices, presumably those mediating executive functions, when performing semantic and episodic memory tasks to compensate for losses attributable to the degenerative process of AD. Importantly, Grady et al. showed that activity in dorsolateral prefrontal and posterior cortical regions was correlated with better task performance. Finally, Daselaar et al.48 utilized an event-related paradigm to investigate separately the contributions of encoding and retrieval to memory. Results demonstrated reductions in MTL activity alongside increases in brain activity in other regions in the subgroup with reduced memory performance, suggesting differences in retrieval strategy and compensation for the encoding deficit. Nevertheless, not all studies support conclusions consistent with increased brain response as compensation.49
With respect to fMRI studies of at-risk groups, contradictory findings across studies (e.g., decreased vs increased BOLD responses) may center on continued uncertainties regarding the mechanisms linking the hemodynamic response to its underlying neuroanatomy and neurophysiology. One particular concern centers on the notion that differences in the “resting state” will influence the amplitude of the BOLD response.50 Such a possibility in AD has been demonstrated,51 and a recent animal model using APP23 transgenic mice demonstrated amyloid plaques to have a direct effect on the hemodynamic response, due in part to compromised cerebrovascular reactivity.52 Other human studies also demonstrate that the hemodynamic response itself changes in an age-related manner.33,53 Thus, future efforts should incorporate other MR-based techniques (e.g., perfusion imaging, structural morphometry, cerebral blood volume mapping) to help parcellate the contributions of the neuroanatomic and neurophysiologic underpinnings to the BOLD signal.54 Nonetheless, fMRI-based findings of possible brain compensation are analogous to neuropsychologic demonstrations of non-episodic memory functions, such as semantic knowledge or visuospatial abilities, remaining intact early in the course of AD.15
In addition, the differing directions in brain response between the left and right MTLs as well as the differential patterns of association between hippocampal BOLD response and learning on the CVLT for the two groups also suggest possible compensatory brain activity. Prior work55 with nondemented adults demonstrates a pattern of association evinced by the ε3 group in our study: a positive correlation between processing novel stimuli in the hippocampus and learning on the CVLT. Although the left hippocampus is generally engaged during encoding of new verbal information, the right hippocampus appears to become increasingly involved with higher CVLT performance.55 This finding further suggests that successfully engaging the right as well as left hippocampus results in greater memory capacity. Again, our data from the ε3 group are in close accord with this pattern, but the ε4 group's negative correlation is in stark contrast to this expected association. Thus, the APOE ε4 group may be invoking bilateral MTL activation of brain resources in an aberrant manner as an attempt to facilitate performance while learning new material.40
Although the present results offer an interesting and plausible methodology for detection of incipient AD, the findings remain speculative given the small sample sizes and until longitudinal follow-up and clinical outcomes of the participants are gathered, which is under way. Furthermore, given the MTL's considerable structural and functional variability across individuals, ROI analyses may be better accomplished in native rather than standard space.56 An additional unanswered question is whether the differential BOLD response demonstrated in APOE ε4 persons represents a predictor of AD development or a normal phenomenon present across the life span. Although our two groups did not demonstrate broad physiologic differences in our control region (i.e., BA 17–primary visual cortex), fMRI study of young- and middle-aged APOE ε4 and non-ε4 adults may be a more direct assessment of the potential for broad physiologic distinctions between APOE genotypes.57
Supplementary Material
Acknowledgment
The authors thank Dr. Christine Fennema-Notestine, Kelly L. Lange, and Philippe Goldin and the UCSD Laboratory of Cognitive Imaging.
Footnotes
Supported by grant PRG 98019 from the Alzheimer's Association (M.W.B.), National Institute on Aging grant R01 AG12674 (M.W.B.), and REAP (G.G.B.) and MIRECC (L.T.E. and G.G.B.) grants from the Medical Research Service of the Department of Veterans Affairs.
References
- 1.Albert MS, Moss MB, Tanzi R, Jones K. Preclinical prediction of AD using neuropsychological tests. J Int Neuropsychol Soc. 2001;7:631–639. doi: 10.1017/s1355617701755105. [DOI] [PubMed] [Google Scholar]
- 2.Bäckman L, Small BJ, Fratiglioni L. Stability of the preclinical episodic memory deficit in Alzheimer's disease. Brain. 2001;124:96–102. doi: 10.1093/brain/124.1.96. [DOI] [PubMed] [Google Scholar]
- 3.Bondi MW, Monsch AU, Galasko D, Butters N, Salmon DP, Delis DC. Preclinical cognitive markers of dementia of the Alzheimer type. Neuropsychology. 1994;8:374–384. [Google Scholar]
- 4.Chen P, Ratcliff G, Belle SH, Cauley JA, DeKosky ST, Ganguli M. Patterns of cognitive decline in presymptomatic Alzheimer disease: a prospective community study. Arch Gen Psychiatry. 2001;58:853–858. doi: 10.1001/archpsyc.58.9.853. [DOI] [PubMed] [Google Scholar]
- 5.Fuld PA, Masur DM, Blau AD, Crystal H, Aronson MK. Object-memory evaluation for prospective detection of dementia in normal functioning elderly: predictive and normative data. J Clin Exp Neuropsychol. 1990;12:520–528. doi: 10.1080/01688639008400998. [DOI] [PubMed] [Google Scholar]
- 6.Grober E, Kawas C. Learning and retention in preclinical and early Alzheimer's disease. Psychol Aging. 1997;12:183–188. doi: 10.1037//0882-7974.12.1.183. [DOI] [PubMed] [Google Scholar]
- 7.Howieson DB, Dame A, Camicioli R, Sexton G, Payami H, Kaye JA. Cognitive markers preceding Alzheimer's dementia in the healthy oldest old. J Am Geriatr Soc. 1997;45:584–589. doi: 10.1111/j.1532-5415.1997.tb03091.x. [DOI] [PubMed] [Google Scholar]
- 8.Bondi MW, Salmon DP, Galasko D, Thomas RG, Thal LJ. Neuropsychological function and apolipoprotein E genotype in the preclinical detection of Alzheimer's disease. Psychol Aging. 1999;14:295–303. doi: 10.1037//0882-7974.14.2.295. [DOI] [PubMed] [Google Scholar]
- 9.Smith GE, Bohac DL, Waring SC, et al. Apolipoprotein E genotype influences cognitive ‘phenotype’ in patients with Alzheimer's disease but not in healthy control subjects. Neurology. 1998;50:355–362. doi: 10.1212/wnl.50.2.355. [DOI] [PubMed] [Google Scholar]
- 10.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 11.Saunders AM, Strittmatter WJ, Schmechel D, et al. Association of apolipoprotein E allele ε4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 12.Soininen H, Partanen K, Pitkanen A, et al. Decreased hippocampal volume asymmetry on MRIs in nondemented elderly subjects carrying the apolipoprotein E epsilon 4 allele. Neurology. 1995;45:391–392. doi: 10.1212/wnl.45.2.391. [DOI] [PubMed] [Google Scholar]
- 13.Plassman BL, Welsh-Bohmer KA, Bigler ED, et al. Apolipoprotein E ε4 allele and hippocampal volume in twin with normal cognition. Neurology. 1997;48:985–989. doi: 10.1212/wnl.48.4.985. [DOI] [PubMed] [Google Scholar]
- 14.Reiman EM, Caselli RJ, Yun LS, et al. Preclinical evidence of Alzheimer's disease in persons homozygous for the ε4 allele for APOE. N Engl J Med. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- 15.Lange KL, Bondi MW, Galasko DG, Delis DC, Salmon DP, Thal LJ. Decline in verbal memory during preclinical Alzheimer's disease: examination of the effect of apolipoprotein E genotype. J Int Neuropsychol Soc. 2002;8:943–955. doi: 10.1017/s1355617702870096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghebremedhin E, Schultz C, Braak E, Braak H. High frequency of apolipoprotein E ε4 allele in young individuals with very mild Alzheimer's disease-related neurofibrillary changes. Exp Neurol. 1998;153:152–155. doi: 10.1006/exnr.1998.6860. [DOI] [PubMed] [Google Scholar]
- 17.Petersen RC, Stevens JC, Ganguli M, Tangalos EG, Cummings JL, DeKosky ST. Practice parameter: early detection of dementia: mild cognitive impairment (an evidence-based review) Neurology. 2001;56:1133–1142. doi: 10.1212/wnl.56.9.1133. [DOI] [PubMed] [Google Scholar]
- 18.Desgranges B, Baron J-C, Eustache F. The functional neuroanatomy of episodic memory: the role of the frontal lobes, the hippocampal formation, and other areas. NeuroImage. 1998;8:198–213. doi: 10.1006/nimg.1998.0359. [DOI] [PubMed] [Google Scholar]
- 19.Bookheimer SY, Strojwas MH, Cohen MS, et al. Patterns of brain activation in people at risk for Alzheimer's disease. N Engl J Med. 2000;343:450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yesavage JA, Brink TL, Rose TL, et al. Development and validation of a geriatric depression screening scale: a preliminary report. J Psychiatr Res. 1983;17:37–49. doi: 10.1016/0022-3956(82)90033-4. [DOI] [PubMed] [Google Scholar]
- 21.Wechsler D. Wechsler Memory Scale–Revised. Psychological Corporation; New York: 1987. [Google Scholar]
- 22.Delis DC, Kramer JH, Kaplan E, Ober BA. The California Verbal Learning Test. Psychological Corporation; New York: 1987. [Google Scholar]
- 23.Stern CE, Corkin S, Gonzaalez RG, et al. The hippocampal formation participates in novel picture encoding: evidence from functional magnetic resonance imaging. Proc Natl Acad Sci. 1996;93:8660–8665. doi: 10.1073/pnas.93.16.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox RW. AFNI. Software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 25.Talairach J, Tournoux P. A coplanar stereotaxic atlas of the human brain. New York, NY: 1988. [Google Scholar]
- 26.Eyler Zorrilla LT, Jeste DV, Paulus M, Brown GG. Functional abnormalities of medial cortex during novel picture learning among patients with chronic schizophrenia. Schiz Res. 2003;59:187–198. doi: 10.1016/s0920-9964(01)00340-1. [DOI] [PubMed] [Google Scholar]
- 27.Nyberg L, Persson J, Habib R, et al. Large scale neurocognitive networks underlying episodic memory. J Cogn Neurosci. 2000;12:163–173. doi: 10.1162/089892900561805. [DOI] [PubMed] [Google Scholar]
- 28.Gabrieli JDE, Brewer JB, Desmond JE, Glover GH. Separate neural bases of two fundamental memory processes in the human medial temporal lobe. Science. 1997;276:264–266. doi: 10.1126/science.276.5310.264. [DOI] [PubMed] [Google Scholar]
- 29.Constable RT, Carpentier A, Pugh K, Westerveld M, Oszunar Y, Spencer DD. Investigation of the human hippocampal formation using a randomized event-related paradigm and Z-shimmed functional MRI. Neuroimage. 2000;12:55–62. doi: 10.1006/nimg.2000.0583. [DOI] [PubMed] [Google Scholar]
- 30.Lancaster JL, Woldroff MG, Parsons LM, et al. Automated Talairach atlas labels for functional brain mapping. Hum Brain Mapp. 2000;10:120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyler Zorrilla LT, Jeste DV, Brown GG. Functional MRI and novel picture-learning among older patients with chronic schizophrenia: abnormal correlations between recognition memory and medial temporal brain response. Am J Geriatr Psychiatry. 2002;10:52–61. [PubMed] [Google Scholar]
- 32.Callicott JH, Ramsey NF, Tallent K, et al. Functional magnetic resonance imaging brain mapping in psychiatry: methodological issues illustrated in a study of working memory in schizophrenia. Neuropsychopharmacol. 1998;18:186–196. doi: 10.1016/S0893-133X(97)00096-1. [DOI] [PubMed] [Google Scholar]
- 33.D'Esposito M, Zarahn E, Aguirre GK, Rypma B. The effect of normal aging on the coupling of neural activity to the BOLD hemodynamic response. NeuroImage. 1999;10:6–14. doi: 10.1006/nimg.1999.0444. [DOI] [PubMed] [Google Scholar]
- 34.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 35.Segonne F, Dale AM, Busa E, et al. A hybrid approach to the skull stripping problem in MRI. Organization for Human Brain Mapping, Brighton, UK. NeuroImage. 2001:S241. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 36.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward BD. Intracranial segmentation. Biophysics Research Institute, Medical College of Wisconsin; Milwaukee, WI: 1999. Available at: http://afni.nimh.nih.gov/afni/index/shtml. [Google Scholar]
- 38.Nelder JA, Mead R. A simplex method for function minimization. Comp J. 1965;7:308–313. [Google Scholar]
- 39.Courchesne E, Chisum HJ, Townsend J, et al. Normal brain development and aging: quantitative analysis at in vivo MR imaging in healthy volunteers. Radiology. 2000;216:672–682. doi: 10.1148/radiology.216.3.r00au37672. [DOI] [PubMed] [Google Scholar]
- 40.Becker JT, Mintum MA, Aleva K, Wiseman MB, Nichols T, DeKosky ST. Compensatory reallocation of brain resources supporting verbal episodic memory in Alzheimer's disease. Neurology. 1996;46:692–700. doi: 10.1212/wnl.46.3.692. [DOI] [PubMed] [Google Scholar]
- 41.Grady CL, McIntosh AR, Beig S, Keightley ML, Burian H, Black SE. Evidence from functional neuroimaging of a compensatory prefrontal network in Alzheimer's disease. J Neurosci. 2003;23:986–993. doi: 10.1523/JNEUROSCI.23-03-00986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sperling RA, Bates JF, Chua EF, et al. fMRI studies of associative encoding in young and elderly controls and mild Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2003;74:44–50. doi: 10.1136/jnnp.74.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durany N, Michel T, Kurt J, Cruz-Sanchez FF, Cervas-Navarro J, Riederer P. Brain-derived neurotrophic factor and neurotrophin-3 levels in Alzheimer's disease. Int J Dev Neurosci. 2000;18:807–813. [PubMed] [Google Scholar]
- 44.Egan MF, Kojima M, Callicott JH, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 45.DeKosky ST, Ikonomovic MD, Styren SD, et al. Upregulation of choline acetyltransferase activity in hippocampus and frontal cortex of elderly subjects with mild cognitive impairment. Ann Neurol. 2002;51:145–155. doi: 10.1002/ana.10069. [DOI] [PubMed] [Google Scholar]
- 46.Rombouts SA, Barkhof F, van Meel CS, Scheltens P. Alterations in brain activation during cholinergic enhancement with rivastigmine in Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2002;73:665–671. doi: 10.1136/jnnp.73.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Machulda MM, Ward HA, Borowski B, et al. Comparison of memory fMRI response among normal, MCI, and Alzheimer's patients. Neurology. 2003;61:500–506. doi: 10.1212/01.wnl.0000079052.01016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daselaar SM, Veltman DJ, Rombouts SA, Raaijmakers JG, Jonker C. Neuroanatomical correlates of episodic encoding and retrieval in young and elderly subjects. Brain. 2003;126:43–56. doi: 10.1093/brain/awg005. [DOI] [PubMed] [Google Scholar]
- 49.Smith CD, Andersen AH, Kryscio RJ, et al. Altered brain activation in cognitively intact individuals at high risk for Alzheimer's disease. Neurology. 1999;53:1391–1396. doi: 10.1212/wnl.53.7.1391. [DOI] [PubMed] [Google Scholar]
- 50.Cohen ER, Ugurbil K, Kim S-G. Effect of basal conditions on the magnitude and dynamics of the blood oxygenation level-dependent fMRI response. J Cereb Blood Flow Metab. 2002;22:1042–1053. doi: 10.1097/00004647-200209000-00002. [DOI] [PubMed] [Google Scholar]
- 51.Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc Nat Acad Sci. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueggler T, Sturchler-Pierrat C, Baumann D, Rausch M, Staufenbiel M, Rudin M. Compromised hemodynamic response in amyloid precursor protein transgenic mice. J Neurosci. 2002;22:7218–7224. doi: 10.1523/JNEUROSCI.22-16-07218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buckner RL, Snyder AZ, Sanders AL, Raichle ME, Morris JE. Functional brain imaging of young, nondemented, and demented older adults. J Cog Neurosci. 2000;12:24–34. doi: 10.1162/089892900564046. [DOI] [PubMed] [Google Scholar]
- 54.Small SA, Chawla MK, Buonocore M, Rapp PR, Barnes CA. Imaging correlates of brain function in monkeys and rats isolates a hippocampal subregion differentially vulnerable to aging. Proc Natl Acad Sci. 2004;101:7181–7186. doi: 10.1073/pnas.0400285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson SC, Saykin AJ, Baxter LC, Flashman LA, McAllister TW, Sparling M. Brain activation on fMRI and verbal memory ability: functional neuroanatomic correlates of CVLT performance. J Int Neuropsychol Soc. 2001;7:55–62. doi: 10.1017/s135561770171106x. [DOI] [PubMed] [Google Scholar]
- 56.Vandenbroucke MWG, Goekoop R, Duschek EJJ, et al. Interindividual differences of medial temporal lobe activation during encoding in an elderly population studies by fMRI. NeuroImage. 2003;21:173–180. doi: 10.1016/j.neuroimage.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 57.Reiman EM, Chen K, Alexander G, et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc Natl Acad Sci. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mattis S. Dementia Rating Scale: Professional Manual. Psychological Assessment Resources; Odessa, FL: 1988. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


